Abstract
Manual Chart Review (MCR) is an important but labor-intensive task for clinical research and quality improvement. In this study, aiming to accelerate the process of extracting postoperative outcomes from medical charts, we developed an automated postoperative complications detection application by using structured electronic health record (EHR) data. We applied several machine learning methods to the detection of commonly occurring complications, including three subtypes of surgical site infection, pneumonia, urinary tract infection, sepsis, and septic shock. Particularly, we applied one single-task and five multi-task learning methods and compared their detection performance. The models demonstrated high detection performance, which ensures the feasibility of accelerating MCR. Specifically, one of the multi-task learning methods, propensity weighted observations (PWO) demonstrated the highest detection performance, with single-task learning being a close second.
Introduction
Conducting research and quality improvement using manual chart review (MCR) remains widely used in traditional observational clinical studies aimed at assessing detailed information on patients to understand disease course or outcomes and is also a primary modality used for quality improvement, epidemiologic assessments, and for graduate and ongoing professional education and assessement1,2–3. Prominent examples of healthcare quality improvement programs include the American College of Surgeons (ACS) National Surgical Quality Improvement Program (NSQIP) and Centers for Disease Control and Prevention (CDC)’s National Healthcare Safety Network (NHSN), which employ MCR to retrospectively measure healthcare quality and patient safety outcomes4. For example, NSQIP surgical clinical reviewers perform MCR to collect and report 21 surgical adverse events and related preoperative, intra-operative, and postoperative clinical data elements. Similarly, NHSN is a required healthcare- associated infection (HAI) tracking system, which also relies significantly on MCR. The reviewers for both of these programs utilize data from the electronic health record (EHR) and any paper records, including results of diagnostic tests, diagnoses information, and narrative text to ascertain outcomes5-6. Though MCR provides high-quality data for further secondary purposes including research, it is very time-consuming and labor-consuming to conduct and may be a bottle-neck step in the research discovery process.
Among all postoperative complications, the most common types are surgical site infection (SSI), pneumonia, urinary tract infection (UTI), and sepsis, accounting for nearly 60% of all complications7. Severe infections could trigger sepsis and even septic shock, particularly in people who are already at risk. Sepsis and septic shock are common and deadly, and CDC has listed “septicemia” as the 11th leading cause of death nationwide8. In addition, postoperative complications are expensive to treat. According to a recent study, 440,000 of these adverse events happen annually and cost overall up to 10 billion dollars per year in the United States9. Given the significant influence on the quality and cost of healthcare, postoperative complications are increasingly and widely viewed as a quality benchmark and are a strong emphasis of national initiatives for infection prevention and control10. However, infection control departments at medical facilities spend considerable time and resources on MCR to collect infection outcome data for public reporting and surveillance, which has greatly increased the burden on already limited infection prevention and quality improvement resources.
To improve the efficiency of MCR, some groups have explored the feasibility of computerizing the process of MCR for reporting postoperative compilations or for automatically collecting necessary data elements in quality improvement program, but these efforts have mainly been based on unstructured data such as physician narratives and nursing notes11-14. In one example, relevant keywords and phrases were extracted from free text documents for adverse event detection13. In another case, statistical and rule-based extractors were developed to automatically abstract data elements such as procedure type and demographic information from clinical notes14. According to standard NHSN definition, SSIs can be categorized into superficial, deep and organ space15. In our previous work, three subtypes of SSI and the overall SSI detection models were developed based on structured EHR data, including lab tests, vital signs, medications, and orders16. These models have high specificity as well as very high negative predictive values, guaranteeing the vast majority of non-SSIs could be eliminated thus significantly reducing the burden on chart reviewers.
In this study, our aim is to build an automated platform for postoperative complications detection based on structured EHR data by using robust modeling techniques. Included in this analysis are the main postoperative complications of three subtypes of SSI (superficial, deep, and organ space), pneumonia, UTI, sepsis, and septic shock. We hypothesized that EHR data would include significant indicators and signals of postoperative complications and that sophisticated machine learning methods might be able to extract these signals, accelerating the MCR process of these adverse events. Compared with the gold standard MCR process, automated application has potential advantages. First, MCR lacks inter-rater reliability, while an automated abstraction system would provide an objective and consistent reporting protocol that can be applied across multiple medical institutions. Second, a successful automated abstraction system would allow for expansion to include other procedures where postoperative surveillance is not being performed currently.
Specifically, we explore several methods for developing postoperative complication detection models. The most straightforward way to detect each type of complications is to build an independent classifier for each of them, which could be viewed as single-task learning, where detecting each complication is a task. When tasks are known to share similar features, we expect the resultant models to be similar. Learning models for these tasks together allows us to introduce inductive bias to make the resultant models similar, providing us with more robust models. Learning models for related tasks together is referred to as multi-task learning. For example, when our task is to identify a particular SSI subtype, a related task can be to detect any SSI (overall SSI). With a overall SSI model in hand, identifying a particular SSI subtype is easier, because the classifier only needs to learn the difference between overall SSI and the particular SSI subtype, rather than the difference between the SSI subtype and any other complication.
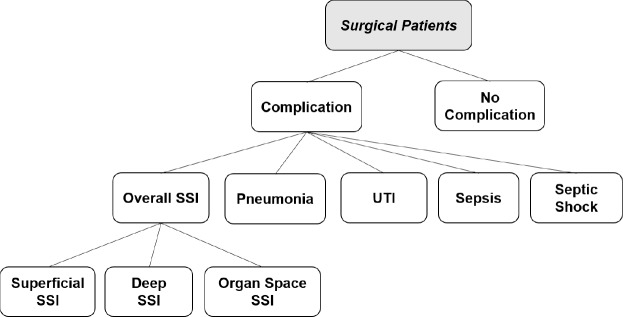
In our application, we have a hierarchy of tasks as shown in Figure 1. The first task is to distinguish patients with infection from those without. Next we distinguish among the various kinds of infections and finally, if the patient happens to have SSI, we distinguish among the three types of SSI. We assume that many infections share some characteristics that other diseases do not; and we further assume that many types of SSI share some characteristics that non-SSI infections do not. Our hypothesis is that by making a task-similarity hierarchy available to the multitask learning methods as domain knowledge, they can utilize these information towards building more robust and better performing detection models.
Figure 1.
A hierarchical structure among postoperative complications
In this work, we compare six methods for developing post-operative complications detection models. First we have single task learning, where predicting each complication is an independent task. We also explore five different methods for multi-task learning, assessing their value in improving the detection performance.
Materials and Methods
The overall methodological approach for this study included four steps: (1) identification of surgical patients and collection of associated data, (2) data preprocessing, (3) supervised single-task and multi-task learning model development, and (4) evaluation of a series of final models using gold standard data from the NSQIP registry. Institutional review board approval (IRB) was obtained and informed consent waived for this minimal risk study.
Data collection
Surgical patients at the University of Minnesota Medical Center are annually collected following strict inclusion and exclusion criteria. Their occurrences of postoperative complications are extracted and documented by chart reviewers, which are used as the gold standard in this study. We first identified surgical patients from 2011 to 2014 who had been selected for inclusion into the ACS-NSQIP. Clinical data for the identified patients were extracted from our clinical data repository (CDR) and their postoperative complication outcomes were retrieved from the NSQIP registry. The dataset was divided into training set (first 2.5 years) for model development and test set (last 1.5 years) for evaluation. The occurrences of postoperative complications in both training and test set are shown in Table 1. Overall SSI included any type of SSI.
Table 1.
Postoperative complications distribution in training and test set
| All Complications | Superficial SSI | Deep SSI | Organ Space SSI | Overall SSI | Pneumonia | UTI | Sepsis | Septic Shock | All Observations | |
|---|---|---|---|---|---|---|---|---|---|---|
| Training set | 571 | 168 | 76 | 105 | 336 | 124 | 140 | 115 | 34 | 5280 |
| Test set | 279 | 59 | 26 | 73 | 157 | 48 | 76 | 30 | 17 | 3629 |
We collected demographic information (e.g., age, gender), laboratory results (e.g., white blood cell count, glucose), microbiological results (e.g., urine culture, blood culture), relevant diagnosis codes (e.g., ICD_9 code 599.0 for UTI, ICD_9 code 995.91 for sepsis), orders and procedures for diagnosis and treatment (e.g., chest radiological exams for making diagnosis of pneumonia, CT guided drainage to treat SSI), antibiotic use, vital signs (e.g., temperature, heart rate), and medications.
Data preprocessing
Data preprocessing generally included data cleaning and missing data imputation. Our previous work on missing data imputation methods suggests that filling in the average value of pateitns without complications (normal value) in the training set and the test set introduces the least bias17. Accordingly, in this research, we chose to follow this imputation method.
For longitudinal lab results and vital records, we used aggregated features. We included the most recent value before the operation as the baseline, and the extreme and mean values from day 3 to day 30 after the operation during follow-up. We assembled a list of relevant antibiotics for each specific outcome, and extracted different classes of antibiotics as features. For relevant orders, procedures, and diagnosis codes, binary variables were created to denote if they were assigned to a patient. Addtionally, we not only considered whether a microbiology test was ordered or not, but also looked at the specific bacterial morphological types (e.g. gram possitive rods, gram negative cocci). Two binary features were built for each test to represent a culture placed or not and a positive/negative result, seperately.
Modeling Methods
After data collection and preprocessing, the six modeling techniques were applied.
When building our detection models, we face three key challenges. The first one is the skewed class distribution. A mere 10% of patients in the training set have any complications and some complications, like septic shock, occur in only. 6% (half a percent) of the patients. The second challenge is the small sample size. Some complications, like septic shock, only have 34 observations in the training and 17 in the test set. While our problem does not appear particularly high dimensional, for these rare complications, the number of predictors (approx. 200) exceeds the number of samples. The third challenge is the heterogeneity of the outcomes. We have 9 outcomes, each having their own specific characteristics and there are also variations among patients who do not have any complications. A successful detection algorithm has to address some of these challenges.
Below, we explain each of the six methods and describe which of the above challenges they address.
Method 1: Single-task learning
The nine outcomes are modeled independently using Lasso-penalized logistic regression. Lasso-penalized regression constructs a sparse model, where some of the coefficients are set to exactly 0. This performs automatic variable selection and also helps with the small sample size for some of the outcomes. This method does not specifically address the other challenges.
Method 2: Hierarchical classification
Let us consider the hierarchical structure among surgical patients as shown in Figure 1. All tasks are divided into three levels and models are constructed in a top-down fashion. The top-most task identifies patients with any postoperative complication. Next, in patients who are predicted to have complications, a 2nd level task is carried out to distinguish between SSI, pneumonia, UTI, sepsis, and septic shock. If a patient is predicted to have SSI, a further 3rd level task is also carried out to identify the SSI type: superficial, deep, and organ space SSI. Each task utilizes Lasso-penalized logistic regression. When more than two classes are possible, the one-vs-all approach is used to break a multi-class classification into a set of binary classifications.
As the method progresses from the top towards the bottom of the hierarchy, it gradually focuses on subpopulations that are enriched in the outcome of interest. This addresses heterogeneity by explicitly ignoring patients without indication of the outcome and also addresses the skewed class distribution. The adoption of LASSO model can overcome the problem of small sample size.
Method 3: Offset method
Similar to the hierarchical method, the classifiers for different tasks are built in a top-down fashion. For the top level task (i.e. complication classifier), a LASSO logistic regression classifier is built directly. For the lower-level tasks, we essentially model the difference between the parent and the child task. For example, the deep SSI classifier models the difference between overall SSI and deep SSI. This is achieved through penalizing the child model against the parent model: the predictions from the parent model are included as an offset term (a term with fixed coefficient of 1) in the child model, which is a LASSO logistic regression classifier. Due to the Lasso penalty, variables that have the same effect in the parent and child model will have a coefficient of 0; and conversely, variables that have non-zero coefficient are the variables in which the parent and child tasks differ. The method addresses the challenge of small sample size in two ways. First, in contrast to method 2, it uses the entire population at each level, thus the problem does not become overly high dimensional. Also the offset biases the child classifier towards the parent model. This method only offers limited ability to address heterogeneity.
Method 4: Propensity weighted observations (PWO)
The propensity weighted observations method also builds classifiers from the top level to the bottom level. The classifier of the top-level task, the complication classifier, is LASSO-penalized logistic regression (same as all of the previous methods). The classifiers for the second level task are built on the entire population, however, the observations (patients) are weighted by their propensity of having a complication. The propensity is obtained from the higher-level (complication) classifier. Patients, who are likely to have a complication receive a relatively large weight, while patients who are unlikely to have a complication receive a small weight. Therefore, patients with complication contribute more to the 2nd level classifiers than those who are unlikely to have complications. Similarly, the 3rd level classifiers, which distinguish between the three kinds of SSI, are also built on the entire population. The weights of the patients are their propensity of having SSI, thus the patients who likely have SSI contribute more to these classifiers than patients who are unlikely to have SSI. Similarly to the offset method, the PWO method uses the entire population, but by applying weights, it reduces outcome heterogeneity (patients with unrelated complications receive small weights) and reduces the skew of the class distribution by enriching the training set with patients having the outcome of interest (these patients receive high weights).
Method 5: Multi-task learning with penalties (MTLP)
Unlike the previous methods, the objective of multi-task learning with penalty (MTLP) method is to learn the regression coefficients βt for all tasks simultaneously. In the MTLP method, we assume that the parent task and its child tasks share some features and the respective models should have similar coefficients for those features. Similarly, to the offset method, this similarity is enforced through penalizing the child model against the parent model. Unlike the offset method, which builds models in a top-down manner, MTLP builds all models simultaneously. Specifically, the objective function is
| (1) |
It consists of three parts, the negative log likelihood of logistic regression l(βt) and two regularization terms, r1(βt) and r2(βt), as shown below.
| (2) |
| (3) |
| (4) |
where T and Nt are the number of tasks and training set for each task, respectively; xt,i and yt,i are the feature vector and the label for the subject i in task t, respectively; βt is the coefficient vector for the task t. The two regularization terms, and r1(βt) and r2(βt), restrict the difference in coefficients between the level 1 parent task and its level 2 child tasks; and the difference between the level 2 parent task and its level 3 child tasks, respectively. Penalizing the difference between the parent and child models make them similar. The MTLP method addresses heterogeneity by explicitly making the parent and child models similar, thereby essentially only modeling the difference between them; and it addresses the small sample size through the use of the entire population and regularization.
Method 6: Partial least squares regression (PLS)
As with the MTLP method, partial least squares (PLS) regression models all tasks simultaneously. PLS regression is similar to principal components regression in the sense that both methods reduce the dimension of input data by projecting the outcomes and predictors into new spaces and then build regression models in those new spaces. PLS differs from MTLP in that the task hierarchy is not explicitly given to the fitting algorithm; the algorithm has to autonomously learn the relationships among the tasks.
Evaluation
Outcomes based on MCR from ACS-NSQIP were used as gold standard to be compared with the results of postoperative complication detection models. The evaluation metric is area under the curve (AUC), which is commonly used to compare detection models. The range for AUC is between. 5 and 1,. 5 indicating a random model and 1 indicating perfect discrimination among the outcomes. We report the cross-validated AUC on the training set. To assess the variability of the detection performances on the test, bootstrap replication was applied and the 95% (empirical) confidiential interval (CI) and mean AUC scores are reported, as well. Since all methods were evaluated on the same bootstrap samples, paired t-test was used to compare each pair of methods and assess the statistical significance of the observed differences in performance.
Results
Evaluation results of six detection methods
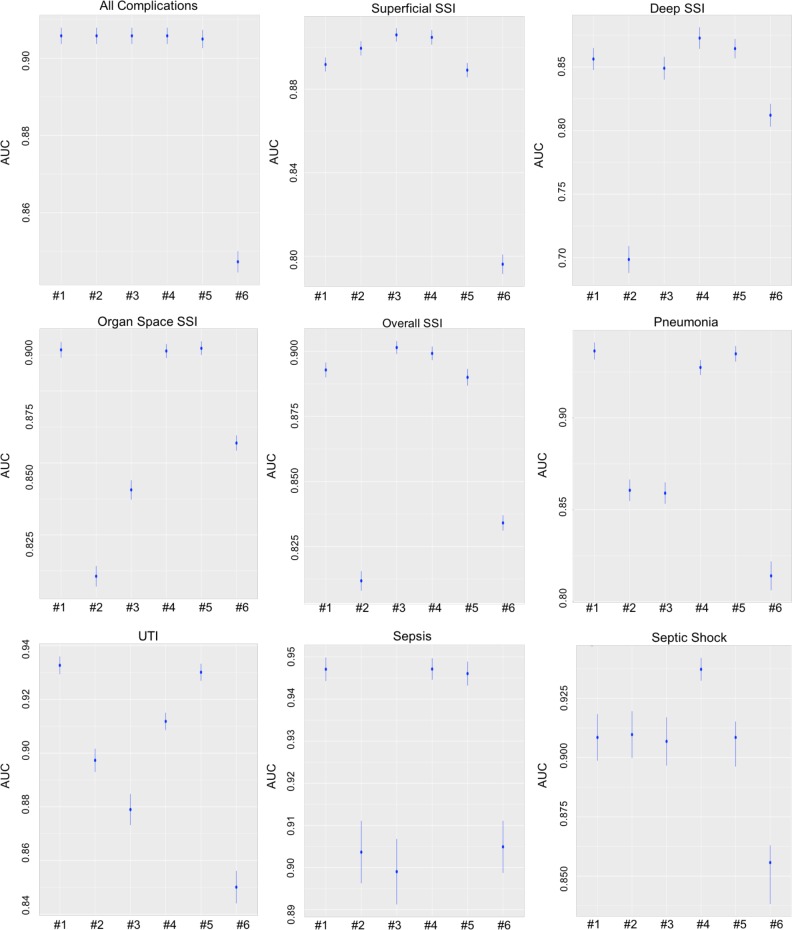
Figure 2 depicts the performances of the six methods. Each plot in Figure 2 corresponds to a task (complication) and each column in each plot corresponds to a method. Methods are numbered in the same order as they appear in the Methods section: #1 corresponds to Single-task, #2 to Hierarchical, #3 to Offset, #4 to Propensity Weighted Observations (PWO), #5 to Multi-Task Learning with Penalty (MTLP), and #6 corresponds to Partial Least Squares (PLS). The vertical axis is AUC. For each method, the mean AUC (across the bootstrapped test samples) is represented by a disk and lines extending out of the disk correspond to the 95% CI.
Figure 2.
Detection performance of six models for all nine tasks, showing the mean and 95% CI
To assess the statistical difference between some of the methods, in Table 2, we show the results of pairwise (paired) t-tests among the various methods. The rows of the table correspond to tasks, the columns to a comparison between two methods. Each cell contains a number, which indicates which method has a significantly better performance and we also provide the p-value in brackets. ‘NS’ means ‘not significant’. The methods are numbered in the same way as above.
Table 2.
Paired t-test results to compare different methods
| Method 1 vs. 2 | Method 1 vs. 3 | Method 1 vs. 4 | Method 1 vs. 5 | Method 2 vs. 3 | |
|---|---|---|---|---|---|
| Superficial SSI | 2 (<2.2e-16) | 3 (<2.2e-16) | 4 (<2.2e-16) | NS | 3 (=1.533e-14) |
| Deep SSI | 1 (< 2.2e-16) | 1 (= 5.073e-07) | 4 (< 9.285e-10) | NS | 3 (< 2.2e-16) |
| Organ Space SSI | 1 (< 2.2e-16) | 1 (< 2.2e-16) | NS | NS | 3 (< 2.2e-16) |
| Overall SSI | 1 (< 2.2e-16) | 1 (< 2.011e-13) | 4 (< 3.572e-15) | NS | 3 (< 2.2e-16) |
| Pneumonia | 1 (< 2.2e-16) | 1 (< 2.2e-16) | 1 (=2.353e-8) | NS | 2 (< 2.2e-16) |
| UTI | 1 (< 2.2e-16) | 1 (< 2.2e-16) | 1 (< 2.2e-16) | NS | 2 (< 2.2e-16) |
| Sepsis | 1 (< 2.2e-16) | 1 (< 2.2e-16) | NS | NS | 2 (< 2.2e-16) |
| Septic Shock | NS | 1 (< 2.2e-16) | 4 (<1.671e-12) | NS | 2 (< 2.2e-16) |
| Method 2 vs. 4 | Method 2 vs. 5 | Method 3 vs. 4 | Method 3 vs. 5 | Method 4 vs. 5 | |
| Superficial SSI | 4 (< 5.879e-16) | 2 (< 2.2e-16) | 3 (=0.001203) | 3 (< 2.2e-16) | 4 (< 2.2e-16) |
| Deep SSI | 4 (< 2.2e-16) | 5 (< 2.2e-16) | 4 (= 1.151e-14) | 5 (= 5.073e-07) | 4 (= 9.285e-10) |
| Organ Space SSI | 4 (< 2.2e-16) | 5 (< 2.2e-16) | 4 (< 2.2e-16) | 5 (< 2.2e-16) | NS |
| Overall SSI | 4 (= 1.607e-14) | 5 (< 2.2e-16) | 3 (= 0.02337) | 3 (= 2.011e-13) | 4 (< 3.572e-15) |
| Pneumonia | 4 (<2.2e-16) | 5 (< 2.2e-16) | 4 (< 2.2e-16) | 5 (< 2.2e-16) | 5 (= 2.353e-8) |
| UTI | 4 (=1.607e-14) | 5 (< 2.2e-16) | 4 (< 2.2e-16) | 5 (< 2.2e-16) | 5 (< 2.2e-16) |
| Sepsis | 4 (<2.2e-16) | 5 (< 2.2e-16) | 4 (< 2.2e-16) | 5 (< 2.2e-16) | NS |
| Septic Shock | 4 (<2.2e-16) | NS | 4 (< 2.2e-16) | NS | 4 (< 2.2e-16) |
For the detection of all complications, Single-task, Hierarchical, Offset, PWO, and MTLP have the same good performance, and are significantly better than PLS. To detect superficial SSI, Offset and PWO have virtually identical performance (difference is not significant) and they perform significantly better than the other four methods. PWO performs best for detecting deep SSI and overall SSI. Single-task, PWO, and MTLP all perform
similarly (no statistically significant difference) in detecting organ space SSI but perform significantly better than the other methods. To detect pneumonia and UTI, Single-task and MTLP are not significantly different from each other but are significantly better than other four methods. Single-task, PWO, and MTLP are the top three in detecting sepsis and PWO is also the best method for detecting septic shock. In general, PWO is the best method for detecting most complications. Single-task and MTLP are close seconds (and they have virtually identical performance) and PLS is the method with the worst overall performance.
Detailed information about the performance of the methods is depicted in Figure 2 and the statistical significance of the pairwise comparisons between the various methods is shown in Table 2.
Significant variables selected
Lasso-penalized regression performs automatic feature selection. In Table 3, we provide a list of the most important features selected by the model that aims to identify whether a patient has a complication. This model is common across most methods. Due to space limitation, we cannot provide a list for all methods and all complications. Below we provide some examples of features selected by the best performing method for each of the complications.
Table 3.
Selected important variables for all complications and their descriptions
| Category | Name | Description |
|---|---|---|
| In/Out patient | Inpatient or outpatient surgery | |
| Diagnosis code | ICD_9 code: 998.59 and 997.32 | Diagnosis code of postoperative SSI and pneumonia |
| Microbiology test order | Abscess culture Blood culture Gram stain culture Sputum culture Urine culture |
These are binary features to indicate if such microbiology test ordered or not during day 3 to day 30 after operation. |
| Microbiology test result | Escherichia. Coli Staphylococcus | These are binary features to indicate if the type of bacteria is positive or not no matter in which kind of microbiology test during day 3 to day 30 after operation. |
| Antibiotic use | Antibiotic_Superficial_SSI Antibiotic_Pneumonia Antibiotic_UTI |
They are binary features to indicate if antibiotics is placed to patients during day 3 to day 30 after operation. |
| Laboratory results | Measurement_CR Measurement PLT Measurement_PREALAB Measurement WBCU |
The number of measurements for creatinine, platelet count test, prealbumin, and urine white blood cells (WBCU). |
Superficial SSI detection model based on Offset and Propensity Weighted Observations methods selected antibiotic use, gram stain ordered, and the ICD_9 code of SSI.
Besides diagnosis codes, antibiotics use, gram stain culture, deep SSI detection model based on PWO selected more features from laboratory results (mean value of creatinine and maximum value of WBC) and two more microbiology tests (tissue and wound culture).
Organ space SSI models based on Single-task, PWO, and MTLP methods have quite similar detection performance and important variables. The selected variables include four features of bacteria type (streptococcus, gram positive cocci, enterococcus, and escherichia. coli), three microbiology cultures (abscess and fluid culture), and the imaging orders of treatment. Interestingly, the diagnosis code of sepsis and imaging orders of sepsis treatment are selected as well. These can be explained by the fact that there are over 30 patients in our cohort have sepsis and organ space SSI together.
Overall SSI detection models based on weighted observation and offset methods perform with no significant difference. The important variables selected are antibiotic use (UTI, superficial and deep SSI), microbiology cultures (abscess, fluid, and wound culture, and the gram stain test), two types of bacteria (escherichia. coli and staphylococcus) and two relevant order features (imaging orders for diagnosis and procedures of treatment).
Besides the diagnosis code and antibiotic use, pneumonia models based on Single-task and MTLP include two features from microbiology test (bronchial and sputum culture), one binary feature of image-guided diagnosis orders, and two aggregate features from lab tests (the mean value of PCAL and PH).
UTI models based on Single-task and MTLP selected diagnosis codes (for UTI), antibiotic use, placement of urine culture, and two bacteria types (proteus and escherichia. coli).
For sepsis models, the top performing methods, Single-task, PWO, and MTLP, selected features including antibiotic use, microbiology cultures (abscess, blood, fluid, and urine culture, and the gram stain), two bacterial types (enterococcus and escherichia. coli), and the image guided orders of treatment.
For septic shock detection, most models only selected diagnosis codes. However, PWO included more variables, such as laboratory tests (maximum value of partial thromboplastic time, PH, mean value of lactate), bacteria types (stenotrophomonas and staphylococcus) and the tracheal culture.
Discussion
Manual chart review for post-operative complications is very resource intensive. In this work, we examined whether EHR-based state-of-the-art predictive modeling approaches can learn characteristics of various types of complications and subsequently detect them reliably. With detection performances (measured as AUC) exceeding 0.8 for all complications and even 0.9 for some complications, the answer is affirmative: machine learning detection models definitely have the potential to help detect post-operative complications automatically. The question is which modeling approach is best suited for this application.
Post-operative complications are heterogeneous; they cover a wide-range of conditions, each having their own diagnostic methods, diagnoses codes, laboratory tests, and diagnostic and therapeutic procedures. They can be organized into a hierarchy and complications on the same level of the hierarchy are more similar to each other than to complications on a higher level of the hierarchy. Multi-task learning methods have the ability to exploit such similarities towards achieving better detection performance and more stable models even when the sample sizes are small.
We compared six approaches to building post-complication detection models. One of them was single-task learning, where we build independent models for each task; four methods were multi-task learning methods that can utilize the hierarchy of complications; and finally, we also utilized Partial Least Squares (PLS), which can simultaneously model multiple outcomes, but it tries to autonomously detect the relationship among the outcomes. PLS thus stands in sharp contrast with the other multi-task learning methods, as PLS automatically infers the relationships among the complications, while the other multi-task learning methods receive this information from an expert.
We found PLS to have the overall worst performance. This is not surprising, since PLS receives less information than the other multi-task learning methods. We expect PLS to bias the models based on the relationships among the outcomes, but we do provide it with these relationships. If PLS infers the relationships among outcomes incorrectly, it will bias the models incorrectly, eroding detection performance. With some of the complications having small sample sizes, it is unsurprising the PLS failed to infer the correct relationships. If we had substantially more samples,
PLS could have inferred the relationship among complications possibly better than what the expert can provide, but our sample size, albeit relatively large, was insufficient for this purpose. Single task learning managed to (significantly) outperform PLS, because we did not “force” it to bias the models beyond applying Lasso-penalty which is virtually mandatory given our sample sizes for some of the complications.
Hierarchical modeling also had disappointing performance. The essence of hierarchical modeling is to build classifiers in a subpopulation that is greatly enriched in the outcome of interest. For example, distinguishing among the three types of SSI is easier in a subpopulation of SSI patients than it is in the general population. The performance of the method did not live up to our expectation for two reasons. First, while these outcomes are rare (deep SSI occurred in 76 patients out of 5280), they still occur in sufficient numbers for a Lasso-penalized logistic regression model. The second reason concerns the way the subpopulations were constructed. If the higher-level classifier (does this patient has SSI?) predicts the patient to be free of SSI, then this patient does not enter the subpopulation and the deep SSI detector has no opportunity to learn from this sample. We could have built the deep classifier on the true SSI patients (rather than the predicted SSI patients), but then the distributions of the training SSI patients (true SSI patients) and the test SSI patients (predicted SSI patients) would be different, leading to degraded detection performance. Our results with the Propensity Weighted Observations method tell us that the concept of enriching patients with SSI for (say) the deep SSI classifier is valid; the hierarchical method simply implemented this concept suboptimally.
The Propensity Weighted Observations (PWO) method achieved the overall highest performance with a margin that is statistically significant. PWO is closely related to the hierarchical method in that it enriches the training sample with patients who have the outcome of interest. In contrast to the hierarchical method, it achieves this enrichment through constructing a new sample, which is a propensity weighted version of the original population. For example, to identify patients with deep SSI, PWO uses the entire population, but patients with high propensity for SSI receive high weight and patients with low propensity for SSI receive low weight. The hierarchical method is a binary version of PWO, where the weights are either 0 or 1. Having the propensity weighted population removes the problem of excluding patients based on an incorrect prediction. Suppose our SSI classifier misclassifies a deep SSI patient as not having SSI. This patient will still be included in the training set for the deep SSI classifier; this observation will receive a slightly lower weight. Admittedly, using propensity score weighing for multi-task learning is rather unusual; we did not expect this method to perform so well.
The offset method, like PWO, always uses the entire population for classification. Its performance falls short of that of PWO, because its ability to remove heterogeneity is limited. It can bias a child model against the parent model, which helps with small sample sizes (it performed well on superficial SSI), but has limited effect on removing the variation in (say) normal patients. PWO is more effective at removing heterogeneity: patients with unrelated complications receive a low weight and contribute to the model only minimally.
MLTP is essentially identical to the offset method, except MLTP optimizes all outcomes simultaneously (as opposed to sequentially in a top-down manner). In a top-down construction scheme, only the parent task can influence the child task; the model from the child task cannot influence the parent model. When all tasks are carried out simultaneously, the child models can influence the parents, as well. As a result, MLTP was either the best or second best method for almost all rare (<3% of patients) outcomes. The caveat of simultaneous optimization is the increased potential for overfitting. Indeed, comparing MTLP’s cross-validated AUC scores on training set to those on the test set, reveal signs of overfitting. For example, to detect sepsis, it has a very high training AUC (>0.96), but the 95% CI of AUC on test set is only (0.8986, 0.9183).
Conclusion
Developing machine learned models to automatically detect post-operative complications definitely has the potential to accelerate the manual chart review process. We found that multi-task learning, specifically, the propensity weighted observations method, statistically significantly outperformed the single-task learning approach. While the difference in detection performance was relatively modest (albeit significant), the additional cost of implementing this method over the standard single-task learning method is minimal. Thus, we would recommend trying both single-task learning and PWO.
Our application was relatively easy: we had sufficiently many samples for Lasso-penalized logistic regression to construct a good model even for the most infrequent outcome. In an application, where fewer samples are available or outcome distributions are more skewed, we would expect the performance gap between multi-task learning and single-task learning to open up, providing a more attractive implementation cost versus detection performance proposition for multi-task learning.
Our future work includes building postoperative complications detection models using both structured and unstructured EHR data. We hypothesize that the combination of structured and unstructured clinical data would include more significant indicators and signals of postoperative complications, and improve the performance of detection. The performance of the models with only structured data and that of the models with both structured and unstructured data will be compared and evaluated.
Acknowledgements
The National Institutes of Health through the National Library of Medicine (R01LM011364 and LM011972-01A1), Clinical and Translational Science Award (8UL1TR000114-02), and University of Minnesota Academic Health Center-Faculty Development Grant supported this work. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health.
Reference
- [1].Matt V, Matthew H. The retrospective chart review: important methodological considerations. Journal of Educational Evaluation for Health Professions. 2013;10 doi: 10.3352/jeehp.2013.10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gearing RE, Mian IA, Barber J, Ickowicz A. A Methodology for Conducting Retrospective Chart Review Research in Child and Adolescent Psychiatry. Journal of the Canadian Academy of Child and Adolescent Psychiatry. 2006;15(3):126–134. [PMC free article] [PubMed] [Google Scholar]
- [3].Kahn MG, Raebel MA, Glanz JM, Riedlinger K, Steiner JF. A Pragmatic Framework for Single-site and Multisite Data Quality Assessment in Electronic Health Record-based Clinical Research. Medical care. 2012;50 doi: 10.1097/MLR.0b013e318257dd67. 010.1097/MLR.0b013e318257dd67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Surveillance for Surgical Site Infection (SSI) Events. http://www.cdc.gov/nhsn/acute-care-hospital/ssi/ [Google Scholar]
- [5].Sarkar S, Seshadri D. Conducting Record Review Studies in Clinical Practice. Journal of Clinical and Diagnostic Research: JCDR. 2014;8(9):JG01–JG04. doi: 10.7860/JCDR/2014/8301.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Adeleke IT, Adekanye AO, Onawola KA, et al. Data quality assessment in healthcare: a 365-day chart review of inpatients’ health records at a Nigerian tertiary hospital. Journal of the American Medical Informatics Association: JAMIA. 2012;19(6):1039–1042. doi: 10.1136/amiajnl-2012-000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Khan NA, Quan H, Bugar JM, Lemaire JB, Brant R, Ghali WA. Association of Postoperative Complications with Hospital Costs and Length of Stay in a Tertiary Care Center. J Gen Intern Med. 2006 Feb;21(2):177–180. doi: 10.1111/j.1525-1497.2006.00319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lawson EH1, Hall BL, Louie R, Ettner SL, Zingmond DS, Han L, Rapp M, Ko CY. Association between occurrence of a postoperative complication and readmission: implications for quality improvement and cost savings. Ann Surg. 2013 Jul.258(1):10–8. doi: 10.1097/SLA.0b013e31828e3ac3. [DOI] [PubMed] [Google Scholar]
- [9].Dimick JB, Pronovost PJ, Cowan JA, Jr, Lipsett PA, Stanley JC, Upchurch JR., Jr Variation in postoperative complication rates after high-risk surgery in the United States. Surgery. 2003 Oct;134(4) doi: 10.1016/s0039-6060(03)00273-3. [DOI] [PubMed] [Google Scholar]
- [10].Biscione FM. Rates of surgical site infection as a performance measure: Are we ready? World Journal of Gastrointestinal Surgery. 2009;1(1):11–15. doi: 10.4240/wjgs.v1.i1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tinoco A, Evans RS, Staes CJ, Lloyd JF, Rothschild JM, Haug PJ. Comparison of computerized surveillance and manual chart review for adverse events. Journal of the American Medical Informatics Association: JAMIA. 2011;18(4):491–497. doi: 10.1136/amiajnl-2011-000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wu ST, Sohn S, Ravikumar KE, et al. Automated chart review for asthma cohort identification using natural language processing: an exploratory study. Annals of allergy, asthma & immunology: official publication of the American College of Allergy. Asthma, & Immunology. 2013;111(5) doi: 10.1016/j.anai.2013.07.022. 10.1016/j.anai.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Murff HJ, FitzHenry F, et al. Automated identification of postoperative complications within an electronic medical record using natural language processing. JAMA. 2011;306(8):848–855. doi: 10.1001/jama.2011.1204. [DOI] [PubMed] [Google Scholar]
- [14].Yetisgen M, Klassen P, Tarczy-Hornoch P. Automating Data Abstraction in a Quality Improvement Platform for Surgical and Interventional Procedures. EGEMS (Wash DC) 2014 Nov 26;2(1):1114. doi: 10.13063/2327-9214.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15]. http://www.hopkinsmedicine.org/heic/infection_surveillance/ssi.html.
- [16].Hu Z, Simon GJ, Arsoniadis EG, Wang Y, Kwaan MR, Melton GB. Automated Detection of Postoperative Surgical Site Infections Using Supervised Methods with Electronic Health Record Data. Stud Health Technology Inform. 2015;216:706–10. (Medinfo 2015) [PMC free article] [PubMed] [Google Scholar]
- [17].Hu Z, Melton GB, Simon GJ. Strategies for Handling Missing Data in Detecting Postoperative Surgical Site Infections; ICHI, 2015, 2015 International Conference on Healthcare Informatics (ICHI), 2015 International Conference on Healthcare Informatics (ICHI); 2015. p. 499. [Google Scholar]