Abstract
Medical data is routinely collected, stored and recorded across different institutions and in a range of different formats. Semantic harmonization is the process of collating this data into a singular consistent logical view, with many approaches to harmonizing both possible and valid. The broad scope of possibilities for undertaking semantic harmonization do lead however to the development of bespoke and ad-hoc systems; this is particularly the case when it comes to cohort data, the format of which is often specific to a cohort’s area of focus. Guided by work we have undertaken in developing the ‘EMIF Knowledge Object Library’, a semantic harmonization framework underpinning the collation of pan-European Alzheimer’s cohort data, we have developed a set of nine generic guiding principles for developing semantic harmonization frameworks, the application of which will establish a solid base for constructing similar frameworks.
Introduction
The quantity, quality and prevalence of digitally represented medical data available for research world-wide is now such that a tipping point has been reached in terms of the ability of that data to inform novel and innovative methods of research1. This so called ‘Big Data’ revolution, where the volume of data available is such that previously unobtainable research results are obtainable will inevitably inform medical research practice moving forwards2,3. That data is universally available in digital form does not however mean that it is available and represented in the same form – data sets large enough to catalyze novel research will inevitably come from a myriad of sources4. These sources, at least in the present, can and will draw from a large number of potential methods of representing data. As such, drawing data from multiple sources and combining it into a form upon which research can be conducted will require a process of either combining data into a single representation or explicitly mapping and translating between items of knowledge from the various sources5.
Semantic harmonization then is the process of combining multiple sources and representations of data into a form where items of data share meaning6. Harmonized data imparts the ability of allowing single given questions to be asked and answered across the data as a whole, without need to modify or adapt queries for a given data source, invaluable as a tool for researchers. The process of harmonizing data is far from trivial though. The technical undertaking of specifying a representation for capturing harmonized knowledge, the person-effort of specifying domain specific mappings between instances of data and the fact that for any given items of knowledge from within the same domain there may not be a universally agreed translation between them combine to make the task of harmonizing data, even for relatively specialized and small domains of knowledge, onerous if not impossible. Regardless of the feasibility of successfully harmonization any given data sets, undertaking the process and building a framework for semantic harmonization is a common and perhaps inevitable part of projects leveraging ‘Big’ medical data7.
Given that the implementation of some form semantic harmonization framework will necessarily form part of a project that utilizes medical data from across multiple sources, it seems natural to ask the question as to what common frameworks are available for enabling these harmonization processes? In answering this question we will draw a distinction between a method of harmonization and a framework for harmonizing data. A harmonization method is the underlying technical or logical means of specifying knowledge or mapping between items of knowledge. Formalisms such as ontological representations and their related reasoning mechanisms8, or openEHR archetypes9 would qualify as harmonization methods. It is uncommon and perhaps unwise for projects dealing with semantic harmonization to go down the path of specifying novel underlying representation formats. A harmonization framework on the other hand would be the overarching infrastructure for enabling harmonization and utilizing its results as well as the specific choice of how to use a chosen harmonization method. While systems such as I2B210 would fall into the category of harmonization frameworks it is still often necessary to develop novel frameworks for capturing data, particularly where the domain the data is being drawn from is focused or specialized. One such area would be that of cohort data. The collection of cohort data in a particular disease area will often focus on items or forms of knowledge specific to that area and the representation and capturing of that knowledge may require methods and formalisms unique to the domain in question.
Regardless, the development of frameworks for enabling semantic harmonization, particularly given the ongoing development of the field of ‘Big Data’ research in the medical informatics space, will continue. Each such system will be different; they will take varying approaches to how to harmonize data and how to enable the use of that data. We believe however that there should be a set of core principles that underlie the development of any such system. Key to these principles are features that inform the separation of concerns in the representation of knowledge and approaches to the stratification of knowledge. These are outlined following a description of the project that informed the development of these principles.
The EMIF Knowledge Object Library
The European Medical Informatics Framework (EMIF) is a large-scale collaboration between academic research institutions, small and medium sized companies and members of the European Federation of Pharmaceutical Industries and Associations (EFPIA), part of the Innovative Medicines Initiative (IMI)11. It aims to develop a suite of tools for enabling the reuse and linkage of healthcare data from across Europe. Specific use cases within EMIF are drawn from the participation of data cohorts from the Alzheimer’s and Metabolics spaces. It is with a focus on the Alzheimer’s space that we have produced the EMIF Knowledge Object Library (EKOL). This is a system for capturing both specific local knowledge from Alzheimer’s disease patient cohorts and mapping this knowledge into a global representation that allows the combination of data from multiple cohorts into a single representation, giving researchers the ability to launch research queries against this unified view of the data. Data recorded by the participating cohorts is universally represented in individualized data schemas and the majority of the data is coded with reference to local vocabularies, when formally coded at all. This led to the need for a system that dealt very explicitly with local data in a form unique to its source, but local data that in turn generally referred to the same set of global concepts.
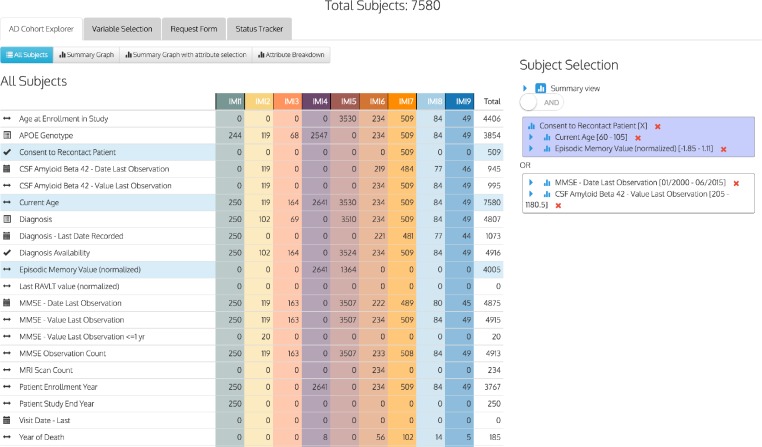
One practical outcome of this work was the application of the Knowledge Object Library as the underlying mechanism used to capture, process and provide data for the EMIF ‘Participant Selection Tool’ (PST). The PST is a web based tool that presents researchers with a list of approximately 20 core criteria present in the majority of EMIF AD cohorts (covering items such as participant age, diagnosis and mental state examination scores), showing counts of patients per cohort that have records covering those items of data. It then allows researchers to select logical combinations of these criteria and their values and dynamically adjusting the count of patients that match the selected criteria. So, for example, we could select ‘Patients over 50 with a diagnosis of Mild Cognitive Impairment OR Patients over 60 with a diagnosis of Alzheimer’s Disease’ and be presented with an updated count of the number of patients across the EMIF cohorts that match this selection. An illustrative screenshot of the PST is shown in Figure 1. The PST is part of a wider tool-chain that ultimately allows for the specification of research studies and the export of associated harmonized data for use by researchers. The information presented by the PST comes from data harmonized using the EKOL harmonization framework.
Figure 1.
A screen from the EMIF Participant Selection Tool, showing and overview of harmonized cohort data filtered by a criteria selection
As part of the process of specifying, designing and developing the EKOL system we explicitly drew out and developed a set of principles for designing semantic harmonization frameworks. These were developed through a combination of analysis of user requirements, an iterated agile approach to the development of the associated software which allowed for philosophical design decisions to be rapidly tested and potentially rejected in real-world settings and analysis of existing systems. These principles, outlined in the next section, both informed the development of the EKOL framework and directed the specification and design and application of the Participant Selection Tool.
The successful harmonization of data for use within the PST and its deployment of as a tool for research offer an empirical justification that the principles we explicated during the development of the EKOL framework at least offer the beginnings of a generalizable approach to the design and development of similar systems. These principles are outlined below.
Principles of Semantic Harmonization
Our work in the development of EKOL has lead to the formulation of the following set of generic guiding principles for developing frameworks for semantic harmonization. Given the range of ways in which knowledge can be defined, represented and captured, we use the generic term knowledge object to describe a singular representation of an item of knowledge, independent of the underlying formalism for capturing that item of knowledge. In this generic sense it is the representation of knowledge objects combined with the ability to the system to capture and specify them that underlies the functionality of any semantic harmonization system.
The nine points, presented below, when taken together are designed to guide the development of frameworks for supporting and enabling the semantic harmonization of medical data from multiple sources.
1. Separate technical from semantic harmonization
Data can be stored in many different file formats or databases. The ability to access and process available data is clearly an essential prerequisite to being able to semantically harmonize that data. We refer to the process of transforming data into the same technical storage implementation or transferring it to a system that can be queried by the same standard query engine as technical harmonization. The process of making all the data available on compatible platforms is a technical problem and should be separated from the semantic harmonization of the data, which is a data content problem - technical harmonization is not semantic harmonization
As such:
-
-
A technical connector to the local data source should be developed only once.
-
-
Technical upload scripts or tools should not be impacted by a change in semantics, such as a change in a vocabulary.
-
-
Conversely changes or additions to technical harmonization infrastructure or process should not impact semantic modelling or representation.
2. Distribute ownership of local and global knowledge objects
In general it is the role and responsibility of a subject matter expert to specify how variables need to be harmonized semantically. Semantics encapsulate meaning and ultimately it is only a subject matter expert, particularly in the medical domain, who can specify such meaning. However, knowledge of how variables and data can be utilised, particularly for research, may fall within another domain of knowledge, such as that of the researcher, whose knowledge and expertise, whilst complimentary to the domain expert, is not necessarily the same. They should be able to describe the variables they require and, if applicable, how normalization or harmonization should be performed. Variables that are used for data analysis are the global representations of knowledge. They are defined independently of any data source. Knowledge of measurement protocols and local variables – how variables are measured and what they really represent – are, on the other hand, available only at the data source. It is therefore the responsibility of the data source custodian to describe these variables semantically and create a local representation of knowledge for each variable they want to make available.
Bridging the gap between local knowledge, as described by local experts, and global knowledge, as requested by researchers, is a joint responsibility and effort and is part of the workflow of a semantic harmonisation process.
As such:
-
-
Ownership of local and global representations of knowledge objects should be distributed according to the location of expertise
-
-
Explicit distinction should be made between what is local knowledge and what is global
-
-
It should be possible to build up and maintain a library of global knowledge objects over time – both global and local concepts should be extensible
-
-
Global knowledge should make only generic reference to local knowledge, but local knowledge should be grounded in terms of global concepts
3. Separate vocabulary from structure
Data integration in the medical field is often achieved by the use of common vocabularies or taxonomies. Examples of well-known taxonomies are SNOMED-CT12 or the NCI thesaurus13. CDISC14 also specifies vocabularies that need to be used for naming variables or for standardizing values for those variables used commonly in clinical research.
Similar to the distinction we have drawn between technical and semantic harmonization we will also highlight a distinction between the act of specifying a vocabulary for grounding the meaning of items of knowledge via explicit reference to vocabularies and the act of structuring items of knowledge such that their internal relationships carry meaning. Whilst structure is often present to varying degrees within formal medical vocabularies and this structure can be leveraged in the formalization of semantic harmonization frameworks, drawing a conceptual distinction between vocabulary and structure can avoid a system relying on the tacit knowledge carried in language to derive its full utility. When the meaning of knowledge is externalized a system can no longer manipulate that knowledge and utility is lost.
So, the concept of vocabularies should be incorporated into the underlying representation of knowledge, since without any reference back to the real world the ultimate meaning of the semantics of knowledge is lost. However, a knowledge object should be a general construct that gets its definition from a link to a vocabulary term, rather than begin inferred from its structure.
As such:
-
-
Knowledge representation structures should be generic, with terminology details and dependencies separated by reference to vocabularies
-
-
Knowledge representations should retain meaning across vocabularies, and maintain meaning and structure in the absence of a vocabulary
-
-
The addition of new vocabularies should be possible with zero, or minimal, change to the structure of global knowledge representation
4. Re-use standard vocabularies where possible
Following on from point 3, we note that many medical vocabularies exist today and a semantic harmonization framework should re-use what is available. This both removes the need to maintain vocabularies and, given the ability to take on board new vocabularies or extend existing ones. The design of a novel semantic harmonization framework does not necessitate the design of a novel vocabulary, these are separate (though clearly related) areas of research, and should not be conflated.
As such:
-
-
Existing vocabularies should always be used in the first instance
-
-
Where there is a need to define new terms not available in a public vocabulary there should be mechanisms in place to accommodate this.
-
-
Such mechanisms should be used sparingly
5. Use declarative mappings
Domain experts are generally better at describing the logic of a mapping between knowledge objects, rather than describing the control flow of the computation of that mapping. Given the need to capture knowledge from domain experts, rule based rather than computation-based systems are better. A domain expert, assuming the role knowledge author, needs to be presented with a mechanism for capturing rich and complex domain knowledge. Declarative rule based systems are best suited for this purpose.
As such:
-
-
Mappings between knowledge objects should be described using rules.
-
-
Mapping rules need to be unambiguous and executable and as such expressed in a formal language
-
-
Such rules should still be simple enough to explain in plain language to other domain experts for verification.
6. Code isolation
The use of rules to specify mappings has the added benefit that a rule can be specified as a stand-alone object, independent of other mapping rules. This contrasts to scripts or other representations of computational flow, where changes to the code can have an impact on code statements further down the control flow. A rule for specifying knowledge should encapsulate a single concept and should be as limited as possible in scope. With this approach rules can be replaced or amended without affecting other rules within the system and the effort of authoring and maintaining rules can be distributed and scaled.
As such:
-
-
A given mapping rule should be assigned to or linked with a single knowledge object
-
-
A rule should contain the instructions on how to calculate a given knowledge object only from upstream knowledge objects
-
-
The scope of rule should be limited solely to the knowledge object it describes and the upstream knowledge objects it is derived from – it should be unaware of any other variables of knowledge objects.
-
-
The impact of changing any given rule on the global structure of knowledge should be minimised.
7. Enable integrated security and provenance
Semantic harmonisation more often than not will need to occur across data sources where the ownership of data (in terms of access control) differs between sources. It is essential that the data custodian for a given data source can control access to the data source variables or local knowledge objects, even when data is ultimately queried at the level of the global knowledge objects. Access to a local knowledge object should be propagated through the dependency graph to global knowledge objects. A knowledge representation system should have constructs in place that can handle this propagation.
Access to data can be restricted based on a number of dimensions:
Users: access is restricted to certain users or research teams, based on the description of their scientific use case.
Variables: the researchers should only have access to the data they need to answer the research question. To avoid having to set permissions on each variable, data source custodians can group variables (as is done for example in CDISC domains) or provide access to the complete data source.
Patients: data source custodians could allow access to data of a subset of patients only, based on inclusion or exclusion criteria or on legal restrictions.
Time: access to source data can be limited in time.
Permission level: access could be granted to the patient-level data or to aggregated knowledge objects only. At the level of the outflow tools it could be restricted to pre-defined reports or could allow custom queries on the data.
As such:
-
-
Items should be grouped as much as possible in order to minimize administrative complexity (for instance users can be grouped in teams, institutions, capability groups etc.)
-
-
In the context of describing security variables should be grouped in domains or by data type (e.g. clinical data vs. high dimensional data).
-
-
Patients should be grouped and categorized by originating data source
-
-
Derived instance values corresponding to knowledge objects should always reference source data instances from which they have been derived, allowing full traceability of every derived value (data provenance).
-
-
The data source of a derived value should be inferable directly from the mapping rules.
8. Separate WHAT is measured from HOW the measurement is done
When mapping data from different data sources we clearly need to make sure that variables that are mapped to a common global knowledge object are compatible, i.e. that they measure the same thing. Using terms of the Ontology for Biomedical Investigations (OBI)15, they need to share the same “quality” that “inheres in” instances of the same “material entity”. If there were no measurement errors (instruments have infinite accuracy), variables that measure the same quality in the same material entity could directly be pooled.
However, the actual measured value is also determined by the measurement protocol and the accuracy of the instruments used. Compatibility of protocols or normalization of values (to factor out the effect of the protocol) is use case dependent. The scientist analysing the data needs to decide if data measured using different protocols can be pooled. Whilst it is not necessarily the case the full details of a given measurement protocol should be modelled, we recommend specifying the independent variables of a protocol using vocabularies so they become comparable between data sources. Figure 2 gives a graphical overview.
Figure 2.
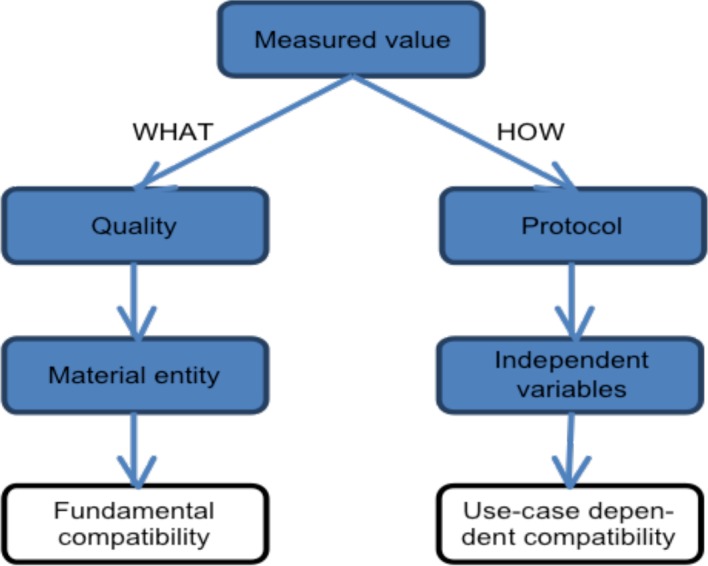
Compatibility of measurement variables depends on what is being measured and on how the measurement was performed (the protocol).
The knowledge object representation should be able to capture the “quality” and “material entity” for the variable it models, but should also capture as many of the protocol’s independent variables as deemed useful. Independent variables can be used as filters at query time, or be retrieved as additional information to the actual measurement value. They can also be moved into the “quality” to make the knowledge object more specific.
A typical example would be a knowledge object for hippocampal volume. The “quality” in this example is “volume”. The “material entity” is the hippocampus (e.g. bilateral). But there are many independent variables in the measurement protocol, such as the type of scanner, manufacturer of the scanner, image acquisition sequence, field strength, segmentation algorithm, digital brain atlas used etc. We could also define a knowledge object for “hippocampal volume as measured on 1.5T MRI using Freesurfer cross-sectional”, which would fix some of the independent variables. Such a knowledge object would measurements of different data sources more comparable but decreases the number of data sources that can deliver the data represented by the knowledge object.
As such:
-
-
A knowledge representation should always distinguish what is measured from how it is measured
-
-
This principle should be applied to measured observations both for continuous and categorical variables, textual information and other coded observations.
9. Balance generic versus specific descriptions
The range of possible variables that can be captured by knowledge objects is very large. For clinical data common use cases at least cover all the domains described in the CDISC SDTM standard. CDISC models each domain differently and specifies columns that are common to (most) domains, but also lists columns that are domain specific. Most CDISC domains specify one column per variable to record measurement values (pivoted data format). The demographics domain, for example, has a separate column for Age, Sex, Race etc. But some domains use one column to record the test that was performed and another for the measured value (unpivoted data format). The lab domain, for example, uses the column LBTEST for the laboratory test name and LBORRES for the (original) measured value. Other common data models such as OMOP take a more general approach and store all clinical observations in a single database table (Observation)16. Patient related information, however, is also stored in tables such as Person or Condition occurrence.
Choosing specific data structures for different types of variables can customize the information that is recorded. A generic data structure may have difficulty recording domain specific details. However, storing information across different tables and columns can result in domain dependent models.
There is no clear guideline on what the optimum generality level of a data model should be. The structure of a knowledge object tries to be sufficiently general to be able to capture the anticipated types of data and its metadata, but over-generalization has the drawback of a possible loss of domain dependent detail.
This challenge is equally recognised in the development of semantic interoperability sources such as detailed clinical models for healthcare information exchange. Discussions about how to balance generic versus very specific clinical models are currently ongoing within the Clinical Information Modeling Initiative (CIMI)17. There is a complementary ongoing debate about the level of detail and completeness that models (and knowledge objects) should aim to specify: should one aim for large very comprehensive representations or smaller fragments that may be combined in different ways.
This final principal is in a sense necessarily vague; in developing a semantic harmonization framework, the representation of knowledge into which data is harmonized needs to be generic enough to harness the benefits of simplicity without losing the ability to capture domain specific structure that can come from a bespoke knowledge representation structure.
As such:
-
-
The design of a knowledge representation scheme for semantic harmonization should be as generic as possible.
-
-
Specificity of representation should only occur where the representation of that knowledge cannot be generalised, i.e. where there are features of the representation not shared by any other objects in the general knowledge representation scheme.
Discussion
Systems or frameworks for harmonizing knowledge from multiple heterogeneous data sources must, where knowledge is being explicitly represented, be designed around that representation of knowledge. In this sense it is crucial that a clear, principled approach to the design of those systems, particularly when it comes to the stratification, specification and codification of knowledge, is taken. In this paper we have outlined a set of guiding principles that we believe when followed will lead to the design of frameworks for semantic harmonization that are robust, future proof and able to capture the full richness of knowledge within a given domain.
The nine principles outlined above can be applied en masse or adopted piecemeal, and conflicting views or implementations are inevitable. We do believe however that to a large extent the nine principles express a clarification of common design philosophy rather than a dictation of choice of approach. The value in outlining the set of principles comes from their explication rather than from the authors having made binary choices on behalf of others.
The focus of the principles outlined here focus specifically on areas of semantic harmonization that address aspects of conceptual modeling as opposed to value modeling (in terms of the ISO 11179 MDR standard18 at the data element concept rather than data element value level). This focus stemmed primarily from the emphasis of this work being driven by the need to harmonize cohort data, where we found that the primary differences between data sources lay in the structuring rather than the value of measurements and results. Adaptation or revision of these principles to account more for value driven harmonization efforts could be an important extension of this work.
We have drawn a distinction in this work between both between methods and frameworks of harmonization and between technical and semantic harmonization and believe that the field as a whole would benefit from applying further similar analysis to the structure of the field itself and particularly application development within it. The field of software development has benefited immensely from a rich and varied analysis of the types of approaches that can be taken to software development itself, leading to myriad approaches to the ‘art’ of the field. We believe that the area of semantic harmonization would benefit similarly from a similar reflective approach being taken.
Acknowledgements
The research leading to these results has received support from the Innovative Medicines Initiative Joint Undertaking under EMIF grant agreement n° 115372, resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007-2013) and EFPIA companies’ in kind contribution.
References
- 1.Murdoch TB, Detsky AS. The inevitable application of big data to health care. JAMA. 2013;309(13):1351–2. doi: 10.1001/jama.2013.393. [DOI] [PubMed] [Google Scholar]
- 2.Bellazzi R. Big data and biomedical informatics: a challenging opportunity. Yearb Med Inform. 2014;9(1):8–13. doi: 10.15265/IY-2014-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneeweiss S. Learning from big health care data. N Engl J Med. 2014;370(23):2161–3. doi: 10.1056/NEJMp1401111. [DOI] [PubMed] [Google Scholar]
- 4.Kuhn KA, Giuse DA. From hospital information systems to health information systems. Problems, challenges, perspectives. Methods Inf Med. 2001;40(4):275–87. [PubMed] [Google Scholar]
- 5.Van Harmelen F, Lifschitz V, Porter B, editors. Handbook of knowledge representation. Elsevier; 2008. Jan 8, [Google Scholar]
- 6.Bowles KH, Potashnik S, Ratcliffe SJ, Rosenberg MM, Shih MN, Topaz MM, Holmes JH, Naylor MD. Conducting research using the electronic health record across multi-hospital systems: semantic harmonization implications for administrators. The Journal of nursing administration. 2013 Jun;43(6):355. doi: 10.1097/NNA.0b013e3182942c3c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin-Sanchez F, Verspoor K. Big data in medicine is driving big changes. Yearb Med Inform. 2014;9(1):1420. doi: 10.15265/IY-2014-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rector A, Horrocks I. Experience building a large, re-usable medical ontology using a description logic with transitivity and concept inclusions. InProceedings of the Workshop on Ontological Engineering, AAAI Spring Symposium (AAAI’97). AAAI Press; 1997; 26 pp. [Google Scholar]
- 9.Martínez-Costa C, Menárguez-Tortosa M, Fernández-Breis JT. An approach for the semantic interoperability of ISO EN 13606 and OpenEHR archetypes. Journal of biomedical informatics. 2010 Oct 31;43(5):736–46. doi: 10.1016/j.jbi.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 10.Murphy SN, Weber G, Mendis M, Gainer V, Chueh HC, Churchill S, Kohane I. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) Journal of the American Medical Informatics Association. 2010 Mar 1;17(2):124–30. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visser PJ, Streffer J. A european medical information framework for Alzheimer’s disease (EMIF-AD). Alzheimer’s & Dementia. The Journal of the Alzheimer’s Association. 2015 Jan 7;11(7):P120–1. [Google Scholar]
- 12.Donnelly K. SNOMED-CT: The advanced terminology and coding system for eHealth. Studies in health technology and informatics. 2006 Jan;(121):279. [PubMed] [Google Scholar]
- 13.Sioutos N, de Coronado S, Haber MW, Hartel FW, Shaiu WL, Wright LW. NCI Thesaurus: a semantic model integrating cancer-related clinical and molecular information. Journal of biomedical informatics. 2007 Feb 28;40(1):30–43. doi: 10.1016/j.jbi.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Kuchinke W, Aerts J, Semler SC, Ohmann C. CDISC standard-based electronic archiving of clinical trials. Methods of information in medicine. 2009;48(5):408–13. doi: 10.3414/ME9236. [DOI] [PubMed] [Google Scholar]
- 15.Brinkman RR, Courtot M, Derom D, Fostel JM, He Y, Lord P, Malone J, Parkinson H, Peters B, Rocca-Serra P, Ruttenberg A. Modeling biomedical experimental processes with OBI. Journal of biomedical semantics. 2010 Jun 22;1(1):1. doi: 10.1186/2041-1480-1-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuemie MJ, Gini R, Coloma PM, Straatman H, Herings RM, Pedersen L, Innocenti F, Mazzaglia G, Picelli G, van der Lei J, Sturkenboom MC. Replication of the OMOP experiment in Europe: evaluating methods for risk identification in electronic health record databases. Drug safety. 2013 Oct 1;36(1):159–69. doi: 10.1007/s40264-013-0109-8. [DOI] [PubMed] [Google Scholar]
- 17.Clinical Information Modeling Initiative (CIMI) 2016. Jul 7, http://www.opencimi.org/ Accessed.
- 18.ISO/IEC JTC1 SC32 WG2 Development/Maintenance, ISO/IEC 11179, Information Technology. Metadata Registries (MDR) 2016. Jul 7, http://metadata-stds.org/11179/ Accessed.