Abstract
Atrial fibrillation (AF) is a common cardiac rhythm disorder, which increases the risk of ischemic stroke and other thromboembolism (TE). Accurate prediction of TE is highly valuable for early intervention to AF patients. However, the prediction performance of previous TE risk models for AF is not satisfactory. In this study, we used integrated machine learning and data mining approaches to build 2-year TE prediction models for AF from Chinese Atrial Fibrillation Registry data. We first performed data cleansing and imputation on the raw data to generate available dataset. Then a series of feature construction and selection methods were used to identify predictive risk factors, based on which supervised learning methods were applied to build the prediction models. The experimental results show that our approach can achieve higher prediction performance (AUC: 0.71~0.74) than previous TE prediction models for AF (AUC: 0.66~0.69), and identify new potential risk factors as well.
Introduction
Atrial fibrillation (AF) is one of the most common clinical arrhythmias, affecting approximately 4 million adults in China1. AF significantly increases the risk of ischemic stroke and other thromboembolism (TE). Moreover, compared to non-AF ischemic stroke, AF related ischemic stroke is more fatal and disabling2. Oral anticoagulation (OAC) including warfarin has shown great efficacy in preventing ischemic stroke and TE for AF patients3. However, because OAC may have severe side effects such as warfarin bleeding, it is normally only recommended to AF patients with high risk of TE in clinical guidelines4,5. Besides, though radiofrequency ablation (RFA) is an effective procedure to treat AF and then reduce the risk of TE, it is still a scarce medical resource in present day China and increases economic burden on AF patients. Therefore, it is critical to accurately predict the risks of TE for AF patients and identify those truly high risk patients that should be treated by OAC and/or RFA.
Current ischemic stroke and TE risk models for AF, such as CHADS26, CHA2DS2-VASc7 and Framingham Score8, were developed to stratify AF patients into categories of high, intermediate, and low risk. The risk factors used in these models, such as age, gender, prior ischemic stroke and TE, hypertension, diabetes, congestive heart failure (CHF), etc., are grounded in previous known evidence and experience, which are well understood and easy to apply. However, these risk models have only moderate prediction performance9 (the area under the receiver operating characteristic curve (AUC) is usually less than 0.77). It is mainly because that some potential risk factors that are highly related to TE occurrence for AF patients were not previously identified and involved in these risk models.
The objective of this study was to build 2-year ischemic stroke and TE prediction models for AF with high prediction ability and interpretability, based on the Chinese Atrial Fibrillation Registry (CAFR) data. The CAFR study started from the year of 2011, and has enrolled more than 17,000 AF patients from 32 hospitals in Beijing, China. The study collected the patients’ demographics, symptoms and signs, medical history, results of physical examination and laboratory test, details of treatments at baseline, and followed up the patients every 6 months. At every follow-up visit, the clinical events such as ischemic stroke and TE were collected.
Many previous works used statistical inference and machine learning methods to build high accuracy risk prediction models for patients with cardiovascular, diabetes and other diseases10,11,12,13. However, it is still a challenging problem to build accurate and clinically interpretable TE prediction models from CAFR data. The first challenge is from the dataset, which is heterogeneous, non-standardized, incomplete and redundant. Much elaborate data curation work has to be done to remedy the data before analysis. Also, feature engineering should be performed to transform data types and reduce the redundancy of features. Another challenge is from the requirement of interpretability. Since we wanted to build human understandable and applicable prediction models, the dimensionality reduction and learning algorithms in which resulting models are difficult to interpret (e.g., principle component analysis and support vector machine) were not preferable.
In this paper, we address these issues by using integrated machine learning and data mining approaches to build TE prediction models for AF patients. We first performed data cleansing and imputation on the raw CAFR dataset to standardize the features and fill-in missing entries. Then a series of feature construction and selection methods were used to identify predictive risk factors and reduce redundancy. Finally, we applied different categories of supervised learning methods that have good interpretability, including generalized linear model, Bayes model and decision tree model, to build TE prediction models for AF. The experimental results show that our approach can achieve higher prediction performance than previous TE risk models, and also identify new potential risk factors that have not been previously identified or commonly used.
Methods
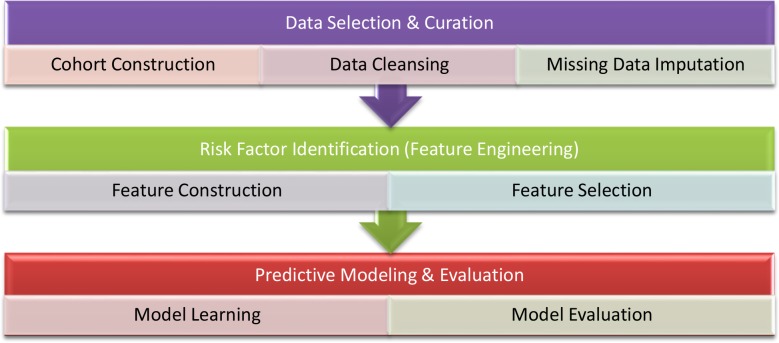
Figure 1 shows our approach pipeline of building ischemic stroke and TE prediction models for AF patients from CAFR data. We first selected and constructed the patient cohort of interest. Then data curation, including data cleansing and missing data imputation, were performed to generate available dataset. After that, we applied a series of feature engineering methods, including feature construction and feature selection, to identify the potential risk factors for predicting TE in AF. Finally, we trained prediction models using different supervised learning algorithms, and evaluated their prediction performance in terms of AUC and the area under the precision recall curve (AUPR) by cross validation and train/test splitting.
Figure 1.
Pipeline of building ischemic stroke and TE prediction models for AF patients
Cohort
The purpose of TE prediction models is to help clinicians identify AF patients with high risk of TE, and then decide to whom relevant interventions such as OAC and RFA should be used. Therefore, in this study the patients of interest are those who had not been treated with OAC (mainly warfarin in our data) or RFA at baseline. From the CAFR data, we identified 1864 AF patients who meet this criteria, where 193 patients (10.4%) are cases who had TE within 2 years after baseline, and 1671 patients are control instances who completed 2-year follow-ups and did not have TE within 2 years. The features used in our analysis include demographics, symptoms and signs, medical history, vital signs, laboratory test results, life styles and treatments.
Data Curation
In the raw CAFR data, more than half of the features have non-standardized and dirty values, and this percentage for the numeric type is particularly higher. Besides, the raw data has significant omissions due to the questionnaire structure, unknown values or errors in data collection. The workload of manually correcting these dirty and missing values could be enormous. Therefore, we preformed automatic data curation, including data cleansing and missing data imputation, before predictive analysis.
1). Data Cleansing
The raw data is heterogeneous and includes different types of data items: binary type (e.g, hypertension history), nominal type (e.g., AF type) and numeric type (e.g., systolic blood pressure, SBP). For different data types, we designed sets of cleansing rules to remedy the non-standardized and dirty values in batch. These cleansing rules can be used to standardize data formats, correct input errors, or discard the values that cannot be recognized as the target types.
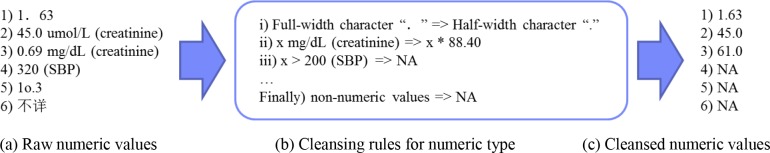
For the numeric features, some values in the raw data are not standardized (e.g, the full-width Chinese character “. ” in Figure 2(a.1)), so we first defined a set of cleansing rules to standardize these values (e.g., Figure 2(b.i) that transforms “. ” to the half-width character “.”). Besides, because the dataset was collected from different hospitals, a numeric feature may have different units (e.g., mg/dL and μmol/L for serum creatinine in Figure 2(a.2-3)). Therefore, we defined cleansing rules to unify these units (e.g., Figure 2(b.ii) that transforms mg/dL to μmol/L for serum creatinine). Finally, we discarded the numeric values that are out-of-range (e.g., the SBP greater than 200 mmHg in Figure 2(a.4)) and the non-numeric values (e.g., the values in Figure 2(a.5-6)) in numeric columns. Figure 2(c) shows the cleansed numeric values of the examples in Figure 2(a). For the binary features and nominal features, we also defined corresponding cleansing rules to standardize the formats and discard the unstructured values.
Figure 2.
Examples of data cleansing for numeric values
2). Missing Data Imputation
Data imputation is the process to remedy missing data, which is usually necessary for building a reasonable prediction model. Since the dataset is derived from questionnaire and the features have interrelationships, we first built a set of imputation rules to infer the missing values from the other relevant features. For example, if a patient’s SBP was greater than 140 mm Hg, then his/her hypertension history can be replaced with “true”. For another example, if the value of a parent question “having heart failure symptoms” is “false”, then all its children questions, such as “cantering rhythm”, can be imputed with “false”.
After that, we statistically imputed the remaining missing values that cannot be inferred from other features. We first discarded the features with too many missing entries, because their distributions are difficult to estimate, which may lead to inaccurate imputation results. Concretely, if a binary feature has more than 80% missing instances, or a numeric/ multi-value nominal feature has more than 60% missing entries, then this feature was removed from the dataset. For the remaining features, every missing value of a numeric feature (e.g., SBP) was replaced with the mean of the feature’s observed values, every missing value of an ordinal feature (e.g., NYHA level) was replaced with the median of its observed values, and every missing value of an unordered nominal feature (e.g., AF type) was replaced with the mode of its observed values. These methods can statistically minimize the impact of the imputed values in predictive modeling.
Risk Factor Identification
Risk factors for ischemic stroke and TE in AF have been previously studied6,7,8. These knowledge-based risk factors are grounded in previous evidence and have good interpretability. However, many other risk factors that are highly related to TE occurrence for AF patients were not previously identified and involved, and the models built from the previously known factors may not have adequate predictive power.
On the other hand, feature selection methods in machine learning14 can be used to automatically test and select predictive features from a large number of candidate features, and discover new risk factors that have not been previously identified. The prediction models built from the automatically identified factors can represent more complex disease progressions, and usually have higher predictive performance than the models derived from the knowledge-based factors10,12. The disadvantage of the data driven methods is that the resulting models may usually be difficult to interpret or apply in real clinical practices, because the original features in the data are not as easy to understand as the knowledge-based factors. To address these problems, we first performed feature construction to transform the original features and combine knowledge-based features. Then feature selection algorithms were applied to identify potential risk factors from the original and knowledge-based features.
1). Feature Construction
We first preformed feature transformation to split each multi-value feature to a set of binary features. After that, a set of formulas provided by clinicians were used to generate knowledge-based combination features, which can also be used as candidate features in feature selection. These knowledge-based features describe high-level clinical concepts, and each of them maps to multiple original features in the CAFR data. For example, in this study, “CHF” is defined by four features: “NYHA level” > 2 or “left ventricular ejection fractions” < 40% or “having heart failure history” or “having heart failure symptoms”. Similarly, “diabetes mellitus” is defined as “glycated hemoglobin (HbAlc)” ≥ 6.5% or “fasting plasma glucose” ≥ 7.0 mmol/L or “having diabetes history”.
2). Feature Selection
Before feature selection, we first performed pre-selection to remove the unreasonable features. We asked the clinicians to select the feature categories of interest, and discarded the uninteresting features (e.g., all subjective features about quality of life were discarded in order to avoid bias). The close-to-constant features, in which 99% of the instances have identical values, were also be removed.
In machine learning, there are three main supervised feature selection strategies: filter, wrapper and embedded optimization14. In this study, we employed and compared these methods in identifying predictive risk factors.
Filter. This category of methods calculates a score to represent the relevancy of a feature (or a group of features) against the outcome, and then filters the features based on the score. In this study, we applied two univariate filter methods, which respectively use the p-value from chi-squared test and the information gain as the relevancy score to filter each feature independently. We also used the correlation-based feature subset selection method15 (CFS), which is a multivariate filter method that evaluates features in a batch way, to obtain the subset of features that are highly correlated with the outcome while having low intercorrelation between the features.
Wrapper. This type of methods utilizes a specific classifier (e.g., logistic regression) to select the subset of features that provides the best performance for a specific metric (e.g., AUC). In this study, we applied the wrapper subset selection method16. This method evaluates a subset of features by the prediction performance of the classifier using cross validation, and uses the best first search strategy to search the subset of features that can achieve optimized performance.
Embedded optimization. These methods incorporate feature selection directly into the learning process of a model. In this study, we used Lasso17 to introduce L1-norm regularization to generalized linear models (e.g., logistic regression), which can achieve feature selection by shrinking the coefficients of low relevant features to zero during model training.
Predictive Modeling
In this study, we applied and compared different categories of machine learning models that have good interpretability, including generalized linear models, Bayes models and decision tree models, to build TE prediction models for AF.
Generalized linear model (GLM). GLM generalizes ordinary linear regression by allowing the linear model to be related to the response variable via a link function. We used logistic regression, which is a GLM with a logit link function and a binomial distribution. It was widely used in both medical statistics and machine learning due to its good performance and interpretability. We also applied Cox proportional hazards model18 in this study, which is a statistical model commonly used in survival analysis. Cox is a semi-parametric GLM that takes into account the time of observations.
Bayes model. Naïve Bayes model19 is a probabilistic classifier based on Bayes theorem with strong independence assumptions between the features. Naïve Bayes models can be trained very efficiently, and have decent prediction performance and interpretability as well.
Decision tree model. The classification and regression tree (CART) method was applied to build tree-based prediction model, which is very easy to interpret. We also employed random forest20, which constructs a multitude of decision trees and outputs the mode of the classes of the individual trees. Compared to other decision tree learning methods, random forest generally has greater performance by reducing the problem of over-fitting, though having worse interpretability.
Results
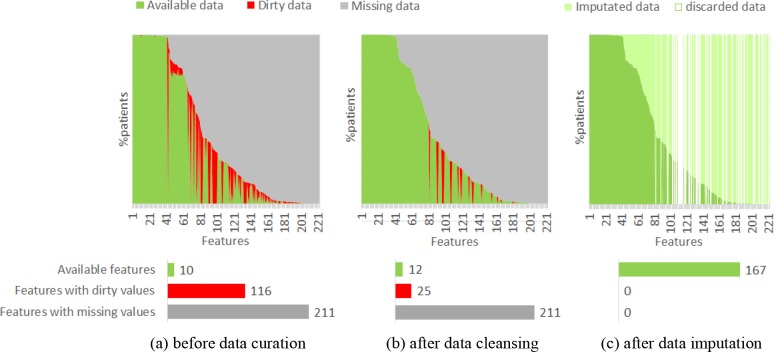
We evaluated the performance of our approaches, including data curation, feature engineering and supervised learning, in building 2-year TE prediction models for AF patients from CAFR dataset. Figure 3 demonstrates the proportions of dirty and missing data in our dataset before and after data curation. In the 221 original features of the raw data, 116 features (55.0%) have non-standardized and dirty values, and 211 features (95.5%) have missing values (Figure 3(a)). After data cleansing, the dirty data in 91 features were standardized and/or corrected (Figure 3(b)). And after data imputation, the missing data in 155 features were filled-in, while 54 features with dirty values and/or missing values that cannot be remedied were discarded (Figure 3(c)). Finally, 167 available features were produced to the following feature engineering step.
Figure 3.
Statistics of data quality before and after data curation
Two standard metrics, AUC and AUPR, were used to evaluate the prediction performance of models. We used AUPR in addition to AUC because in our case of imbalanced dataset, AUC may provide an overly optimistic view of performance, while AUPR can provide a more informative assessment under this situation21. Note that the baseline of AUPR for our dataset is 0.104, which is the average precision of randomly predicting the risk (i.e., an AUPR of 0.208 means the average precision is doubled than that of random prediction).
To compare the performance of the feature engineering methods, we built logistic regression models on different feature sets generated by different feature construction and feature selection methods, and evaluated the mean and standard deviation of AUC and AUPR of each model on 5 different 10-fold cross validation partitions of the data. As shown in Table 1, all the feature selection algorithms can significantly improve the AUC and AUPR of logistic regression, where the multivariate filter method (CFS) and the wrapper method achieved better prediction performance than the univariate filter and embedded methods on our dataset. Besides, when using multivariate filter, wrapper and embedded selection methods, combining knowledge-based features in predictive modeling can also improve the prediction performance to some extent.
Table 1.
Mean and standard deviation of AUC and AUPR of the logistic regression models built on different feature sets, evaluated by cross validation. The standard deviation of every AUC and AUPR is less than 0.01.
| Candidate Features: | Original features | Original + knowledge-based features | ||||
|---|---|---|---|---|---|---|
| Selection Method | No. | AUC | AUPR | No. | AUC | AUPR |
| None | 107 | 0.649 | 0.176 | 117 | 0.644 | 0.177 |
| Chi-squared filter (p < 0.001) | 21 | 0.711 | 0.214 | 25 | 0.709 | 0.209 |
| Information gain (IG > 0.001) | 36 | 0.697 | 0.200 | 44 | 0.690 | 0.196 |
| CFS | 16 | 0.722 | 0.230 | 10 | 0.734 | 0.243 |
| Wrapper for AUC | 23 | 0.759 | 0.241 | 21 | 0.759 | 0.244 |
| Lasso (C = 0.1) | 22 | 0.716 | 0.213 | 20 | 0.719 | 0.224 |
To evaluate the performance of different supervised learning algorithms, we built different learning models on various feature sets (the combination of original and knowledge-based features were used as candidate features in this and the following experiments). As shown in Table 2, GLM methods (logistic regression and Cox) achieved the best performance on our dataset after performing feature selection. Naïve Bayes also got decent AUC and AUPR and their trends are similar with GLM. The decision tree method CART did not work well on our dataset. In comparison, random forest achieved the best performance when applied on all candidate features, but its performance cannot be stably increased by feature selection (except wrapper).
Table 2.
Average AUC and AUPR of different learning models on different feature sets, evaluated by cross validation. The standard deviation of every AUC and AUPR is less than 0.02.
| Selection: | None | Chi-squared filter | CFS | Wrapper for AUC | ||||
|---|---|---|---|---|---|---|---|---|
| Learning Models | AUC | AUPR | AUC | AUPR | AUC | AUPR | AUC | AUPR |
| Logistic regression | 0.645 | 0.177 | 0.709 | 0.209 | 0.734 | 0.243 | 0.759 | 0.244 |
| Cox | 0.621 | 0.173 | 0.707 | 0.208 | 0.735 | 0.243 | 0.755 | 0.239 |
| Naïve Bayes | 0.689 | 0.194 | 0.706 | 0.226 | 0.726 | 0.232 | 0.745 | 0.240 |
| CART | 0.653 | 0.171 | 0.645 | 0.173 | 0.651 | 0.173 | 0.667 | 0.185 |
| Random forest | 0.696 | 0.203 | 0.708 | 0.210 | 0.666 | 0.174 | 0.757 | 0.235 |
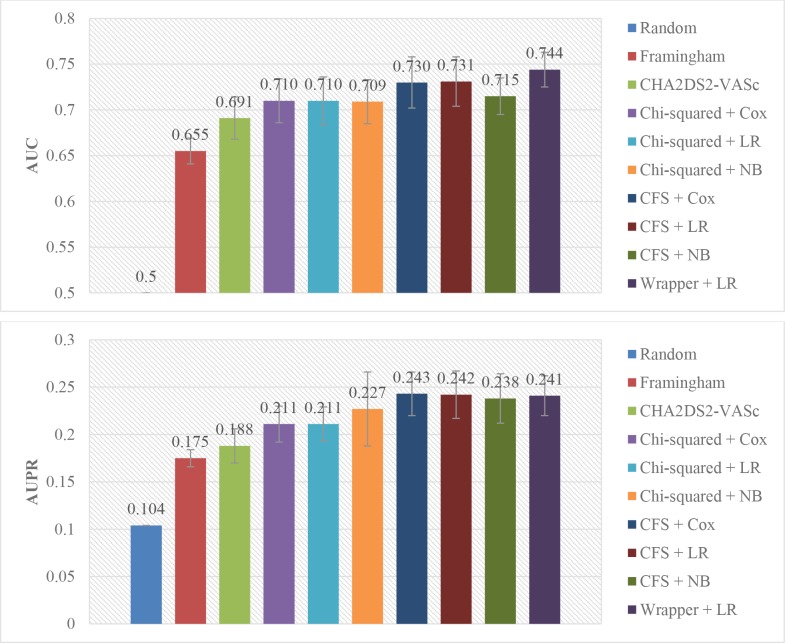
We also compared the performance of our approaches to the state-of-the-arts risk models: Framingham8 and CHA2DS2-VASc7 scores. We randomly split the dataset to a training set with 60% instances and a testing set with remained instances, and selected feature engineering and supervised learning algorithms based on the above experiments to train our prediction models on the training set. Then we applied both previous models and our trained models on the same testing set, and computed the AUC and AUPR of each model. This process was repeated 5 times, and the means and standard deviations of AUC and AUPR of the models were compared. As shown in Figure 4, the prediction performance of our models outweigh the Framingham and CHA2DS2-VASc models.
Figure 4.
AUC and AUPR of different models, evaluated on the same randomly split testing sets. LR = logistic regression, NB = Naïve Bayes
Discussion
In this study, we compared several feature selection and supervised learning methods in building TE prediction models for AF patients. The GLM (logistic regression and Cox) and Naïve Bayes methods did not work well on the whole feature set, but their prediction performance can be significantly improved by appropriate feature selection. This is probably because both GLM and Naïve Bayes have the assumption of no multicollinearity between the features, but the whole feature set is highly redundant and intercorrelated, which negatively affects the prediction performance of GLM and Naïve Bayes. The feature selection algorithms, especially the CFS algorithm that minimizes the intercorrelation and the wrapper algorithm that directly optimizes the AUC, can reduce the redundancy of features and therefore increase prediction performance. As a whole, the decision tree methods (CART and random forest) did not achieve satisfactory performance on our dataset, probably due to the problem of over-fitting. However, by implicitly embedding feature selection, random forest worked relatively well on the whole feature set with high intercorrelation. Besides, in this study, the wrapper selection method achieved the best prediction performance in terms of AUC and AUPR, because the method directly optimizes the performance metric of the specific learning models. However, the time complexity of wrapper selection is very high, which is not practicable for larger datasets. In addition, because the knowledge-based combination features provided by clinicians can describe high-level clinical concepts, adding them during feature engineering can increase the prediction performance while reducing model complexity, when appropriate feature selection is performed.
In addition to achieving higher predictive performance than existing TE prediction models for AF patients, our approach also identified potential risk factors that had not been commonly used. Table 3 shows the risk factors in the previous Framingham and CHA2DS2-VASc models, as well as the factors that were most commonly selected by multiple feature selection methods in this study. The identified risk factors and their odds ratios in logistic regression were verified by clinicians, concluding that the majority of new risk factors, including cardiovascular problem histories, disease durations, relevant electrocardiography and laboratory tests, as well as medications, are interpretable and reasonable to clinicians.
Table 3.
Risk factors in different models
| Framingham score | CHA2DS2-VASc score | Commonly selected risk factors |
|---|---|---|
| Age | Age | Age |
| Prior stroke/TIA | Prior TE | Prior TE |
| Sex | Congestive heart failure | Ischemic stroke confirmed by CT or MRI |
| Diabetes mellitus | Hypertension | Congestive heart failure |
| Systolic blood pressure | Diabetes mellitus | Left ventricular posterior wall thickness |
| Vascular disease | Left ventricular ejection fraction | |
| Sex | Total cholesterol Myocardial infarction Intracranial hemorrhage Drug use for ventricular rate control Years since last TIA Years since diabetes diagnosis Years since paroxysmal supraventricular tachycardia |
Despite the promising results in building TE prediction models for AF patients, there are still several aspects of the approach that could be improved. First of all, in this study, we combined knowledge-based features in feature construction, improving the performance of the resulting models. Besides this, there are also other forms of domain knowledge could be used to enhance the predictive power and/or interpretability of models. For example, clinicians have some knowledge and common sense about the impact of features on a target outcome, from their experience or literature. These knowledge could be built as constraints in the modeling algorithms, which could reduce the bias of a specific dataset and then improve the applicability of the models.
Secondly, we applied the state-of-the-arts feature selection algorithms in machine learning to identify risk factors and used performance metrics AUC and AUPR to evaluate their performance. However, the statistical significance (p-value) of the factors, which is critical for a model to be published and adopted in real clinical practices, was not considered. Though the traditional stepwise selection methods can ensure the significance of selected factors, their prediction performance is usually not good enough. Therefore, a new wrapper-based algorithm that optimizes both prediction power and statistical significance would be a practical method to build more interpretable models.
Thirdly, in this study, we only used original features of CAFR data and knowledge-based combinations as candidate features for building prediction models. There are some frequent pattern mining and pattern abstraction methods22 could be used to discover more complex co-occurrence or temporal patterns, which could be used as combination features in building more effective and interpretable prediction models.
Lastly, in order to use our prediction models in practice, we are also developing a mobile phone app which is named Health Risk Advisor. The TE prediction models can be integrated into the app to provide risk assessment to AF patients, and alert physicians once their patients’ risk levels are changed. A clinical pilot trial for the prediction models could also be conducted in the future based on Health Risk Advisor.
Conclusion
AF significantly increases the risk of ischemic stroke and TE, and accurate prediction of TE for AF patients is critical for early intervention and prevention. In this study, we used integrated machine learning approaches, including data curation, feature engineering and supervised learning, to build TE prediction models for AF patients from CAFR data. The experimental results show that our approach can achieve significantly better prediction performance than previous TE risk models for AF, and identify new potential risk factors as well.
References
- 1.Zhou Z, Hu D. An epidemiological study on the prevalence of atrial fibrillation in the Chinese population of mainland China. J Epidemiol. 2008;18(5):209–216. doi: 10.2188/jea.JE2008021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin HJ, Wolf PA, Kelly-Hayes M, Beiser AS, Kase CS, Benjamin EJ, et al. Pryor TA, Gardner RM, Clayton RD, Warner HR. Stroke severity in atrial fibrillation. The Framingham Study. Stroke. 1996;27:1760–1764. doi: 10.1161/01.str.27.10.1760. The HELP system. J Med Sys. 1983;7:87-101. [DOI] [PubMed] [Google Scholar]
- 3.Verheugt FW, Granger CB. Oral anticoagulants for stroke prevention in atrial fibrillation: current status, special situations, and unmet needs. Lancet. 2015;386:303–10. doi: 10.1016/S0140-6736(15)60245-8. [DOI] [PubMed] [Google Scholar]
- 4.Camm AJ, Lip GY, De Caterina R, Savelieva I, Atar D, Hohnloser SH, et al. 2012 focused update of the ESC Guidelines for the management of atrial fibrillation: An update of the 2010 ESC Guidelines for the management of atrial fibrillation. Eur Heart J. 2012;33:2719–2747. doi: 10.1093/eurheartj/ehs253. [DOI] [PubMed] [Google Scholar]
- 5.January CT, Wann LS, Alpert JS, Calkins H, Cigarroa JE, et al. AHA/ACC/HRS guideline for the management of patients with atrial fibrillation: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and the Heart Rhythm Society. Circulation. 2014. 2014;130:199–267. doi: 10.1161/CIR.0000000000000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gage BF, Waterman AD, Shannon W, et al. Validation of clinical classification schemes for predicting stroke: results from the National Registry of Atrial Fibrillation. JAMA. 2001;285:2864–70. doi: 10.1001/jama.285.22.2864. [DOI] [PubMed] [Google Scholar]
- 7.Lip GY, Nieuwlaat R, Pisters R, et al. Refining clinical risk stratification for predicting stroke and thromboembolism in atrial fibrillation using a novel risk factor-based approach: the euro heart survey on atrial fibrillation. Chest. 2010;137:263–72. doi: 10.1378/chest.09-1584. [DOI] [PubMed] [Google Scholar]
- 8.Wang TJ, Massaro JM, Levy D, et al. A risk score for predicting stroke or death in individuals with new-onset atrial fibrillation in the community: the Framingham Heart Study. JAMA. 2003;290(8):1049–1056. doi: 10.1001/jama.290.8.1049. [DOI] [PubMed] [Google Scholar]
- 9.Van Staa TP, Setakis E, Di Tanna GL, Lane DA, Lip GY. A comparison of risk stratification schemes for stroke in 79,884 atrial fibrillation patients in general practice. J Thromb Haemost. 2011;9:39–48. doi: 10.1111/j.1538-7836.2010.04085.x. [DOI] [PubMed] [Google Scholar]
- 10.Khosla A, Cao Y, Lin CC, Chiu HK, Hu J, Lee H. An integrated machine learning approach to stroke prediction; Proceedings of the 16th ACM SIGKDD international conference on Knowledge discovery and data mining; 2010 Jul 25. pp. 183–192. [Google Scholar]
- 11.Neuvirth H, Ozery-Flato M, Hu J, Laserson J, Kohn MS, Ebadollahi S, Rosen-Zvi M. Toward personalized care management of patients at risk: the diabetes case study; Proceedings of the 17th ACM SIGKDD international conference on Knowledge discovery and data mining; 2011 Aug 21. pp. 395–403. [Google Scholar]
- 12.Sun J, Hu J, Luo D, Markatou M, Wang F, Edabollahi S, et al. Combining knowledge and data driven insights for identifying risk factors using electronic health records; AMIA Annu Symp Proc; 2012. pp. 901–10. [PMC free article] [PubMed] [Google Scholar]
- 13.Ogunyemi O, Kermah D. Machine learning approaches for detecting diabetic retinopathy from clinical and public health records; AMIA Annu Symp Proc; 2015. pp. 983–90. [PMC free article] [PubMed] [Google Scholar]
- 14.Tang J, Alelyani S, Liu H. Feature selection for classification: A review. Data Classification: Algorithms and Applications. 2014. p. 37.
- 15.Hamilton HZ. New Zealand: 1998. Correlation-based feature subset selection for machine learning. [Google Scholar]
- 16.Kohavi R, John GH. Wrappers for feature subset selection. Artificial intelligence. 1997 Dec 31;97(1):273–324. [Google Scholar]
- 17.Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society Series B. (Methodological) 1996:267–88. [Google Scholar]
- 18.Cox DR. Regression models and life-tables. Journal of the Royal Statistical Society Series B. Methodological. 1972 Jan;34(2):187–220. [Google Scholar]
- 19.John GH, Langley P. Estimating continuous distributions in Bayesian classifiers; Proceedings of the Eleventh conference on Uncertainty in artificial intelligence; 1995. Aug. pp. 338–345. [Google Scholar]
- 20.Breiman L. Random forests. Machine learning. 2001 Oct.45(1):5–32. [Google Scholar]
- 21.He HB, Garcia EA. Learning from imbalanced data. IEEE transactions of knowledge and data engineering. 2009;21(9):1263–84. [Google Scholar]
- 22.Wang F, Liu C, Wang YJ, Hu JY, Yu GQ. A graph based methodology for temporal signature identification from EHR; AMIA Annu Symp Proc; 2015. pp. 1269–78. [PMC free article] [PubMed] [Google Scholar] [Retracted]