Abstract
Natural Language Processing (NLP) is essential for concept extraction from narrative text in electronic health records (EHR). To extract numerous and diverse concepts, such as data elements (i.e., important concepts related to a certain medical condition), a plausible solution is to combine various NLP tools into an ensemble to improve extraction performance. However, it is unclear to what extent ensembles of popular NLP tools improve the extraction of numerous and diverse concepts. Therefore, we built an NLP ensemble pipeline to synergize the strength of popular NLP tools using seven ensemble methods, and to quantify the improvement in performance achieved by ensembles in the extraction of data elements for three very different cohorts. Evaluation results show that the pipeline can improve the performance of NLP tools, but there is high variability depending on the cohort.
Introduction
Extracting concepts from narrative clinical notes using Natural Language Processing (NLP) techniques is essential to enhance cohort identification processes. Researchers have developed many clinical NLP concept extraction tools (NLP tools), such as cTAKES1 (clinical Text Analysis and Knowledge Extraction System) and MetaMap.2 An NLP tool may be suitable and powerful for certain concept extraction tasks; there is hardly an NLP tool that is general enough to deal with all extraction tasks. This issue becomes more challenging when the types of concepts to be extracted are numerous and very diverse. For example, in the task of extracting data elements (important concepts related to certain medical conditions, such as encounter information, laboratory tests, imaging tests, and medications) from clinical notes, there are usually several types of data elements that need to be extracted (e.g., 183 data elements in our three cohorts), and different categories of data elements may be better extracted by different NLP tools because of the difference in concept dictionaries or extraction algorithms/pipelines. To address this issue, we propose to apply ensemble methods to integrate NLP tools. Ensemble methods have been proven to be effective to boost the performance of classifiers.5 The superiority of ensemble methods has been shown empirically in a wide range of data mining and NLP competitions.6, 7, 8, 9, 10 Many ensemble methods are available, including basic methods (union and intersection) and advanced ones (Binary Relevance (BR),15 Multi-Label K-Nearest Neighbor (MLkNN),16 Instance-Based Logistic Regression for Multi-Label (IBLR-ML),17 Random k-Labelsets (RAkEL),18 and Ensemble of Classifier Chains (ECC)19), spanning a large spectrum of sophistication. Advanced ensemble methods can weigh the importance of component NLP tools by applying machine learning to learn the best set of weights.
Although ensemble methods are empirically known to improve classification performance in several problems, it is not clear to what extent ensembles of NLP tools improve the extraction of numerous and diverse data elements from clinical notes. Several recent studies applied ensemble methods to extract concepts from clinical text.11, 12, 13, 48, 49, 50 However, most of them focus on the extraction of a few concept types, such as identifying 6 types of medication information,48 predicting 3 types of concepts (problem, test, or treatment),11, 49, 50 classifying 9 types of radiology concepts,12 or determining 8 concept types related to heart disease.13 It is unclear whether these methods are suitable for dealing with numerous and diverse concepts such as the 183 data elements from our project.
Phenotyping (i.e., characterization of disease states using electronic health records) relies heavily on structured data items, as well as on NLP for data element extraction from narrative text, and is a critical component of precision medicine.29, 30, 31, 32, 33 Our goal was to quantify the improvement in performance achieved by ensembles of popular NLP tools in the phenotyping of three very different cohorts for (1) congestive heart failure (CHF), (2) weight management/obesity (WM/O), and (3) Kawasaki disease (KD). These three conditions are use cases for the pSCANNER (patient-centered SCAlable National Network for Effectiveness Research) clinical data research network, a stakeholder-governed federated network that utilizes a distributed, service-oriented architecture to integrate data from multiple health systems20.
In order to evaluate the ensemble approach, we developed an NLP ensemble pipeline to integrate two popular NLP tools, cTAKES1 and MetaMap.2 Our pipeline can integrate these NLP tools using the basic and advanced ensemble methods mentioned previously, and allows us to evaluate the performance of NLP tools when they are applied alone and when they are integrated using different ensemble methods.
Methods
NLP Ensemble Pipeline
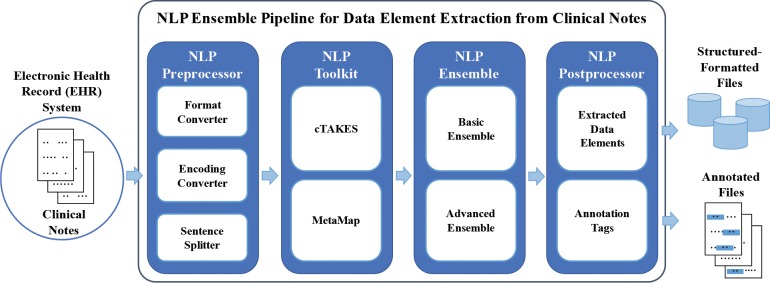
Figure 1 provides an overview of our pipeline. Inputs are clinical notes, which are preprocessed before they are ready for NLP tool extraction of data elements. The output of the NLP tools is integrated by the ensemble methods, and either structured-formatted files or annotated files serve as outputs. The output formats of our pipeline include extraction results such as structured data that are ready to be exported in a Common Data Model (CDM) format (such as the one pSCANNER uses, the Observational Medical Outcomes Partnership CDM (http://omop.org/CDM)), as well as annotation tags in the format used by the Brat Rapid Annotation Tool (BRAT),24 a Web-based graphical interface for text annotation that allows users to visualize and correct outputs if necessary.
Figure 1.
System diagram of the NLP ensemble pipeline for the pSCANNER project. The basic ensemble methods include union and intersection. The advanced ensemble methods include Binary Relevance (BR), Multi-Label K-Nearest Neighbor (MLkNN), Instance-Based Logistic Regression for Multi-Label (IBLR-ML), Random k-Labelsets (RAkEL), and Ensemble of Classifier Chains (ECC).
In this paper, we focus on three cohorts of pSCANNER: CHF, WM/O, and KD. Subject matter experts identified important data elements for research on patients with these conditions. To extract these data elements, we integrated two NLP tools: cTAKES,1 an NLP tool for concept extraction from free text clinical notes in an EHR; and MetaMap,2 a tool for recognizing UMLS23 concepts in text. These tools cover a wide range of applications of NLP for clinical note concept extraction and clinical note processing.
Data Elements and Mapping Tables
We worked with subject matter experts to identify 183 common data elements related to each cohort (50 for CHF, 96 for WM/O, and 37 for KD). The resulting data elements for CHF, WM/O and KD are shown in Tables 1, 2 and 3, respectively. PR and CNH mapped all data elements to the most specific standard codes (SNOMED-CT,26 LOINC,27 RxNorm,28 or UMLS23) using the BioPortal web service34 and created a data-element mapping table for each condition. We used the data-element mapping tables to normalize the output formats of the NLP tools that we integrated. The output standard codes of cTAKES include SNOMED-CT and RxNorm. MetaMap outputs UMLS. These standard codes were mapped to unique data elements in the table. If the output of any tool contained multiple standard codes for an extracted concept, we mapped all standard codes to the unique data elements. This way the outputs of all NLP tools were normalized and ready to be inputs for the ensemble methods.
Table 1.
Common data elements for congestive heart failure (CHF)
| Category | Data Elements |
|---|---|
| Terms | Congestive Heart Failure |
| Encounter Information | First Outpatient Appointment Date, Days Since Symptom Onset, Date Admitted, Date Discharged |
| Other Information | Height, Weight, Body Mass Index |
| Laboratory Tests | Blood Urea Nitrogen, Brain Natriuretic Peptides, Lipids, Serum Creatinine, Red Blood Cell Count, Serum Albumin, N-Terminal of the Prohormone Brain Natriuretic Peptide, Troponin |
| Imaging Tests | Chest X-Ray, Cardiac Magnetic Resonance Imaging, Angio Computed Tomography, Cardiac Nuclear Medicine Study, 2D Echo |
| Medications | Angiotensin-Converting Enzyme Inhibitor, Angiotensin Receptor Blocker, Antiarrhytmics, Warfarin, Apixaban, Edoxaban, Fondaparinux, Rivaroxaban, Enoxaparin, Heparin, Bivalirudin, Lepirudin, Argatroban, Dabigatran, Diuretics, Beta-Blocker |
| History and Progress | Chief Complaint, Past Medical History, Allergy |
| Comorbidities | Hypertension, Diabetes Mellitus, Atherosclerotic Disease, Obesity |
| Implants and Procedures | Valve Replacements, Coronary Angioplasty, Implantable Cardioverter-Defibrillator, Implantable Pacemaker, Aneurysm Surgery, Cardiac Resynchronization Therapy |
Table 2.
Common data elements for weight management/obesity (WM/O)
| Category | Data Elements |
|---|---|
| Terms | Obesity, Overweight, Morbid Obesity, Abnormal Weight Gain, Obesity By Adipocyte Growth Pattern, Hyperplastic Obesity |
| Encounter Information | First Outpatient Appointment Date, Days Since Symptom Onset, Date Admitted, Date Discharged |
| Other Information | Height, Weight, Body Mass Index, Health Status, Disability, Smoking Status, Nutrition, Alcohol, Physical Activity |
| Laboratory Tests | Triglyceride, Glycerol, Cholesterol, Apolipoprotein, Lipoprotein, High-Density Lipoprotein, Low-Density Lipoprotein, Homocystine, C-Reactive Protein, Thyroid Function, Liver Function Tests, H. Pylori Testing, Fasting Glucose, Hemoglobin A1c, Lipids, Serum Creatinine, Vitamin-D |
| Imaging Tests | Hip X-Ray, Knee X-Ray, Spine X-Ray, Hip Magnetic Resonance Imaging, Knee Magnetic Resonance Imaging, Spine Magnetic Resonance Imaging, Angiography, Right Upper Quadrant Ultrasound, Cholangiogram, Polysomnogram |
| Medications | Nonsteroidal Anti-Inflammatory Drug, Phentermine, Contrave Naltrexone, Contrave Bupropion, Qsymia, Belveq, Xenical, Metformin, Statins, Diethylpropion, Phendimetrazine, Benzphetamine, Liraglutide, Probiotics |
| History and Progress | Chief Complaint, Past Medical History, Allergy |
| Comorbidities | Hypertensive Disorder, Diabetes Mellitus, Hyperlipidemia, Obstructive Sleep Apnea, Cardiovascular Disorder, Intracranial Hypertension, Depressive Disorder, Binge Eating, Degenerative Arthritis, Congestive Heart Failure, Nonalcoholic Steatohepatitis, Cancer, Human Immunodeficiency Virus |
| Surgical Procedures | Bariatric Surgery, Laparoscopic Surgery, Gastric Bypass, Roux-En-Y, Lap-Band, Gastroplasty, Sleeve Gastrectomy, Duodenal Switch |
| Comorbidities Surgical Procedures | Knee Arthroplasty, Cholecystectomy, Aortocoronary Bypass |
| Vital Signs | Temperatue, Blood Pressure, Pulse, Respiratory Rate, Pain |
| Demographic Information | Address, Languages Spoken, Socioeconomic Status |
| Enrollment in Care-Coordination | Home Telehealth Monitoring |
Table 3.
Common data elements for Kawasaki disease (KD)
| Category | Data Elements |
|---|---|
| Terms | Kawasaki Disease |
| Encounter Information | Fever Days at Admission, Date Admitted, Date Discharged |
| Other Information | Height, Weight, Age |
| Laboratory Tests | Erythrocyte Sedimentation Rate, C Reactive Protein, White Blood Cell, Hemoglobin, Platelet, Absolute Neutrophil Count, Gamma-Glutamyl Transpeptidase, Alanine Transaminase, Albumin, Electrolytes, Urinalysis |
| Imaging Tests | Echo, Cardiac Magnetic Resonance Imaging, Angio Computed Tomography |
| Medications | Steroids, Intravenous Immunoglobulin, Naprosyn, Antiplatelets-Abciximab, Acetylsalicylic Acid, Clopidogrel, Anticoagulants-Heparin, Warfarin, Enoxaparin, Direct Thrombin Inhibitor, TNF-Alpha Antagonists-Infliximab |
| Echo | Left Main Coronary Artery, Left Anterior Descending, Right Coronary Artery, Left Circumflex Artery, Ejection Fraction |
Ensemble Methods
Our pipeline can integrate NLP tools using two basic ensemble methods: Union, which extracts the data element if any of the NLP tools detects the data element; and Intersection, which extracts the data element if both NLP tools detect the data element.
Although basic ensemble methods can combine the results of multiple NLP tools, the relationships (e.g., category, similarity, shared procedure or component) among data elements are not considered. To explicitly consider the relationships among data elements, we further converted the ensemble problem into a binary Multi-Label Classification (MLC) task, where each instance is a sentence in a note, features are the binary extraction results of each data element for each NLP tool, and labels are the binary ground truth of whether the data element exists in that sentence or not. For example, suppose there are 10,000 sentences in the data repository, 50 data elements to be extracted, and 2 NLP tools for the ensemble, then the corresponding MLC problem consists of 10,000 instances, 50 labels, and 100 (50 * 2) features.
To solve the MLC problem, we used five well-known algorithms to build our advanced ensemble: BR,15 which trains a binary classifier for each label independently; MLkNN,16 which is an instance-based method that extends the k- Nearest Neighbor (kNN) method to multi-label data; IBLR-ML,17 which is based on a formalization of instance-based classification as logistic regression and takes the correlation among labels into account; RAkEL,a which randomly selects subsets of the label powerset (treating each distinct combination of labels as a different class) and combines them using a voting scheme; and ECC,19 which trains several classifiers in random order on a random subset of data, and combines them by voting. All advanced methods, except BR, learn relationships among labels (data elements) during training process.
Test Corpus
We tested the above mentioned NLP tools and ensemble methods on 130 clinical notes (100 from the public domain and 30 private).
For public clinical notes, TTK, PR and CNH collected 45,136 notes from MT Samples,35 i2b2 Challenges 2006, 2008 - 2012,36, 37, 38, 39, 40, 41, 42, 43 and ShARe CLEF eHealth Tasks 2013 Task 1 and 2, and 2014 Task l.44, 45, 46 Then, for each condition, the notes were selected based on keyword combinations: we used “congestive heart failure” for CHF, “weight management AND obesity” for WM/O, and “Kawasaki OR (fever AND rash AND red AND child) for KD. It should be noted that these rules only serve as simple filters for sampling notes, thus we did not consider synonyms or stemming. After narrowing down the search by keywords, we randomly selected notes for the evaluation from the filtered notes: 33 notes for CHF, 34 notes for WM/O, and 33 notes for KD. The total number of public domain notes was 100.
For private clinical notes, TTK and CNH randomly sampled 30 notes, from a pool of 381 notes collected by JDC for KD14, 22 (from Rady Children’s Hospital and the Emory University; Institutional Review Boards approved this study).
For the 130 notes, PR and CNH manually annotated 6,914 mentions of data elements (1,885 for CHF, 1,728 for WM/O, 1,678 for KD-Public, and 1,623 for KD-Private. TTK and CNH applied Stanford CoreNLP47 to split the 130 notes into 9,320 sentences (3,045 for CHF, 1,778 for WM/O, 2,824 for KD-Public, and 1,673 for KD-Private). We tested a total of four datasets: CHF (33 notes), WM/O (34 notes), KD-Public (33 notes), and KD-Private (30 notes). For each dataset, we randomly selected 50% of these notes for training, and held out the remaining 50% for performance reporting. We used 10-fold cross validation in training.
Evaluation Metrics
We considered a data element extraction to be correct using two different levels: corpus-level and sentence-level. For corpus-level, the prediction of a data element in a clinical note is considered correct if the data element does appear in the ground truth annotations in the same note (not necessarily in the same sentence). For sentence-level, the prediction is considered correct if the data element does appear in the ground truth annotations in the same sentence. It should be noted that the predictions are binary for both levels, thus multiple concepts of a corrected predicted data element would only count as a single true positive.
We computed Precision, Recall and F1-Scores for each method as our evaluation measures, and computed the corpus-level and sentence-level of each metric. The corpus-level F1-Score is defined as where corpus-level precision , and corpus-level recall . The sentence-livel Fl-Scoreis defined as where N = # of sentences, for eachsentence i, Sentence-level Fl-Score is defined as , and sentence-level recall .
Implementation
We utilized the implementation of the MLC algorithms available in the MULAN21 package for ensembles, and applied J4825 as the base learner for the MLC algorithms. For cTAKES,1 we utilized the Dictionary Lookup Fast Pipeline51 and the built-in concept dictionary, which was a subset of UMLS23 containing SNOMED-CT,26 RxNorm,28 and all of the synonyms. The system was implemented in Java, Python, and Shell Scripts. Also, we released the ensemble component in our pipeline. In this component, there were three inputs: (1) ground truth annotations in BRAT format, (2) annotations generated by an individual NLP tool (also in BRAT format), and (3) the beginning and ending position of each sentence in notes (generated by the sentence splitter). This component can perform basic and advanced ensembles, compare the ground truth annotations, and output the corpus- and sentence-level evaluation results. The code is available at https://github.com/tsungtingkuo/ensemble.
Results
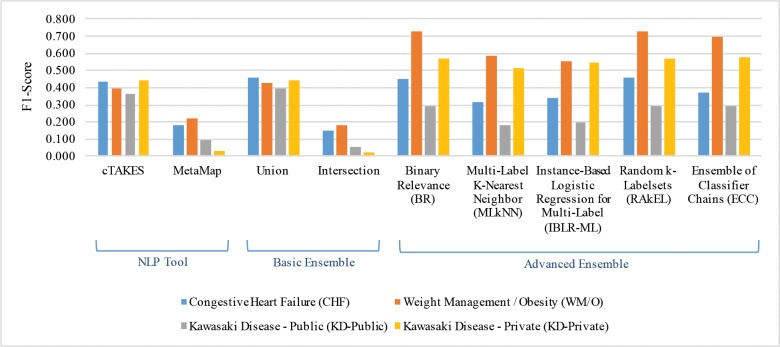
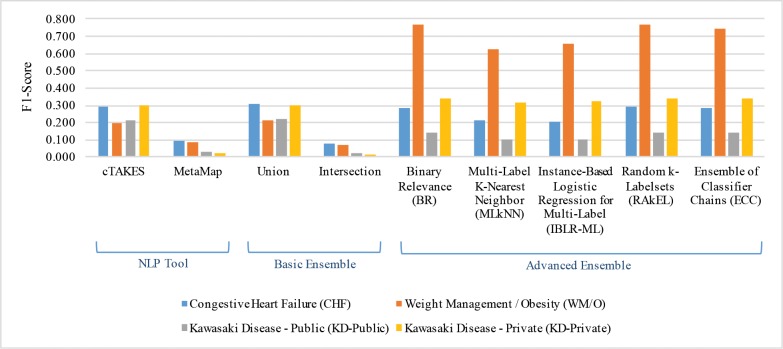
Figures 2 and 3 show corpus- and sentence-level F1-Scores for the four datasets using the two NLP tools, two basic ensemble methods, and five advanced ensemble methods. The detailed Precision (P), Recall (R), and F1-Scores (F) for corpus- and sentence-level results are shown in Tables 4 and 5. In general, the scores of corpus-level evaluation are higher than those of sentence-level. We believe this is due to the aggregation of corpus-level results from sentence-level results, as sentence-level extraction is more challenging than corpus-level extraction.
Figure 2.
Corpus-level F1-Scores for NLP tools, basic ensemble, and advanced ensemble methods.
Figure 3.
Sentence-level FI-Scores for NLP tools, basic ensemble, and advanced ensemble methods.
Table 4.
Corpus-level Precision (P), Recall (R) and Fl-Score (F) of NLP tools, basic ensemble, and advanced ensemble methods. Numbers highlighted as blue underlined text indicate the best scores for each evaluation trial.
| Category | Method | Congestive Heart Failure (CHF) | Weight Management / Obesity (WM/O) | Kawasaki Disease - Public (KD-Public) | Kawasaki Disease - Private (KD-Private) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | R | F | P | R | F | P | R | F | P | R | F | ||
| NLP Tool | cTAKES | .882 | .285 | .431 | .619 | .292 | .397 | .918 | .227 | .364 | .951 | .286 | .440 |
| MetaMap | .842 | .102 | .181 | .902 | .126 | .222 | .920 | .052 | .098 | 1.000 | .015 | .029 | |
| Basic Ensemble | Union | .890 | .307 | .456 | .634 | .320 | .425 | .925 | .249 | .393 | .951 | .289 | .443 |
| Intersection | .809 | .080 | .146 | .916 | .099 | .179 | .867 | .029 | .057 | 1.000 | .012 | .024 | |
| Advanced Ensemble | Binary Relevance (BR) | .794 | .317 | .453 | .738 | .715 | .726 | .848 | .175 | .291 | .891 | .422 | .573 |
| Multi-Label K-Nearest Neighbor (MLkNN) | .797 | .199 | .318 | .715 | .499 | .588 | .849 | .101 | .181 | .835 | .375 | .518 | |
| Instance-Based Logistic Regression for Multi- Label (IBLR-ML) | .370 | .321 | .344 | .557 | .550 | .553 | .505 | .124 | .199 | .681 | .459 | .549 | |
| Random k-Labelsets (RAkEL) | .836 | .313 | .455 | .737 | .717 | .727 | .848 | .175 | .291 | .894 | .417 | .569 | |
| Ensemble of Classifier Chains (ECC) | .431 | .321 | .368 | .729 | .670 | .698 | .848 | .175 | .291 | .887 | .427 | .577 | |
Table 5.
Sentence-level Precision (P), Recall (R) and F1-Score (F) of NLP tools, basic ensemble, and advanced ensemble methods. Numbers highlighted as blue underlined text indicate the best scores for each evaluation trial.
| Category | Method | Congestive Heart Failure (CHF) | Weight Management / Obesity (WM/O) | Kawasaki Disease - Public (KD-Public) | Kawasaki Disease - Private (KD-Private) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | R | F | P | R | F | P | R | F | P | R | F | ||
| NLP Tool | cTAKES | .354 | .268 | .293 | .224 | .203 | .197 | .273 | .193 | .213 | .425 | .257 | .296 |
| MetaMap | .132 | .077 | .091 | .125 | .073 | .086 | .062 | .026 | .034 | .034 | .017 | .020 | |
| Basic Ensemble | Union | .369 | .280 | .306 | .237 | .216 | .209 | .281 | .204 | .223 | .432 | .263 | .303 |
| Intersection | .115 | .065 | .076 | .101 | .060 | .070 | .047 | .015 | .021 | .027 | .011 | .013 | |
| Advanced Ensemble | Binary Relevance (BR) | .329 | .281 | .285 | .791 | .793 | .767 | .208 | .128 | .144 | .412 | .315 | .341 |
| Multi-Label K-Nearest Neighbor (MLkNN) | .268 | .200 | .211 | .673 | .621 | .629 | .147 | .088 | .102 | .385 | .289 | .312 | |
| Instance-Based Logistic Regression for Multi- Label (IBLR-ML) | .211 | .273 | .209 | .683 | .656 | .654 | .144 | .100 | .103 | .364 | .326 | .321 | |
| Random k-Labelsets (RAkEL) | .350 | .276 | .294 | .790 | .793 | .767 | .208 | .128 | .144 | .413 | .314 | .341 | |
| Ensemble of Classifier Chains (ECC) | .325 | .278 | .282 | .778 | .760 | .743 | .208 | .128 | .144 | .413 | .319 | .343 | |
For basic ensemble methods, Union generally improved performance over a single NLP tool, indicating that coverage is a critical concern for these data element extraction tasks (Figures 2 and 3). The importance of coverage can also be seen in Tables 4 and 5, where single NLP tools show high precision but relatively low recall. This also explains why Intersection consistently performs worse than all other methods (even worse than single NLP tools).
For advanced ensemble methods, although no method consistently performed the best, we did observe an interesting phenomenon: these MLC ensemble algorithms boosted performance on WM/O and KD-Private datasets, but they did not perform well on CHF and KD-Public datasets. This is related to label density, which is the number of ground truth annotations per sentence. For example, for WM/O the label density was (1,678 annotations) / (1,778 sentences) = 0.94, while for CHF the label density was only (1,885 annotations) / (3,045 sentences) = 0.62. Since the advanced ensemble methods applied multi-label learning, the datasets with higher label density (WM/O and KD-Private) provided better training examples for the classifiers and better recall (as shown in Tables 4 and 5). It should be noted that although we only use two NLP tools in our experiment, advanced ensemble methods may still be very useful to improve the extraction results. For example, consider we are extracting CHF data elements from this sentence: “Mr. X is being discharged on Lasix, Digoxin and Toprol daily.” One NLP tool may successfully extract “Lasix” as the data element “Diuretics,” while the other tool may successfully extract “Digoxin” as “Antiarrhythmics,” but both tools may fail to extract “Toprol” as “Beta-Blocker.” In this scenario, neither union nor intersection can improve the extraction results. However, if these three data elements (“Diuretics,” “Antiarrhythmics,” and “Beta-Blocker”) are usually mentioned together in the training clinical notes, MLC ensemble algorithms may be able to recover “Beta-Blocker” even if both NLP tools miss the mention “Toprol.” We believe this is the reason why advanced ensemble methods are able to largely improve the results compared to individual NLP tools.
Also, the performance of the advanced ensemble methods is bounded by the limited availability of the annotated clinical notes. We anticipate that, when more annotated clinical notes become available for training and testing, the performance improvement of the advanced ensemble methods will be more consistent and obvious, especially for data with high label density. Additionally, in some settings (such as clinical notes from primary care providers with patients facing multitudes of conditions), the number of data elements might be on the order of hundreds of thousands, and thus more training examples are required for learning MLC models.
We also conducted qualitative analysis of our results. For each dataset, the data elements with highest and lowest F1-Score, extracted using the Union ensemble method (because it consistently performed better), are listed in Table 6 to illustrate which data elements were the most or the least successful in the extraction. A comparison of data elements with the highest and lowest F1-Scores suggests that, in general, items in the category history and progress are harder to accurately extract than those in comorbidities. However, for some categories such as medications and laboratory tests, the extraction performance varies for each data element. This observation also indicates that adding more diverse tools (e.g., specifically designed to extract history and progress, or to extract the data elements of medications or laboratory tests) may further boost overall performance. Our average processing time (seconds per note) is 1.14 for cTAKES, 44.29 for MetaMap, 0.19 for basic and 2.10 for advanced ensembles.
Table 6.
Results for data elements with highest and lowest F1-Scores using the Union ensemble method. The data elements are ordered according to their F1-Scores (or the number of matches to break a tie).
| Fl-Score | Dataset | Category | Data Element |
|---|---|---|---|
| Highest | Congestive Heart Failure (CHF) | Medications | Heparin |
| Comorbidities | Diabetes Mellitus | ||
| Laboratory Tests | Serum Albumin | ||
| Weight Management / Obesity (WM/O) | Surgical Procedures | Roux-En-Y | |
| Comorbidities | Diabetes Mellitus | ||
| Hyperlipidemia | |||
| Kawasaki Disease – Public (KD-Public) | Medications | Acetylsalicylie Acid | |
| Anticoagulants-Heparin | |||
| Intravenous Immunoglobulin | |||
| Kawasaki Disease – Private (KD-Private) | Laboratory Tests | Gamma-Glutamyl Transpeptidase | |
| Albumin | |||
| Medications | Intravenous Immunoglobulin | ||
| Lowest | Congestive Heart Failure (CHF) | Medications | Angiotensin-Converting Enzyme Inhibitor |
| Warfarin | |||
| History and Progress | Chief Complaint | ||
| Weight Management / Obesity (WM/O) | Vital Signs | Pulse | |
| Laboratory Tests | Serum Creatinine | ||
| History and Progress | Chief Complaint | ||
| Kawasaki Disease – Public (KD-Public) | Medications | Warfarin | |
| Clopidogrel | |||
| Imaging Tests | Angio Computed Tomography | ||
| Kawasaki Disease – Private (KD-Private) | Imaging Tests | Echo | |
| Terms | Kawasaki Disease | ||
| Laboratory Tests | Platelet |
Conclusion
We developed an NLP ensemble pipeline to extract data elements from clinical notes, using state-of-the-art NLP tools and ensemble methods. We tested our pipeline on public and private notes for CHF, WM/O and KD. The results indicate that the Union ensemble method provides consistent improvement, while the MLC-based ensemble methods may be useful on datasets with higher density of concepts in the clinical notes. However, our study was limited to data elements selected for three cohorts only and limited by the size of clinical notes. Additionally, we used ensembles of only two NLP tools, thus our current evaluation results might not generalize to other pairs of systems nor to larger sets of systems. Nevertheless, we demonstrated that our proposed ensemble pipeline can improve the performance of NLP tools, and it may provide a practical solution for the extraction of data elements from clinical notes.
In the future, we plan to test our pipeline on a larger number of clinical notes, include additional data elements for these three and additional cohorts to increase generalizability, wrap additional NLP tools into the pipeline, and add more concept dictionaries to improve coverage (e.g., dictionaries that include acronyms). Furthermore, we will add additional ensemble methods to improve the overall extraction accuracy and sensitivity of our pipeline (adding more NLP tools enables us to use voting and include more complex ensemble algorithms). We also plan to perform additional analysis to explain the differences in the corpora and the performance of the ensemble methods (e.g., learning curve, execution time, and tool-category performance analysis), adopt parallel computing to increase scalability, and release annotations for the public domain clinical notes. We will disseminate our methods to other researchers, and test our processes in other clinical data research networks like pSCANNER.
Acknowledgements
This work is funded by PCORI contract CDRN-1306-04819. We thank the following domain experts for identifying and refining the data elements for CHF, WM/O and KD: Howard Taras, MD, UCSD; Zhaoping Li, MD, UCLA; Jane Burns, MD, UCSD; Adriana Tremoulet, MD, UCSD; Michael Ong, MD, PhD, UCLA; and Paul A. Heidenreich, MD, MS, Stanford and Palo Alto VA. Part of the de-identified clinical records used in this research were provided by the i2b2 National Center for Biomedical Computing funded by U54LM008748 and were originally prepared for the Shared Tasks for Challenges in NLP for Clinical Data organized by Dr. Ozlem Uzuner, i2b2 and SUNY. The computational infrastructure was provided by the iDASH National Center for Biomedical Computing funded by U54HL108460 and managed by the Clinical Translational Research Institute CTSA Informatics team led by Antonios Koures, PhD, funded in part by UL1TR001442.
References
- 1.Savova Guergana, Masanz James, Ogren Philip, Zheng Jiaping, Sohn Sunghwan, Kipper-Schuler Karin, Chute Christopher. 2010. Mayo Clinic Clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. JAMIA. 2010;17:507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aronson AR, Lang F-M. An overview of MetaMap: historical perspective and recent advances. JAMIA. 2010;17(3):229–236. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garvin Jennifer H, et al. Automated extraction of ejection fraction for quality measurement using regular expressions in Unstructured Information Management Architecture (UIMA) for heart failure. JAMIA. 2012;19(5):859–866. doi: 10.1136/amiajnl-2011-000535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denny Joshua C, et al. Identifying QT prolongation from ECG impressions using a general-purpose Natural Language Processor. International journal of medical informatics. 2009;78:S34–S42. doi: 10.1016/j.ijmedinf.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dietterich Thomas G. Multiple Classifier Systems. Springer Berlin Heidelberg; 2000. Ensemble methods in machine learning; pp. 1–15. [Google Scholar]
- 6.Lo Hung-Yi, et al. Learning to improve area-under-FROC for imbalanced medical data classification using an ensemble method. ACM SIGKDD Explorations Newsletter. 2008;10(2):43–46. [Google Scholar]
- 7.Lo Hung-Yi, et al. An ensemble of three classifiers for KDD cup 2009: Expanded linear model, heterogeneous boosting, and selective naive Bayes. JMLR W&CP. 2009;7 [Google Scholar]
- 8.Hsiang-Fu Yu, et al. Feature Engineering and Classifier Ensemble for KDD Cup 2010. KDD Cup 2010 WS [Google Scholar]
- 9.Chen Po-Lung, et al. A Linear Ensemble of Individual and Blended Models for Music Rating Prediction. JMLR-W&CP. 2012 [Google Scholar]
- 10.McKenzie Todd G, et al. Novel Models and Ensemble Techniques to Discriminate Favorite Items from Unrated Ones for Personalized Music Recommendation. JMLR-W&CP. 2012 [Google Scholar]
- 11.Torii M, Wagholikar K, Liu H. Using machine learning for concept extraction on clinical documents from multiple data sources. Journal of the American Medical Informatics Association. 2011 Sep 1;18(5):580–7. doi: 10.1136/amiajnl-2011-000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esuli A, Marcheggiani D, Sebastiani F. An enhanced CRFs-based system for information extraction from radiology reports. Journal of biomedical informatics. 2013 Jun 30;46(3):425–35. doi: 10.1016/j.jbi.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Chen Q, Li H, Tang B, Wang X, Liu X, Liu Z, Liu S, Wang W, Deng Q, Zhu S, Chen Y. An automatic system to identify heart disease risk factors in clinical texts over time. Journal of biomedical informatics. 2015 Dec 31;58:S158–63. doi: 10.1016/j.jbi.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doan S, Maehara CK, Chaparro JD, Lu S, Liu R, Graham A, Berry E, Hsu CN, Kanegaye JT, Lloyd DD. Ohno- Machado L. Building a Natural Language Processing Tool to Identify Patients with High Clinical Suspicion for Kawasaki Disease from Emergency Department Notes. Academic Emergency Medicine. 2016 Jan 1; doi: 10.1111/acem.12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsoumakas Grigorios, Katakis Ioannis. Dept. of Informatics, Aristotle University of Thessaloniki. Greece: 2006. Multi-label classification: An overview. [Google Scholar]
- 16.Zhang Min-Ling, Zhou Zhi-Hua. ML-KNN: A lazy learning approach to multi-label learning. Pattern recognition. 2007;40(7):2038–2048. [Google Scholar]
- 17.Cheng Weiwei, Hüllermeier Eyke. Combining instance-based learning and logistic regression for multilabel classification. Machine Learning. 2009;76(2-3):211–225. [Google Scholar]
- 18.Tsoumakas Grigorios, Vlahavas Ioannis. Machine learning: ECML 2007. Springer Berlin Heidelberg; 2007. Random k-labelsets: An ensemble method for multi-label classification; pp. 406–417. [Google Scholar]
- 19.Read Jesse, et al. Classifier chains for multi-label classification. Machine learning. 2011;85(3):333–359. [Google Scholar]
- 20.Ohno-Machado Lucila, et al. pSCANNER: patient-centered Scalable National Network for Effectiveness Research. Journal of the American Medical Informatics Association. 2014;21(4):621–626. doi: 10.1136/amiajnl-2014-002751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsoumakas Grigorios, et al. Mulan: A java library for multi-label learning. The Journal of Machine Learning Research. 2011;12:2411–2414. [Google Scholar]
- 22.Hsu Chun-Nan, Doan Son, Chaparro Juan D, Maehara Cleo K, Liu Ruiling, Lu Sisi, Berry Erika K, Chhabra Divya, Tremoulet Adriana H, Burns Jane C. Natural language processing to screen for Kawasaki disease; Abstract accepted for poster presentation in Pediatric Academic Societies Annual Meeting (PSA); San Diego, CA. 2015. Apr 27, [Google Scholar]
- 23.Bodenreider Olivier. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic acids research. 2004;32(1):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stenetorp Pontus, et al. Association for Computational Linguistics. 2012. BRAT: a web-based tool for NLP-assisted text annotation. Proceedings of the Demonstrations at the 13th Conference of the European Chapter of the Association for Computational Linguistics. [Google Scholar]
- 25.Hall Mark, et al. The WEKA data mining software: an update. ACM SIGKDD explorations newsletter. 2009;11(1):10–18. [Google Scholar]
- 26.Stearns Michael Q, et al. SNOMED clinical terms: overview of the development process and project status. Proceedings of the AMIA Symposium. American Medical Informatics Association. 2001 [PMC free article] [PubMed] [Google Scholar]
- 27.McDonald Clement J, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clinical chemistry. 2003;49(4):624–633. doi: 10.1373/49.4.624. [DOI] [PubMed] [Google Scholar]
- 28.Liu Simon, et al. RxNorm: prescription for electronic drug information exchange. IT Prof. 2005;7(5):17–23. [Google Scholar]
- 29.Kotfila C, Uzuner ö. A systematic comparison of feature space effects on disease classifier performance for phenotype identification of five diseases. Journal of biomedical informatics. 2015;58:S92–S102. doi: 10.1016/j.jbi.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Q, Melton K, Lingren T, Kirkendall E.S, Hall E, Zhai H, Ni Y, Kaiser M, Stoutenborough L, Solti I. Phenotyping for patient safety: algorithm development for electronic health record based automated adverse event and medical error detection in neonatal intensive care. Journal of the American Medical Informatics Association. 2014;21(5):776–784. doi: 10.1136/amiajnl-2013-001914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shivade C, Raghavan P, Fosler-Lussier E, Embi P.J, Elhadad N, Johnson S.B, Lai A.M. A review of approaches to identifying patient phenotype cohorts using electronic health records. Journal of the American Medical Informatics Association. 2014;21(2):221–230. doi: 10.1136/amiajnl-2013-001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gundlapalli A.V, Redd A, Carter M, Divita G, Shen S, Palmer M, Samore M.H. Validating a strategy for psychosocial phenotyping using a large corpus of clinical text. JAMIA. 2013;20(e2):e355–e364. doi: 10.1136/amiajnl-2013-001946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu S, Liao K.P, Shaw S.Y, Gainer V.S, Churchill S.E, Szolovits P, Murphy S.N, Kohane I.S, Cai T. Toward high-throughput phenotyping: unbiased automated feature extraction and selection from knowledge sources. Journal of the American Medical Informatics Association. 2015;22(5):993–1000. doi: 10.1093/jamia/ocv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whetzel Patricia L, et al. BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic acids research. 2011;39(2):W541–W545. doi: 10.1093/nar/gkr469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MT Samples. http://www.mtsamples.com.
- 36.Uzuner Ö, Juo Y, Szolovits P. Evaluating the state-of-the-art in automatic de-identification. JAMIA. 2007 doi: 10.1197/jamia.M2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uzuner Ö, Goldstein I, Luo Y, Kohane I. Identifying patient smoking status from medical discharge records. JAMIA. 2008;15(1):15–24. doi: 10.1197/jamia.M2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uzuner Ö. Recognizing obesity and co-morbidities in sparse data. JAMIA. 2009 Jul;16(4):561–570. doi: 10.1197/jamia.M3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uzuner Ö, Solti I, Xia F, Cadag E. Community annotation experiment for ground truth generation for the i2b2 medication challenge. JAMIA. 2010 doi: 10.1136/jamia.2010.004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uzuner Ö, Solti I, Cadag E. Extracting medication information from clinical text. JAMIA. 2010 doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uzuner Ö, South B, Shen S, DuVall S. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. JAMIA. 2011;18:552–556. doi: 10.1136/amiajnl-2011-000203. Published Online First: 16 June 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uzuner Ö, et al. Evaluating the state of the art in coreference resolution for electronic medical records. JAMIA. 2012;19(5):786–791. doi: 10.1136/amiajnl-2011-000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun Weiyi, Rumshisky Anna, Uzuner Ozlem. Evaluating temporal relations in clinical text: 2012 i2b2 challenge. JAMIA. 2013;20(5):806–813. doi: 10.1136/amiajnl-2013-001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suominen Hanna, et al. Multilinguality, Multimodality, and Visualization. Berlin Heidelberg: Springer; 2013. Overview of the ShARe/CLEF eHealth evaluation lab 2013. Information Access Evaluation; pp. 212–231. [Google Scholar]
- 45.Kelly Liadh, et al. Multilinguality, Multimodality, and Interaction. Springer International Publishing; 2014. Overview of the ShARe/CLEF eHealth evaluation lab 2014. Information Access Evaluation; pp. 172–191. [Google Scholar]
- 46.Goldberger AL, Amaral LAN, Glass L, Hausdorff JM, Ivanov PCh, Mark RG, Mietus JE, Moody GB, Peng CK, Stanley HE. PhysioBank PhysioToolkit, and PhysioNet: Components of a New Research Resource for Complex Physiologic Signals. Circulation. 2000 Jun 13;101(23):e215–e220. doi: 10.1161/01.cir.101.23.e215. [DOI] [PubMed] [Google Scholar]
- 47.Manning Christopher D, Surdeanu Mihai, Bauer John, Finkel Jenny, Bethard Steven J., McClosky David. The Stanford CoreNLP Natural Language Processing Toolkit; Proceedings of 52nd Annual Meeting of the Association for Computational Linguistics: System Demonstrations; 2014. pp. 55–60. [Google Scholar]
- 48.Doan S, Collier N, Xu H, Duy PH. Phuong TM. Recognition of medication information from discharge summaries using ensembles of classifiers. BMC medical informatics and decision making. 2012 May 7;12(1):1. doi: 10.1186/1472-6947-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang N, Afzal Z, Singh B, Van Mulligen EM, Kors JA. Using an ensemble system to improve concept extraction from clinical records. Journal of biomedical informatics. 2012 Jun 30;45(3):423–8. doi: 10.1016/j.jbi.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 50.Kim Y, Riloff E. A stacked ensemble for medical concept extraction from clinical notes. AMIA Jt Summits Transl Sci Proc. 2015 [Google Scholar]
- 51.Health NLP. http://healthnlp.github.io/examples.