Abstract
Observational data recorded in the Electronic Health Record (EHR) can help us better understand the effects of therapeutic agents in routine clinical practice. As such data were not collected for research purposes, their reuse for research must compensate for additional information that may bias analyses and lead to faulty conclusions. Confounding is present when factors aside from the given predictor(s) affect the response of interest. However, these additional factors may not be known at the outset. In this paper, we present a scalable literature-based confounding variable discovery method for biomedical research applications with pharmacovigilance as our use case. We hypothesized that statistical models, adjusted with literature-derived confounders, will more accurately identify causative drug-adverse drug event (ADE) relationships. We evaluated our method with a curated reference standard, and found a pattern of improved performance ~ 5% in two out of three models for gastrointestinal bleeding (pre-adjusted Area Under Curve ≥ 0.6).
Introduction
Confounding is present when factors aside from given predictor(s) affect the response of interest. For example, one may wish to understand the association between cigarette smoking and the risk of developing cancer while asbestos exposure or genetic predispositions may be present 1. Stuart Mill noted a related disparity between laboratory conditions and raw observation in nature in his System of Logic (1943) 2. Since confounding variables may not be known at the outset, we propose a scalable and computationally inexpensive method for confounding variable discovery (CVD). In this paper, we apply our method to pharmacovigilance (PV). However, confounding is a challenge to multiple fields including epidemiology, biosurveillance and pharmacogenomics.
Pharmacovigilance and FAERS. Some 770,000 adverse drug events (ADEs) occur annually in the United States alone, resulting in morbidity, mortality, and increased cost 3,4. PV aims to address the set of challenges posed by ADEs, including those detected after drugs are released to market. Recognizing the need to systematically monitor adverse effects of drugs, regulatory agencies, such as the US Food and Drug Administration (FDA), have implemented spontaneous reporting systems, through which physicians and administrators of clinical trials can report potential adverse events as they are observed. However, spontaneous reporting systems (e.g., FAERS 5) have limitations, such as incomplete clinical information, under-reporting of side-effects, and unacknowledged sources of bias 3.
Confounding in EHR. Clinical text (notes) recorded in EHRs are another potential primary source of ADE data, yet drawing reliable conclusions from routinely collected clinical data is notoriously difficult 6,7 In the PV literature, the term “signal” refers to data that are pertinent to the therapies or outcomes under study. Unlike the case of clinical trials where subjects are deliberately monitored for side-effects, clinical data were not collected for the purpose of pharmacovigilance, and are often beset with redundancy or missing data, use of non-standard abbreviations, misspellings, and so on. In addition, clinical data contain confounding variables 8. By developing more powerful methods to identify and adjust for confounding variables, we should be able to discriminate better between drug-ADE signal and noise. This in turn would facilitate the timely compilation of more comprehensive pharmaceutical risk profiles and improve public safety.
Background
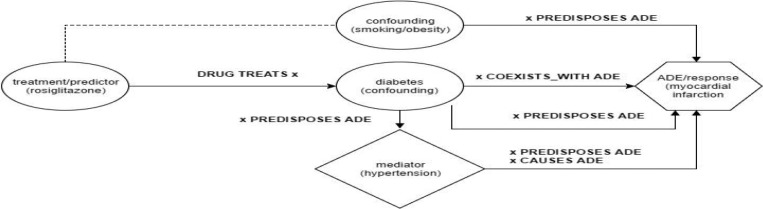
Definitions. A confounding variable influences or biases the magnitude of correlation between a predictor variable (e.g., drug exposure/treatment) and a response variable (i.e. outcome/ADE). In the context of creating models for PV, when a confounding relationship exists between a falsely associated drug-ADE pair and adjustments are made to account for its influence, the association strength for that relationship, should be diminished. For example, given a set of observational clinical notes, it is observed that fish oil intake is highly correlated with acute liver injury (ALI). However, after adjusting the model for the presence of known causal agents of ALI, (e.g., acetaminophen, liver cirrhosis, hepatitis c), that correlation should approach zero. Let us consider another example of an acetaminophen exposure (predictor) and a hepatitis B infection (predictor) where the patient subsequently suffers ALI (response). In this case, each of these predictors, independently, are sufficient preconditions for ALI. As they occur together, these two predictors confound each other. When the association of either predictor is adjusted in the absence of the other predictor, the association may be diminished, but not as dramatically as in the first example. Li et al. introduced a taxonomy of confounding in PV with the following categories: confounding by indication (e.g., preexisting conditions), confounding by comorbidity (e.g., diabetes), and confounding by co-medication (e.g., aspirin) 8.
A mediating variable (also called an intervening variable), by contrast, lies distinctly along the causal pathway between the predictor variable and the response variable themselves and may be neither necessary nor sufficient to cause an ADE by itself. Mediators may sometimes be thought of as “risk factors” or as aspects of the etiology of the ADE itself 8,9. Examples of mediators include bile duct obstruction for ALI or hypertension for myocardial infarction (MI). Mediators tend to be collinear with both the predictor and the response variable, which is to say they tend to be observed together. For more detailed discussion of mediation, see Pearl 9.
Confounding control in PV. The two most common approaches for the control of confounding are control by design (which entails the use of case control matching, for example, where patients are matched by practice type and sometimes demographic cohort) and control by analysis 6. The main thrust of this paper will be the latter (while still others exist, such as counterfactual intervention, they are beyond our scope) 6,10–13. Control by analysis often implies the application of domain knowledge and experience to identify proxy variables in order to control for confounding. Traditionally, statisticians have depended upon the knowledge of domain experts to identify relevant confounding factors. While this is likely to result in the identification of confounding variable candidates (CVCs) pertinent to an individual study, it would be financially intractable to hire the quantity and diversity of experts needed to conduct PV across large numbers of marketed drugs and each of their potential ADEs. Li et al. recently utilized extensive search for CVCs (of the “comorbidity” subtype in PV) in observational clinical data found in EHR using penalized multivariate regression methods, specifically lasso regression 8. Lasso regression is a variable selection technique that shrinks multivariate predictor coefficients that fall beneath a threshold down to zero 14. In general, the lasso produces results that are easy to interpret and parsimonious and, in this case in particular, the results of this study are very encouraging. However, Least Angle Regression (LAR), the algorithm that is most commonly used to perform lasso regression, can be computationally expensive, depending on input, being either quadratic O(n2) or cubic O(n3) in its computational complexity, though recent innovations such as cyclic coordinate descent, have produced improvements in this regard 15,16. Other innovative signal detection approaches have involved combining multiple data sources via meta-analysis, or applying omic or biochemical substantiation techniques to verify the plausibility of the causal mechanisms that may underlie putative drug-ADE associations 8, 17–19.
The methods of CVD mentioned above are to varying degrees financially intractable or computationally expensive, and it has been argued that brute force methods miss the “causal story” in the observational data 20. The purpose of our current work is to assess the extent to which feasibly-sized sets of CVCs that have been observed in the literature can be used to identify concepts for the task of confounding adjustment of clinical data. In doing so, we aim to find a middle ground between human-intensive expert-guided CVD, and computationally intensive selection of such variables based on empirical data alone.
Literature-based Discovery (LBD) is an idea first developed by Don Swanson as a means of using the biomedical literature to discover therapeutically useful associations 20,21. Swanson’s approach involves finding implicit relationships between concepts that suggest an as-yet undiscovered therapeutic relationship. Recent work has incorporated semantic relations extracted from the literature using natural language processing to constrain the search space of associations 22,23. For example, the pattern of semantic relationships “drug INHIBITS x; x ASSOCIATED_WITH disease” may indicate a therapeutic relationship between this drug and this disease. These patterns of relationships are known as “discovery patterns” 23,24. Our work in this area leverages high-dimensional vector space representations of the concept-by-predicate-by-concept relationships concerned to facilitate rapid search and efficient inference, using an approach called Predication-based Semantic Indexing (PSI) 25.
To recap, our operative assumption is that if we can identify plausible therapeutic relationships in a knowledge base of literature (arguably the main focus of LBD work to date), then we can also identify confounding associations, such as associations between concepts that are exogenous to the etiology of an ADE of interest. Such concepts may be predictive of entities that tend to co-occur along with drug-ADE pairs of interest in observational clinical data. We hypothesize that statistical models of drug-ADE association will more accurately identify causative drug-ADE relationships in observational clinical data after adjustments have been made for the influence of CVCs.
Methods
Derivation of CVCs from the literature. We developed an LBD-based method that automates CVD using “false discovery patterns” (FDPs), leveraging existing NLP tools and knowledge resources.
From Predications to FDPs. MedLEE was used to perform automated coding of concepts of interest in our EHR collection. MedLEE has been shown to perform accurately on clinical notes (for example recall of .77, and precision: ~.89 for the task of extracting clinical concepts, and coding them with concept unique identifiers from the Unified Medical Language System (UMLS) 26,27,28. The purpose of SemRep, a publicly available NLP system developed intramurally at the US National Library of Medicine (NLM), is to identify and normalize predications - relationships in which pairs of concepts are connected by predicates (or “verbs”, e.g., “TREATS”, “CAUSES”) in biomedical literature 29. SemRep operates with low recall, but high precision: Kilicoglu et al. reports precision of.745, recall of.640, and an F-score of .689 30. SemMedDB, used as our knowledge base of biomedical literature, is a publicly-available NLM product that contains the output of SemRep processing of the entirety of MEDLINE, where concepts and predicates have been recognized and normalized, and all extracted propositions can be retrieved from a MySQL database in the following form: ARGUMENT0 + PREDICATE + ARGUMENT1 31,32. This representation disallows semantic constructs such as degree, tense, narrator reliability, or verb ditransitivity. From a practical perspective, such predication representations are computable, since the concepts and predicates are normalized, and they facilitate our method’s inferential power.
Vector Symbolic Architectures. To process SemMedDB, which is used as our biomedical knowledge repository, we apply a representational technique called Predication-based Semantic Indexing, or PSI 25. PSI owes its existence ultimately to a contentious debate within the cognitive and neuroscience community in the 1980s over what became known as the parallel distributed processing (PDP) or connectionist paradigm 33–34. A number of scalable approaches to the problem of efficiently representing nested compositional structures (such as those encountered in natural language) in neural networks emerged following Smolensky’s seminal work, which utilized the tensor product as a compositional operator (Holographic Reduced Representations [HRR], Binary Spatter code, Multiply Add Permute [MAP]) 35–38. This is the origin of vector symbolic architectures (VSAs), described in detail elsewhere 39.
PSI for efficient inferences. VSA theory provides the infrastructure for PSI. In PSI, concepts are represented by randomly generated real, binary, or complex values of high dimensionality (the more dimensions that are used generally, the better the recall and precision of the model, with a tradeoff against computational efficiency). These are called “elemental vectors”. These elemental vectors can be superposed upon each other to generate composite semantic structures called semantic vectors 39–45.
Random vectors are an effective way to represent elemental components, since there is a high probability of mutual near-orthogonality, particularly in higher dimensions. Semantic vectors on the other hand are composed as superpositions of the “bound products” of the elemental vectors of predicate-argument pairs (as extracted from literature using SemRep). The binding operator, which varies in implementation across VSAs, is a multiplication-like operator that provides the means to encode additional information, such as the nature and context of a relationship, into the resulting vector space. Since the same predication can be encountered in multiple documents, PSI can be thought of as a distributional model of predications. Critical to PSI, semantic representations of concepts are built up as vectors from relations found in the literature. PSI facilitates rapid search for, and retrieval of, concepts that are related to one another in particular ways (i.e., through particular predicates). As such a space is distributional, concepts in which a relationship of interest occur more frequently will be retrieved first (analogous to the way in which other information retrieval systems facilitate ranked results). In the current work, PSI is used to facilitate rapid retrieval of concepts related to other concepts in specific ways. PSI can be used to retrieve the most strongly associated concepts (called “bridging terms”) across any particular predicate pathway of interest. A discussion of predicate pathways that indicate FDPs will follow. PSI and its applications are discussed in detail elsewhere 25,39–41.
SemRep Predications. For the current work, we used a PSI vector space derived from the version 24_32 of SemMedDB (processed June 20th, 2014 with version 1.5 of SemRep), containing 23.9 million citations and 70.4 million semantic predications 31. The PSI space was built using the Semantic Vectors package (version 5.9), with 48,000-dimensional binary vectors as the elemental vectors 44. A small number of predicates were excluded, indicating negation (such as DOES_NOT_TREAT). Terms with occurrence ≥ 500,000 were excluded.
FDPs for CVD. A browser interface called EpiphaNet has been developed for querying PSI-based representation of SemMedDB using a query language with meta variables that specify predicate vectors, elemental vectors, semantic vectors along with binding and superposition operators 40,41-44,45. We can query our PSI vector space model of SemMedDB to identify CVCs (the “bridging terms” mentioned above) for each drug-ADE pair with PSI queries that represent FDPs. Below in Table 1, we present the FDP queries that we used to evaluate our CVD method. We identified these FDPs while studying how expert users of the EpiphaNet interface interpret results of drug-ADE queries, when we noticed that EpiphaNet would generate reasoning pathways that suggest confounding relationships. Also, note that “DRUG TREATS x; x COEXISTS_WITH ADE” is referred to as a “double predicate” in that it is composed of two predicates that yield CVCs that link to both the drug and ADE cue terms.
Table 1.
FDPs that were used to identify CVCs and how they may relate to different types of confounding.
| FDP | PV Confounding Type | Examples (drug: allopurinol, ADE:liver failure) |
|---|---|---|
| “x CAUSES ADE” | co-medication, comorbidity | transplantation, embolism, cirrhosis |
| “x PREDISPOSES ADE” | co-medication, comorbidity | infection, sodium |
| “DRUG TREATS x; x COEXIST S_WITH ADE” | comorbidity | gout, kidney failure, pericarditis |
In our evaluation, In order to exclude spurious associations (as all vectors in the space are a measurable distance apart) from our CVCs, we made use of a frequency threshold, such that only bridging terms with association strengths ≥ 2.5 standard deviations were included.
Data and Analysis
Reference Set and Data Collection. To derive our data set, we used a reference set of curated drug-ADE associations that was developed by Patrick Ryan and his colleagues as a standard for evaluating PV methods 46. This reference set includes 399 medications/ADE pairs and 4 ADEs with both positive (drug-ADE relationships supported by the literature and other sources, including package labeling events) and negative (drug-ADE relationships without support) control groups per ADE. The four ADEs are as follows: acute kidney injury (AKI), acute liver injury (ALI), gastrointestinal bleeding (GIB), and acute myocardial infarction (AMI). These ADEs were chosen for their importance to PV space, their diagnostics, and their impact on financial and personal cost.
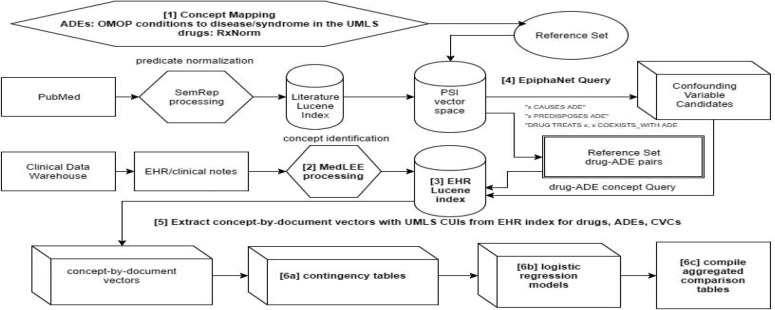
Analysis of Observational Clinical Data. The steps that we took for evaluation of our method are as follows both in the text below and in the corresponding items in Figure 2:
Map and expand drug/ADE synonyms to expose data of interest in the EHR index for extraction and additional processing [1]. We used RxNorm to identify drug synonym CUIs in the UMLS 47. For ADEs, we mapped the reference set’s OMOP terms (several are provide for each ADE of interest) to concepts in the UMLS, using exact string match of the preferred name field 28,48.
Process EHR with MedLEE to represent concepts (drugs, ADEs, CVCs) as CUIs in the UMLS 27,28. This is a subset (~2.1 million documents from ~364,000 in- and outpatients in the Houston metropolitan area between 2004 and 2012) of records from the UTH BIG 49 clinical data warehouse.
Build Lucene EHR index (software that efficiently tabulates the concept co-occurrence so as to facilitate rapid document retrieval) 50.
Query PSI vector space for CVCs given each drug-ADE pair for CVCs and extract data from index.
Extract drugs, ADEs, and CVCs from the EHR index (as lists of document IDs where respective concepts are in evidence) into concept-by-document vectors.
Establish baseline and CVC-adjusted scores for co-occurrence of drug-ADE pairs by constructing contingency tables [6a] and by computing area under curve (AUC) from receiver operator characteristic (ROC) curves from ranked-order of coefficients from logistic regression models [6b]. In descending order of co-occurrence count between drug, ADE, and CVC concepts (with an intersection threshold of 10), construct contingency tables as input for statistical models (using forward stepwise logistic regression), and calculate aggregated performance statistics from AUCs of ROCs. Finally, compile results into tables [6c].
Figure 2.
This graph illustrates our workflow.
In the course of building models iteratively in step 6 above, when or if a model that has incorporated a new CVC statistically converges, that CVC would be incorporated into subsequent models. This process continued for each drug-ADE pair until CVCs have been exhausted for that FDP. When such models fail to converge, the offending concept is added to a list of exclusions for that pair so that it would not be included in subsequent builds and for manual investigation of interesting patterns. If none of the models for a drug-ADE pair converge using CVCs, then the score from the unadjusted, or baseline, logistic regression coefficient is used to calculate AUC.
Results and Discussion
There are two groupings of result data in Table 2, labeled Complete Results and Constrained Results. Complete Results indicates that the AUCs have been calculated from the full data set without imposing any additional criteria. In the Constrained Results, the following criteria were applied to calculate performance metrics values for each field per ADE/FDP row: all logistic regression models must have converged, the count for drug instances was ≥ 100, the count for intersections was ≥10 between med-ADR pair, and that the calculations derive exclusively from cases where CVCs were included in the logistic regression models. As a result, the count of positive and negative controls for the same ADE will vary, since CVCs differ between FDPs.
Table 2.
This table presents the baseline and adjusted AUCs from ROCs that were calculated from ranked order of drug coefficients from logistic regression models from each ADE and FDP combination. caus=“x CAUSES ADE”, pred=“x PREDISPOSES ADE”, tcoe=“DRUG TREATS x; x COEXISTS_WITH ADE”. Counts=number of positive/negative examples. AUCs in bold indicate that an adjusted model drug coefficient is higher than baseline.
| Complete Results | Constrained Results | ||||||
|---|---|---|---|---|---|---|---|
| ADE | FDP | Counts (+/-) | Baseline | Adjusted | Counts (+/-) | Baseline | Adjusted |
| AKI | caus | 24 / 64 | .584 | .5853 | 11 / 13 | .6573 | .6643 |
| pred | .584 | NA | NA | NA | |||
| tcoe | .6126 | 18 / 40 | .6972 | .6125 | |||
| ALI | caus | 81 / 37 | .5167 | .515 | 44 / 14 | .5536 | .5568 |
| pred | .5297 | 50 / 24 | .4992 | .4825 | |||
| tcoe | .492 | 58 / 25 | .509 | .5303 | |||
| GIB | caus | 24 / 67 | .653 | .6418 | 20 / 48 | .6073 | .5792 |
| pred | .699 | 20 / 49 | .5949 | .6571 | |||
| tcoe | .7189 | 20 / 50 | .5964 | .69 | |||
| AMI | caus | 36 / 66 | .5112 | .5158 | 24 / 41 | .6026 | .6148 |
| pred | .5196 | 30 / 47 | .5319 | .5574 | |||
| tcoe | .5032 | 27 / 45 | .5687 | .5835 | |||
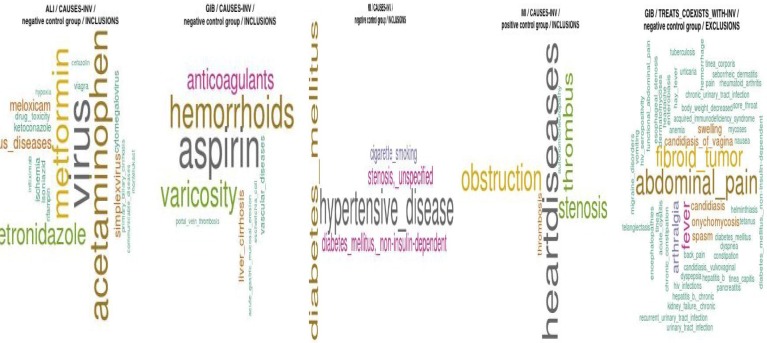
Wordclouds. We have generated word clouds below in Figure 3. The first two word clouds represent the CVCs of ALI and GIB with the “x CAUSES ADE” pathway. The third and fourth show the word clouds for the AMI groups using the “x CAUSES ADE” FDP. The fifth word cloud represents the CVCs that were excluded in building the logistic GLMs for GIB using “DRUG TREATS x; x COEXISTS_WITH ADE” FDP. The reader will notice that CVCs from single predicate FDPs (e.g., “x CAUSES ADE”) that were associated only with the ADE were of both the comorbidity and co-medication confounding subtypes, whereas CVCs from “DRUG TREATS x; x COEXISTS_WITH ADE” are constituted exclusively by comorbidities. The fourth word cloud from the left is interesting in that the most prevalent CVCs are comorbidities and likely mediators of myocardial infarction.
Figure 3.
These word clouds represent CVCs for ADEs and FDPs with font size proportional to aggregated term frequency (across all drugs for a group [positive or negative control] of a given ADE and FDP) in the EHR index.
Observations. There were ~55,000 instances in our EHR index for each of ALI, AMI, and GIB. AKI was the outlier with only ~5,000 instances. In the case of AKI for the “x PREDISPOSES ADE” pathway, no co-occurring CVCs were identified, so no adjustment could be made. In the case of ALI, one might reason that the set is heavily weighted toward the positive examples, so little gain is to be had by adjusting with CVCs.
Analysis of Complete Results. Our method performed the best when there was sufficient baseline support (AUC ≥ 0.6). For example, gastrointestinal bleeding had the best baseline AUCs, and there were notable gains in both Complete Results and Constrained Results for both “x PREDISPOSES ADE” and “DRUG TREATS x; x COEXISTS_WITH ADE” FDPs. In general, CVCs that derived from the double predicate “DRUG TREATS x; x COEXISTS_WITH ADE” FDP were more effective than those from single predicate FDPs. Such FDPs discover concepts with associations with both predictor and response variables, what Turing award winner Judea Pearl, a seminal influence on the field of causal reasoning, refers to as “true confounders” 9.
Analysis of Constrained Results. In the Constrained Results, for models that include CVC adjustments, performance improved in 8 of 11 cases. The “DRUG TREATS x; x COEXISTS_WITH ADE” FDP improved performance in 3 of 4 cases, while same can be said in 2 of 4 for “x PREDISPOSES ADE”. In only one case, AKI, was there an improvement using the “x CAUSES ADE” FDP.
Error Analysis. While using the single predicate FDPs that imply causation or risk factors, e.g., “x CAUSES ADE”, “x PREDISPOSES ADE”, respectively, make sense in terms of their ostensible causal association for adjusting negative controls, this intuition was not supported by the results of our analysis. Such FDPs uncover concepts that exist in the gray area between mediators, risk factors (Pearl’s “indirect effects” predictors), and concepts which could manifest confounding effects, e.g., smoking with respect to a positive control drug and AMI 9. For example, the following examples of mediation-like CVCs from this FDP were identified: stenosis, obstruction, thrombosis, and thrombus. Such concepts are suggestive of mediating concepts that relate to the causal mechanisms for AMI. Since mediators tend to be collinear with response variables, inclusion of such CVCs may be detrimental to performance.
One perplexing CVC from “DRUG TREATS x; x COEXISTS_WITH ADE” FDP for acute myocardial infarction is fibroid tumor. Fibroid tumors of the gastrointestinal tract are relatively rare and usually appear on the uterus. However, review of the predication database suggested that at times anti-inflammatory agents may occur in TREATS relationships with fibroid tumors, as they are used to control the pain associated with this condition. Though the “DRUG TREATS x; x COEXISTS_WITH ADE” FDP is intended to retrieve terms that are associated with both the drug and the ADE, the underlying implementation involves vector superposition - so though we would anticipate terms that are bilaterally connected being retrieved first, terms that are unilaterally connected may still meet the threshold. Such spurious CVCs could be eliminated by using a higher threshold of associational strength than the 2.5 standard deviation level and by making bilateral connection a prerequisite for retrieval.
Future Work. In future iterations of our research in this area, we hope to explore more FDPs, to put in place temporal feasibility constraints, and to experiment with other reference sets 51,52. We plan to incorporate data from other sources (e.g., FAERS, plausibility models), apply causal Bayesian networks instead of regression models, explore automatically inferring FDPs from strongly associated False Positives, and experiment with interactive refinement of literature-derived CVCs.
Conclusion
With the aim of surmounting the obstacle of confounding, a phenomenon which diminishes the validity of information that can be extracted from observational data, we have proposed a scalable and computationally inexpensive LBD-based CVD method. Our results show when there is sufficient support above random for an ADE, i.e., AUC ≥ .6, that statistical models that incorporate adjustments for the influence of dual-predicate FDP-derived CVCs exhibit modest ( .05 AUC or higher) performance gains for the task of re-identifying drug-ADE pairs from observational clinical data. While in the current paper we have presented a use case for our method in the domain of PV, we posit that our method is of potentially broader applicability as a tool complementary to other tools for tasks that involve enhancing signal detection from observational data within biomedicine.
Figure 1.
This graph illustrates EpiphaNet queries in the context of our method.
Acknowledgments
This work is supported by NIH/NLM Grant 1R01LM011563, the Brown Foundation, NIH/NIGMS R01 GM103859, NIH/NCATS grants UL1 TR000371 and UL1 TR001105 and NSF grant III 0964613. We would like to thank Dr. Swaroop Gantela for proofreading this manuscript.
References
- 1.Stolley PD. When genius errs: R. A. Fisher and the cancer controversy. Am J Epidemiol. 1991;133(5):416–25. doi: 10.1093/oxfordjournals.aje.a115904. [DOI] [PubMed] [Google Scholar]
- 2.Mill JS. London: John W. Parker; 1843. A system of logic, ratiocinative and inductive. [Google Scholar]
- 3.Diaz-Garelli J-F, Bernstam EV MSE, Rahbar MH, Johnson T. Rediscovering drug side effects: the impact of analytical assumptions on the detection of associations in EHR data; AMIA Summits on Translational Science Proceedings; 2015. pp. 51–5. [PMC free article] [PubMed] [Google Scholar]
- 4.Hersh WR, Weiner MG, Embi PJ, et al. Caveats for the use of operational electronic health record data in comparative effectiveness research. Med Care. 2013;51:S30–7. doi: 10.1097/MLR.0b013e31829b1dbd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.FAERS. http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Surveillance/AdverseDrugEffects.
- 6.Aronson J, Talbot J. Stephens’ detection and evaluation of adverse drug reactions: principles and practice. In: Talbot J, Aronson JK, editors. 6. Oxford: Oxford University Press; 2012. p. 2012. [Google Scholar]
- 7.Shang N, Xu H, Rindflesch T, Cohen T. Identifying plausible adverse drug reactions using knowledge extracted from the literature. J Biomed Inform. 2014;52:293–310. doi: 10.1016/j.jbi.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Salmasian H, Vilar S, Chase H, Friedman C, Wei Y. A method for controlling complex confounding effects in the detection of adverse drug reactions using electronic health records. J Am Med Inform Assoc. 2014;21(2):308–14. doi: 10.1136/amiajnl-2013-001718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearl J. 2. New York: Cambridge University Press; 2009. Causality: Models, reasoning, inference. [Google Scholar]
- 10.Armstrong BG. Effect of measurement error on epidemiological studies of environmental and occupational exposures. Occup Environ Med. 1998;55(10):651–6. doi: 10.1136/oem.55.10.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenland S, Robins JM, Pearl J. Confounding and collapsibility in causal inference. Statist Sci. 1999;14:29–46. [Google Scholar]
- 12.Greenland S, Hal M. Confounding in health research. Ann Rev Public Health. 2001;(22):189–212. doi: 10.1146/annurev.publhealth.22.1.189. [DOI] [PubMed] [Google Scholar]
- 13.Brookhart M, Stürmer T, Glynn R, Rassen J, Schneeweiss S. Confounding control in healthcare database research: challenges and potential approaches. Med Care. 2010;48:S114–20. doi: 10.1097/MLR.0b013e3181dbebe3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tibshirani R. Regression shrinkage and selection via the lasso. J Royal Statist Soc B. 1996;1(58):267–88. [Google Scholar]
- 15.Efron B, Hastie T, Johnstone I, Tibshirani R. Least angle regression. Ann Statist. 2004;2(32):407–99. [Google Scholar]
- 16.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Soft. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Ryan PB, Wei Y, Friedman C. A method to combine signals from spontaneous reporting systems and observational healthcare data to detect adverse drug reactions. Drug Saf. 2015 Oct.38(10):895–908. doi: 10.1007/s40264-015-0314-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harpaz R, DuMouchel W, Shah NH, Madigan D, Ryan P, Friedman C. Novel data-mining methodologies for adverse drug event discovery and analysis. Clin Pharmacol Ther. 2012;91(6):1010–21. doi: 10.1038/clpt.2012.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nasir M, Keiser MJ, Vilar S, Hripcsak G, Tatonetti NP. Systems pharmacology augments drug safety surveillance. Clin Pharmacol Ther. 2015;97(2):151–8. doi: 10.1002/cpt.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearl J, Glymour M, Jewell NP. Chichester, UK: Wiley; 2016. Causal inference in statistics: a primer. [Google Scholar]
- 21.Swanson DR. Fish oil, Raynaud’s syndrome, and undiscovered public knowledge. Perspect Biol Med. 1986;30(1):7–18. doi: 10.1353/pbm.1986.0087. [DOI] [PubMed] [Google Scholar]
- 22.Swanson DR, Smalheiser NR. An interactive system for finding complementary literatures: a stimulus to scientific discovery. Artif Intell. 1997;91:183–203. [Google Scholar]
- 23.Hristovski D, Friedman C, Rindflesch TC, Peterlin B. Exploiting semantic relations for literature-based discovery; AMIA Annu Symp Proc; 2006. pp. 349–53. [PMC free article] [PubMed] [Google Scholar]
- 24.Bruza P. A broad-coverage natural language processing system. In: Bruza P, Weeber M, Friedman C, editors. Proceedings of the AMIA Symposium; 2000. pp. 270–4. Literature-based Discovery 2008. [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen T, Widdows D, Schvaneveldt RW, Davies P, Rindflesch TC. Discovering discovery patterns with predication-based semantic indexing. J Biomed Inform. 2012;45(6):1049–65. doi: 10.1016/j.jbi.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedman C. A broad-coverage natural language processing system; Proceedings of the AMIA Symposium; 2000. pp. 270–4. [PMC free article] [PubMed] [Google Scholar]
- 27.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc. 2004;11(5):392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.UMLS. https://www.nlm.nih.gov/research/umls/
- 29.Rindflesch T, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Inform. 2003;36(6):462–77. doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Kilicoglu H, Fiszman M, Rosemblat G, Marimpietri S, Rindflesch TC. Arguments of nominals in semantic interpretation of biomedical text; Proceedings of the 2010 Workshop on Biomedical Natural Language Processing; 2010. pp. 46–54. [Google Scholar]
- 31.SemMedDB. http://skr3.nlm.nih.gov/SemMedDB/
- 32.MEDLINE. http://www.ncbi.nlm.nih.gov/pubmed.
- 33.Rumelhart DE, McCelland JL. Parallel distributed processing: explorations in the microstructure of cognition; 1986. [DOI] [PubMed] [Google Scholar]
- 34.Fodor JA, Pylyshyn ZW. Connectionism and cognitive architecture: a critical analysis. Cognition. 1988;1(28):3–71. doi: 10.1016/0010-0277(88)90031-5. [DOI] [PubMed] [Google Scholar]
- 35.Smolensky P. Tensor product variable binding and the representation of symbolic structures in connectionist systems. Artif Intell. 1990;46:159–216. [Google Scholar]
- 36.Plate TA. Stanford, Calif: CSLI Publications; 2003. Holographic Reduced Representation: Distributed Representation for Cognitive Structures. [Google Scholar]
- 37.Gayler RW. Vector symbolic architectures answer Jackendoff’s challenges for cognitive neuroscience. In: Slezak Peter., editor. ICCS/ASCS international conference on cognitive science; University of New South Wales; Sydney, Australia. 2004. pp. 133–8. [Google Scholar]
- 38.Kanerva P. The spatter code for encoding concepts at many levels. In: Marinaro M, Morasso P. G, editors. ICANN ‘94, Proceedings of the International Conference on Artificial Neural Networks (Sorrento, Italy); Springer-Verlag; London. 1994. pp. 226–9. [Google Scholar]
- 39.Widdows D, Cohen T. Reasoning with vectors: a continuous model for fast robust inference. Logic J of IGPL. 2014 Nov.:jzu028. doi: 10.1093/jigpal/jzu028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen T, Schvaneveldt R, Widdows D. Reflective random indexing and indirect inference: a scalable method for discovery of implicit connections. J Biomed Inform. 2010;2(43):240–56. doi: 10.1016/j.jbi.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 41.Cohen T, Widdows D, Schvaneveldt RW, Rindflesch T. Logical leaps and quantum connectives: forging paths through predication space; Quantum Informatics for Cognitive, Social, and Semantic Processes: Papers from the AAAI Fall Symposium; 2012. pp. 40–47. [Google Scholar]
- 42.Semanticvectors package on github. https://github.com/semanticvectors/semanticvectors.
- 43.Kanerva P, Kristoferson J, Holst A. Random Indexing of Text Samples for Latent Semantic Analysis; Proceedings of the 22nd Annual Conference of the Cognitive Science Society; 2000. p. 1036. [Google Scholar]
- 44.Cohen T, Whitfield GK, Schvaneveldt RW, Mukund K, Rindflesch T. EpiphaNet: An Interactive Tool to Support Biomedical Discoveries. J Biomed Discov Collab. 2010;5:21–49. [PMC free article] [PubMed] [Google Scholar]
- 45.EpiphaNet. http://epiphanet.uth.tmc.edu.
- 46.Ryan PB, Schuemie MJ, Welebob E, Duke J, Valentine S, Hartzema A. Defining a reference set to support methodological research in drug safety. Drug Saf. 2013;36:S33–47. doi: 10.1007/s40264-013-0097-8. [DOI] [PubMed] [Google Scholar]
- 47.RxNorm. Observational Medical Outcomes Partnership. https://www.nlm.nih.gov/research/umls/rxnorm/
- 48.(OMOP) http://omop.org/
- 49.UTH BIG. https://redcap.uth.tmc.edu/cdwstats/stats-mpi.htm.
- 50.Lucene. https://lucene.apache.org/
- 51.Coloma PM, Avillach P, Salvo F, Schuemie MJ, Ferrajolo C, Pariente A, et al. A reference standard for evaluation of methods for drug safety signal detection using electronic healthcare record databases. Drug Saf. 2013;36(1):13–23. doi: 10.1007/s40264-012-0002-x. [DOI] [PubMed] [Google Scholar]
- 52.Osokogu OU, Fregonese F, Ferrajolo C, et al. pediatric drug safety signal detection: a new drug–event reference set for performance testing of data-mining methods and systems. Drug Saf. 2015;38(2):207–17. doi: 10.1007/s40264-015-0265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]