Abstract
Personal health records available to patients today suffer from multiple limitations, such as information fragmentation, a one-size-fits-all approach and a focus on data gathered over time and by institution rather than health conditions. This makes it difficult for patients to effectively manage their health, for these data to be enriched with relevant information from external sources and for clinicians to support them in that endeavor.
We propose a novel conceptual architecture for person-centered health record information systems that transcends many of these limitations and capitalizes on the emerging trend of socially-driven information systems. Our proposed personal health record system is personalized on demand to the conditions of each individual patient; organized to facilitate the tracking and review of the patient’s conditions; and able to support patient-community interactions, thereby promoting community engagement in scientific studies, facilitating preventive medicine, and accelerating the translation of research findings.
Introduction
Recent, significant investments in health information technology (health IT) have resulted in an unprecedented increase in the “digitization” of the healthcare system in the US. However, the digitization of healthcare information has not automatically translated into increased utility and usability for patients and providers. Indeed, according to a recent report from the Office of the National Coordinator for Health Information Technology (ONC), “there is much work to do to see that every individual and their care providers can get the health information they need in an electronic format when and how they need it to make care convenient and well-coordinated …”1.
An example of the practical limitations common today is the fact that patients often must interact with more than one patient portal, especially when they see clinicians in multiple health systems. This makes it difficult for them to review and synthesize their health information, and act efficiently and effectively on it. A second limitation is that the health information captured about patients is organized using a one-size-fits-all model. For instance, even in electronic systems male patients are routinely asked whether they are pregnant or not. Thus, in the words of the ONC report, health information is not easy for “individuals, their families, and care providers to send, receive, find, and use in a manner that is appropriate, secure, timely, and reliable”1. Against the backdrop of these limitations, the increasing penetration of health IT, coupled with our - now commonplace - expectation that we can manage almost all aspects of our lives electronically, presents an unprecedented opportunity to shape the health sector via a socially-driven, digital approach for person health record (PHR) management. This paper discusses some of the gaps preventing the mainstream adoption of PHRs and proposes a framework that can help bridge these gaps. The goal of this framework is the automated generation of a PHR customized to individual patients and their health and disease conditions. Through this framework, patients will have the opportunity to manage their own health information based on their specific medical conditions rather than a generic one-size-fits-all model. The expected benefits include:
-
-
consolidating and organizing the underlying data in manner that makes information easily understandable and accessible to patients and health care providers;
-
-
enabling targeted preventive medicine tailored to individual patients in an effective and efficient manner; and
-
-
facilitating large scale studies, and promoting the active engagement of the patient in local, regional and national research.
The Health Information Digitization Roadmap
The roadmap for the digitization of the health sector emphasizes three main initiatives: 1) migrating legacy ad hoc electronic health records (EHR) into standardized enterprise-level EHR systems, 2) facilitating EHR-to-EHR interoperability and exchanges of health information, and 3) enabling EHR-to-Patient/PHR exchanges of this health information.
The success of the first initiative, the migration to standardized EHR systems, has resulted in:
-
-
93% of non-Federal acute care hospitals adopting certified EHR systems by 2014 according to the ONC2; and
-
-
200 million patients in the US expected to be covered by the Epic3 EHR system once all of its ongoing system deployments are completed.
With regard to the second initiative, EHR-to-EHR interoperability, the federal government launched an unprecedented nationwide effort in 2009 to help evolve a digital, interoperable health care system in the US4. This effort resulted in more than 50% of office-based professionals and 80% of hospitals using EHRs through which they are now able to electronically exchange standardized patient information in a basic fashion5, 6. A similar state-wide EHR interoperability endeavor is the Indiana Network for Patient Care (INPC), which was started by the Regenstrief Institute in 1998 and is now operated by the Indiana Health Information Exchange. Today, the INPC connects 25,000 physicians, 106 hospitals, 110 clinics, surgery centers and many other healthcare organizations7, and maintains information about over 14 million patients.
In connection with the third initiative, EHR-to-Patient/PHR information exchange, it was estimated that as of 2012, 57% of healthcare providers had a patient portal solution in place allowing patients to access all or part of their health information8. Moreover, in order to investigate the challenges of supporting two-way information exchange between patients and health care providers (Figures1 and 2), the ONC conducted a pilot through the National Association for Trusted Exchange (NATE)9. This pilot focused on data flow and exchange of information from the provider to the PHR (EHR-to-PHR) and vice-versa (PHR-to-EHR). The NATE pilot9 bi-directionally connected EHRs to PHRs using Blue Button10 and HealthVault11. Blue Button allows patients to electronically access their own health information from providers such as health plans, pharmacies, and hospitals. HealthVault is a PHR that allows patients to connect to health providers via Blue Button and download their health related information into a cloud- based account.
Figure 1.
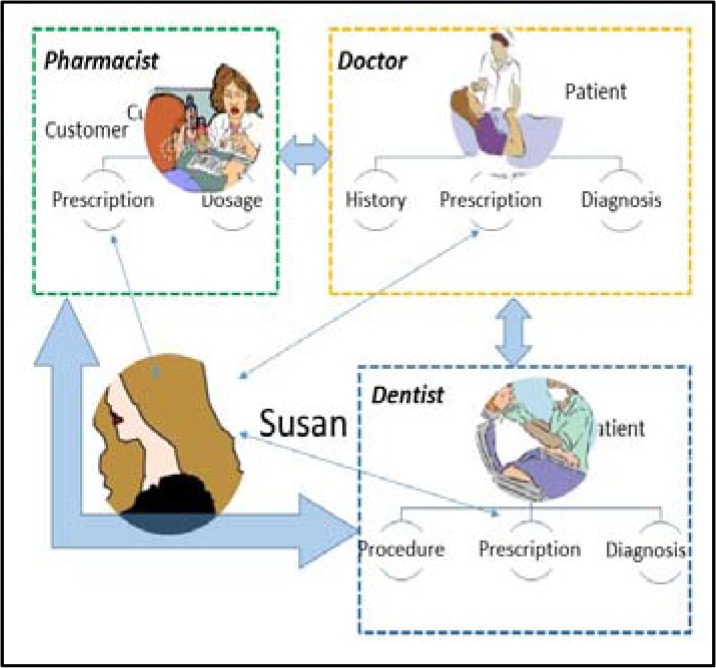
Sample PHR scenario.
Figure 2.
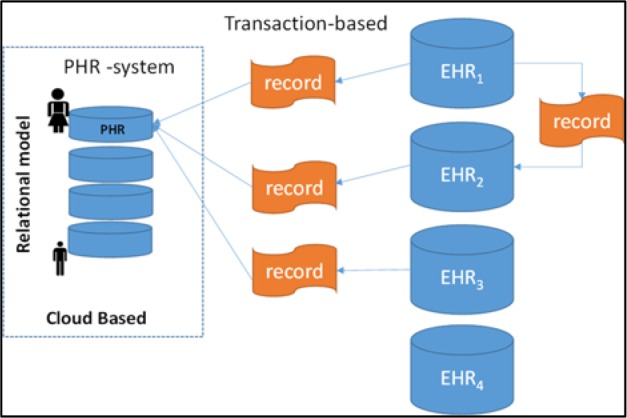
Current framework for PHR information systems.
The three groundbreaking initiatives mentioned above are essential enablers to support the difficult “last mile” effort that will deliver much anticipated benefits of PHRs to the patient. Currently several patient-oriented software applications12 are being offered as solutions to bridge the last mile. However, the limitations of these solutions are still significant enough to have prompted the ONC to articulate its recent 10-year vision. This vision encourages “last mile” solutions that a) enable individuals to manage their own health information, b) share it with their health care providers and c) enable health care providers to individualize diagnosis and therapy and adapt it, as needed, to the patient’s condition in real-time by 20251.
In the following section, the features as well as the limitations of some of the existing patient-focused solutions are discussed in order to establish a baseline and motivate the need for a PHR framework that is capable to meet the ONC 2025 vision.
Current Patient-focused Software Applications
Different vendors offer patient-focused software applications12 such as personal health data-marts, process flow and decision support systems. We briefly review these different types of software applications in this section.
HealthVault, Google Health13 and Indivo-Dossia14 can be categorized as personal health data-marts. HealthVault provides the ability to organize patient health information (PHI), register medical devices (for acquisition of parameters such as blood pressure and glucose) and aggregate family accounts. Figure 3 shows the specific one-size-fits-all data model used by HealthVault. Under this model, patients have a “health” account on the cloud that is used to house their records retrieved from different health care providers’ EHRs, which are, in the overwhelming number of cases, completely independent of each other.
Figure 3.
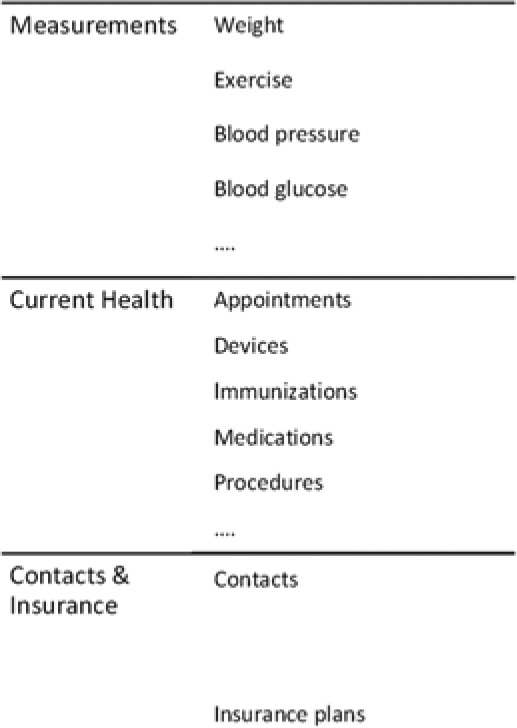
PHR structured relation.
Google Health was an early PHR that used a model similar to that of HealthVault. It was discontinued in 2011. Indivo is an open source PHR which is supported by several large corporations including WalMart. It allows a limited level of personalization through the selection of apps such as immunization tracker and medication manager. These pioneering tools use a relational model with a focus on collecting data from different sources (Figure 2 and 3).
An additional feature of Indivo-Dossia is the ability to automate the process flow between the health service provider and the patient. For instance, a patient may request an examination by using Indivo. The doctor can then review the patient’s health profile and update it by entering a diagnosis or ordering a laboratory test. The request for the laboratory test is forwarded to the information management system of the corresponding laboratory. Once the test is completed, the results are entered and the doctor is notified. Indivo uses ontologies to semantically disambiguate concepts that may be expressed differently in the participating sub-systems.
MeTree15 and Health Heritage16 are decision support systems. MeTree focuses on collecting and organizing family history in order to help support primary care decision-making. Health Heritage focuses on matching the family history with recent scientific research in order to personalize care and prevention plans.
Given the variety and multitude of these types of patient-focused software applications, some of which have been in existence for nearly a decade, the obvious question is: Why is widespread community adoption lacking? The result of a survey of PHR penetration rates in the US indicated that, in 2010, only 7% of the survey participants had ever used a PHR and less than half of these continued using it17. Among veterans, 71% of which utilize the Internet, about 20% reported using a PHR in a 2012 survey18.
In an attempt to answer the above question and by using a process of elimination, cost can be excluded since most of the above-mentioned patient-focused software applications are free. Privacy concerns can also be excluded based on the findings of an earlier study19. This study indicated that while a reasonable level of privacy is expected, a large percentage of the participants were willing to share their health information. The answer can then hypothetically be attributed to 1) the community interest level or 2) ease of use and the added benefit trade-off. In the next section, we present the results of a study that shows that community interest level can also be excluded. This conclusion is supported in the literature20, 21, 22 with a consensus around the fact that the adoption of PHRs is low in spite of the anticipated benefits. Some of the identified reasons20 for the low adoption include the need for additional education and training but most importantly for a better PHR model that can leverage technological advances while meeting the needs of patients and health care professionals.
Community Interest
In order to assess the potential interest level, we conducted a simple study to measure the current community engagement for a representative social media platform, Twitter. Twitter activities related to three common diseases, namely cancer, asthma and diabetes, were investigated for three consecutive semesters from 7/1/2014 to 12/31/2015. The results were obtained by querying the tweets for the keywords a) “cancer”, b) “asthma” and “asthmatic”, and c) “diabetes” and “diabetic”. In the case of cancer, the raw data was preprocessed to eliminate irrelevant tweets (e.g., tweets related to the astrological sign cancer).
For each period and disease, Table 1 shows the number of tweets and the number of followers for a given handle (i.e., user) that posted a tweet related to the specific disease during the period. Duplicate entries were removed such that the number of followers for a given handle are counted only once during any given semester. The last column shows the percentage of the total number of followers shared by the top three handles for each disease type.
Table 1.
Number of tweets and followers per disease from the second half of 2014 until the end of 2015.
| (7/1 - 12/31) 2014 | (1/1 - 6/30) 2015 | (7/1 - 12/31) 2015 | % of followers for the top three handles across all periods | ||||
|---|---|---|---|---|---|---|---|
| #tweets | #followers | #tweets | #followers | #tweets | #followers | ||
| Cancer | 34,587 | 41,022,634 | 141,615 | 100,957,640 | 141,086 | 122,399,740 | 11% |
| Diabetes | 5,663 | 5,876,809 | 55,960 | 19,919,609 | 24,793 | 30,487,220 | 39% |
| Asthma | 3,239 | 2,238,906 | 11,049 | 7,524,524 | 10,491 | 13,585,026 | 30% |
Table 1 highlights the following general trends:
-
-
The number of followers show a steady increase for all three disease types.
-
-
Cancer shows the highest level of activity, followed by diabetes and asthma. The number of people in the US impacted by cancer (20.3m), diabetes (21.0m) and asthma (17.7m)23 are relatively close. However, the percentages of total deaths24 attributed to cancer and diabetes in 2013, are 22.5% and 2.9%, respectively. Asthma is not ranked as a leading cause of death24. This comparison could possibly hint at a correlation between the activity level and the mortality rate.
-
-
The top three handles account for 11%, 39% and 30% of the total number of followers for cancer, diabetes and asthma, respectively. This indicates that there are some influencers with large numbers of followers that have the potential to define the landscape of health social media.
The choice of Twitter as a social media platform was only meant as a representative example. Other social media platforms exhibit similarly high health-related activities. Examples of health-focused social platforms include healthboards.com25 and thepatientforum26. Based on the above results, and the fact that Twitter is a widely used social media platform, the interest level can be excluded as a cause for the lack of widespread adoption of PHR systems. Ease of use and the perceived added benefit by the patient seem to be the determining factors in the mainstream adoption of PHR systems.
Conceptual Architecture
As discussed above, one main reason why current PHRs have not yet been able to deliver the benefits articulated in the 2025 ONC vision is likely to be ease of use. Despite the fact that the user-friendly nature of a software application is often associated with the personalization level of the application27, most current PHRs have adopted a one-size-fit-all model. The tailor-made, on-demand experience that the community is now expecting from online services (e.g., on-demand TV, personalized marketing and personalized banking) should be reflected in the services provided by the PHR. Furthermore, a key factor in the success of this personalization is the adoption of a condition-driven health data model rather than the widely used event- and institution-based data model. One of the drawbacks of the event-based data model is that, when reviewing, for instance, a patient’s diabetic condition within current PHRs, clinicians must find and navigate to measurements, appointments, medications and procedures among a potentially large set of data points over time. The condition-driven model allows the individualized PHR to be organized around a) the conditions of concern to each individual, b) the potential correlation between these conditions, and c) the potential relation between the manifestations of these specific conditions in the PHRs of the patient’s relatives.
Figure 4 shows the general framework of the proposed PHR framework. The main modules of this framework consist of a) personalization and registration, b) data management, c) services and updates, and d) notification and authorization modules. Each of these modules is discussed next.
Figure 4.
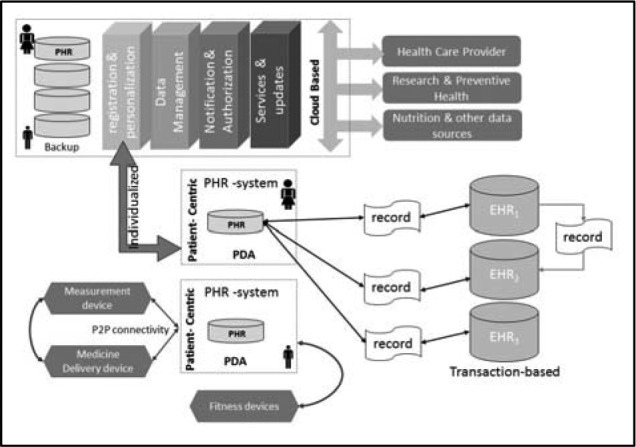
Proposed Person-Centered PHR Architecture.
a) Personalization and registration
The concept of personalizing the PHR by using a condition-based model is sustained throughout the life-cycle of the PHR. When patients initially register, the personalization and registration module will dynamically and interactively create a set of questions enabling the rapid capture of the patient’s health status. For instance, knowledge of the gender of the patient will eliminate, in either case, a large number of gender-irrelevant questions. Similarly, entry of the date of birth will allow the registration to be tailored to health conditions pertinent to the appropriate age group. In addition to its efficiency, this approach offers the possibility to drill down and gather information for important health conditions comprising historical information related to a given disease, family members suffering from the same disease, and previous and current health care providers related to this condition.
The interactive and dynamic registration questionnaire is only the first step of the personalization process. The next step consists of generating a personalized PHR that will organize the health information provided both by the patient as well as that obtained through the healthcare provider in a semantically meaningful way. The details of this personalization are covered throughout the remainder of the paper.
b) Data management
Our proposed data management scheme is based on MongoDB28, a NoSQL document database. MongoDB stores data as documents by using an extension of JavaScript Object Notation (JSON) called Binary JavaScript Object Notation (BSON) which includes primitive data types such as floating point. Most of the current EHR-EHR data exchanges today transmit data directly through HL7 or use the Extensible Markup Language (XML). The debates about the suitability of XML versus JSON or its variant BSON is ongoing. Our choice of BSON for both data representation and data exchange is based on the fact that the latter supports arrays which is an essential data structure for the representation of repetitive measurements especially when the data has a high velocity (e.g., reading of glucose levels multiple times a day). Furthermore, the conversion from XML to BSON is straightforward and can be easily accommodated to retrieve PHI from existing EHRs.
The use of the label “document” in MongoDB is a misnomer. In this paper, the term object is used instead. The proposed data model is designed around basic primitive objects (i.e., documents in MongoDB technical terms). These primitive objects include basic information, measurement, procedure, lab test, prescription and relation. The primitive objects are subject to update and extension without incurring any disruption to the operation of the application. This is a unique feature of MongoDB which simplifies the personalization process. For instance, it is conceivable in the future to add a meal object that will support the nutrition aspect of the PHR as well as the individualization of the managed delivery of insulin for a diabetic. Another possible extension is a physical exercise object that will be used to support fitness activities.
Figure 5 shows a sample instantiation of four of the primitive objects, namely basic information, measurement, observation and relation. These primitive objects are patient-specific and generated by the registration and personalization module (Figure 4). All of the objects reference a “uid”. This is a unique health id for the patient. In a previous study19, participants indicated that they did not object to the use of a unique health id. Furthermore, most of the current users of social media platforms are accustomed to having a unique identifier. For instance, in Twitter this identifier is called a handle and it was used to generate the statistics of Table 1. The health care provider is also associated with a unique identifier (Figure 5–2), with the existing National Provider Identifier being the most logical choice. Moreover, all measurements, observations and scanned documents also have a unique identifier. These latter identifiers are constructed by prepending the uid of the patient to a unique sequence for each element type.
Figure 5.
Sample Primitive Objects
Several of the features of the primitive objects are worth noting:
-
-
The measurement type includes a potential list of mappings. These mappings are added as needed to each individual patient record in order to support interactions with health care providers that use different vocabularies. Mapping between different vocabularies can be performed by using a metathesaurus such as the UMLS29. Furthermore, on a longer term basis, this field can also be used to capture any vulgarization of different measurement names. In general, this functionality reduces the difficulties associated with semantic mapping.
-
-
The primitive measurement object includes an aggregation type field which indicates how the data should be aggregated (e.g., sampling, monthly averages, cumulative averages, etc.). The aggregation type allows the definition of the most appropriate summative method for high velocity data. The method is customizable to each individual, condition and measurement. For instance, different conditions in the same patient may rely on different aggregations of weight or blood pressure readings.
-
-
The reading sequence is a sequence of tuples consisting of a timestamp and a value. The sequence is updated with every reading associated with a measurement. Furthermore, a new measurement object will be instantiated if any of the underlying fields such as the ordering party or the device id are changed.
-
-
The observation object is used to capture the results of an encounter between a patient and a health care provider. Depending on the result of the observation, the PHR may be restructured to highlight a new condition or the observation may be linked to an already existing health condition.
-
-
The relation object will help identify the type of relation (e.g., parent, sibling, child, etc.) as well as the associated uid of the relative. The underlying application will then perform regular updates based on routine updates to the relative’s PHR or based on newly discovered research to establish the association among any of the individual’s health conditions and those of a relative. Furthermore, a scoring mechanism is established and dynamically refined to indicate the strength of this association with each of the relatives and for each of the specific health conditions. Several indexing mechanisms are available in MongoDB that support this type of association.
c) Services and updates
Each individual interacts with the server and the health care provider through her personal digital assistant (PDA). Both the server application and the PDA-application have a three-tier architecture: 1) the front-end is implemented using HTML, CSS and JavaScript, 2) the middle layer is based on GoLang, and 3) the back-end uses Mongo-DB. On the server, Mongo-DB is used to store all the data related to the registration, personalization, authentication, services and updates. It also manages user profiles and displays data according to each user profile.
There are two main types of profiles: the patient profile and the health provider profile. The health provider profile is outside the scope of this paper. It essentially classifies the health providers into roles including physician, nurse, insurance, institution, etc. The physician profile allows the user to interact with the patient through messaging and enables the physician to organize the patients into different groups (e.g., a “critical watch list.”) The physician, once authorized by the patient, can also place the patient’s measurements under alert. This configurable functionality will enable the physician to receive an alert, for example, when the glucose level of the patient exceeds a certain level. The interaction between the health provider and the individual described above represents one layer of communication. The other communication layer consists of the interaction of the individual with the community.
One of the functionalities of the services and updates module in Figure 4 is to extract health information from different sources of specific interest to the patient. This information can be the result of recent research, a nutrition database or preventive health information. This functionality is key as it will help the timely translation of scientific findings into immediate benefit to the patient thereby promoting retention and mainstream engagement.
The services and updates module will also be responsible for recruiting participants for large scale studies. In this case, a given uid will send a request to the server for the distribution of a recruitment form to participants. The server will review the request and based on the uid’s trust level and a pre-authorized protocol will either decide to forward the request to the target participants or request additional endorsement/validation. Once this authorization step is completed, the server application will generate a list of qualified participants. Additional restrictions may apply during this step. For instance, the requesting party may be asked to refine the underlying query if the number of participants that qualify exceeds or is below a given threshold leading to resource exhaustion or privacy violation, respectively. This automated data validation process is currently under investigation.
Once the validation process is completed, the services module will contact the participants using a push notification technology and will register the uid of the users that accept to participate in the study. The anonymized and sanitized data corresponding to the participants will be extracted and forwarded to the requesting investigator. The investigator will be required to disclose the result of the study which will be made available to the participants and to the community at large. Furthermore, the users will be able to score these findings in terms of benefit to their health, thus impacting the accumulated contribution score of the individual investigator. This type of engagement allows the patients to feel empowered and is critical to their retention.
Most of the activities related to the engagement of the patient in community studies and research are handled by the server application without any involvement from the patient beyond the authorization and the feedback steps. In order to further facilitate this process, the user may decide to setup general rules that are applicable to all requests in a given category. An example rule may be to accept requests for participation for all studies sponsored by a given institution or a given individual. This type of rules promotes “brand” loyalty and facilitates stronger research-community relationships.
Typically, HIPAA prevents direct researcher-patient contact and mediation through a health care provider is mandated unless the participant clearly opted in to direct contact. In the proposed architecture, the patient has to provide prior authorization. Beyond this step, the exchange of information is mediated by the services and updates module which will have to be under the administrative management of a “trusted party”. The policy implication of this data exchange model and other models proposed in the paper is the subject of future investigation.
d) The notification and access authorization module
As previously mentioned the structure of the communication of the proposed framework consists of the combination of two layers. The first layer is a cloud server and supports the communication between the patient and the community.
This layer has a centralized architecture and includes the notification and access authorization module shown in Figure 4 which is responsible for:
-
-
The authentication of all users, including patients, service providers and researchers.
-
-
The management of patient-health care provider access authorizations which are given on a per condition basis by the patient to the health care provider. Connection rules are managed through the front-end and data access rules are managed through the back-end in MongoDB.
-
-
The management of access authorizations required for the participation of the patient in research studies.
The second communication layer is a Peer-to-Peer network as illustrated in Figure 6. Peer-to-Peer networks are becoming increasingly popular in social computing (e.g., Napster). In this layer, peers are autonomous and self-organize in a private sub-network that co-exists with other patient-centric or provider-centric networks. Once a user is authenticated by the notifications and access authorization module in the first layer, she can exchange data directly with her selected peers without the involvement of the server. This mode of communication is used for the exchange of private data between the patient and the health care provider. In addition to scalability and congestion control, the Peer-to-Peer network offers better data exchange security.
Figure 6.
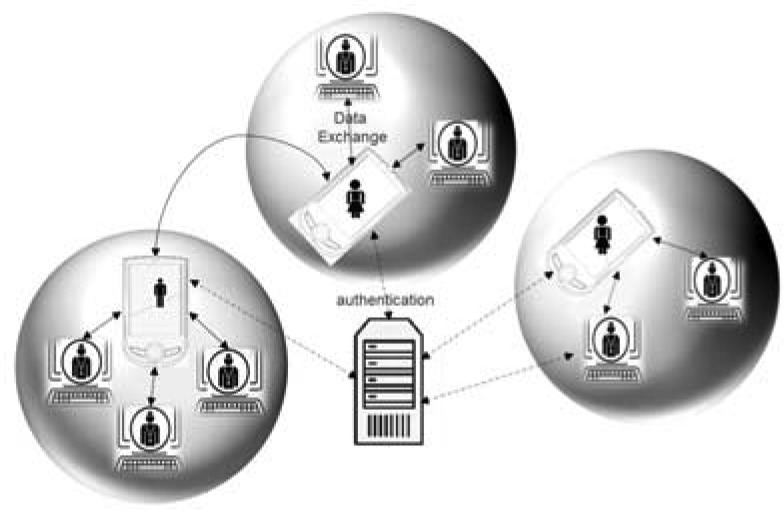
Peer-to-Peer Network
The Personalized PHR
The role of the registration and personalization module is to generate the specific meta-model for the PHR and to populate it with the appropriate data by using the other modules including the services and updates module mentioned above. Figure 7 shows a high level representation of this model for a specific individual. The skeleton of the model is generated by the registration and personalization module when the patient first interacts with the platform. This model is then enhanced and updated as it is enriched by the user, the health care provider and the users’ network.
Figure 7.
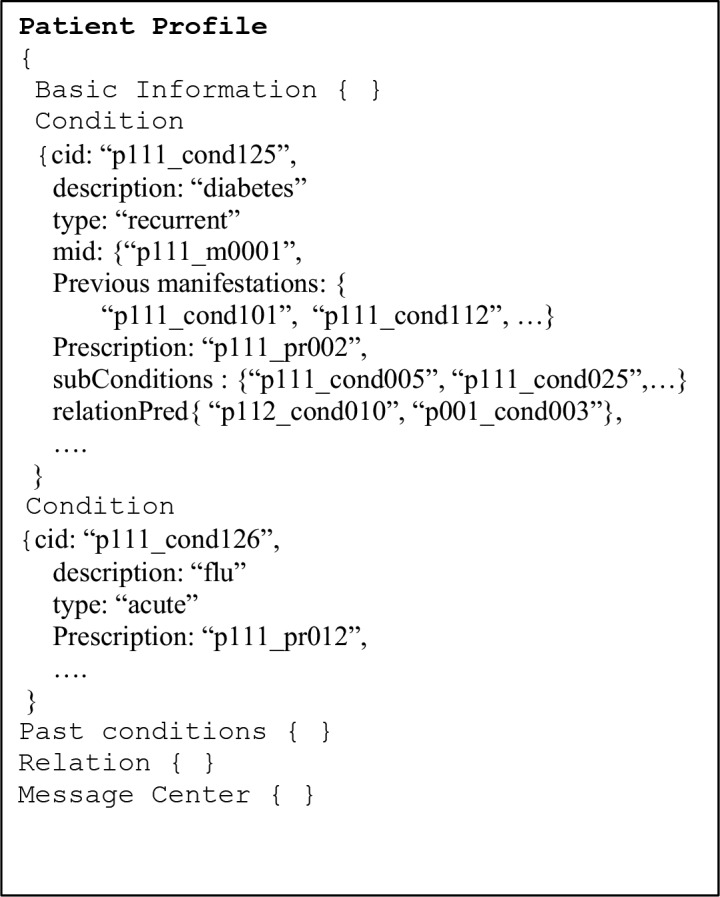
PHR data model
The services and updates module will rely on available application program interfaces (APIs) in order to extract the information needed from different EHRs and research data sites. The APIs can be triggered on a regular basis to update the PHR record of each individual patient. These data are stored in the MongoDB database by using the primitive objects and consolidated based on the uid. A copy of these data and the meta-model are available as part of the back-end database of the client application on the patient’s PDA.
In most traditional client-server applications, the client-side application retrieves data on-demand from the server. In the proposed architecture, the client and the server are both responsible for different types of data exchanges and services. The server application is responsible for the exchange of information between the patient and the community. The client application is responsible for the management of the patient’s private health data. For instance, direct capture of measurement from a home health device and its delivery to the health provider will be handled by the PDA through the patient-centric Peer-to-Peer network; whereas, the delivery of health data necessary for a study will be handled by the server application once patient approval is granted. This delineation allows the patient to focus on the health management issues that are of most concern to his condition. Furthermore, it facilitates managed control of time-critical exchanges with the patient and his personalized private health network (Figure 6).
Data Retrieval and Indexing
One of the major challenges of the proposed application is the ability to provide ease of access for a specific individual as well as across a large set of individuals. Most current database applications either provide one or the other. For instance, organizations often segregate among the two types of accesses by using a transactional database (e.g., EHR), as well as a data warehouse for analysis and reporting purposes. Combining the two is the subject of ongoing research30. One of the emergent application of this combination is social computing. The social media model is relatively simple and based on relations such as “is a friend”, “is a follower”, etc. The proposed PHR model also needs to support both transaction-based interactions (e.g., health care provider - patient) as well as analytics-based processing for large scale studies. However, the health context makes the data retrieval and management complex. Therefore, communication has been segregated into two layers, services have been carefully assigned to the client or the server and data management has been associated with the most appropriate application depending on the context.
Patient local data are essentially managed by the client application and then bi-directionally synchronized with the server as needed. For high velocity data, aggregation can help reduce the size of the data that needs to be stored on the PDA. A detailed view of the data can be obtained on-demand through the server application. Although MongoDB is not an SQL database, it does support aggregation which can be used to implement the above functionality. The other type of data that may represent a burden for the client application is the document type (e.g., images). In this case, the original copy of the document can be stored on the server and the client application can access it on demand from the server using a hyperlink.
Because of the potentially large volume of data on the server, proper indexing becomes a critical requirement. Data on the server is organized by using collections of primitive objects (e.g., basic information, measurements, observations, relation, etc.). These collections are for all individuals and indexed by the uid. In order to facilitate large- scale queries, several other indexes (e.g., indexes by condition or geolocation) need to be maintained. Furthermore, compound indexes that are composed of more than one predicate will be developed overtime as the system matures and is enriched by complex user queries. These indexes may eventually hold the most interesting answer to large scale research questions as they will be able to associate, for example, individuals with different phenotypes or genotypes.
Tree and geospatial indexes can also be used to organize the relation of an individual’s PHR to that of her relatives’ PHRs, thus facilitating condition-based distance scoring. Finally, the Time-to-Live (TTL) index will be used to adequately manage the data in MongoDB. The TTL index is customarily used to automatically delete old data from the system. In this case, the TTL index will be used to archive data to a secondary database. This multi-tier database stack allows a) patient centric data to be close to the patient on his PDA, b) community current data to be close to the researchers and c) historical data to be available for long term studies.
Conclusion
The proposed platform will help close important gaps in the healthcare research, clinical practice and patient continuum by allowing patients to better manage their health, and benefit from up-to-date research efficiently and effectively. The proposed framework will link a patient’s health information to relevant health data about their family members, automatically determine the applicability of current medical evidence or recommendations to the patient, and enable the patient to enrich his record from external data sources. We believe that these features will facilitate mainstream adoption and, more importantly, sustained engagement by the patient in managing his health.
The innovative aspects of the proposed framework and its departure from the limitations of current approaches center on:
-
-
developing methods to custom-build on-demand applications that will manage data for each individual patient in a personalized and focused manner; and
-
-
identifying mechanisms that can translate research findings into practical and systematic methods for organizing patient health information.
We believe that this level of quality of service will keep patients engaged and interested in the management of their data. Furthermore, it will encourage patients to become willing participants in large-scale medical studies that can further improve research. Our goal is to leverage the viral effect of social networks towards a customizable solution for patient-centered health record and a cost-effective mechanism to provide answers to important questions that are unattainable through traditional research studies.
References
- 1.ONC. A 10-Year Vision to Achieve an Interoperable health IT Infrastructure. Office of the National Coordinator for Health Information Technology. 2015.
- 2.Charles D, Gabriel M, Searcy T. Adoption of Electronic Health Record Systems among U.S. Non-Federal Acute Care Hospitals 2008-2014 ONC Data Brief no. 23 2015 [Google Scholar]
- 3.Eisen M. Epic Systems: Epic Tale. Isthmus . 2008 [Google Scholar]
- 4.Thompson TG, Brailer DJ. The decade of health information technology: delivering consumer-centric and information-rich health care - framework for strategic action.; Office of the National Coordinator for Health Information Technology; 2004. [Google Scholar]
- 5.ONC. Data Analytics Update. Office of the National Coordinator for Health Information Technology. 2014.
- 6.ONC. U.S. Hospitals’ Capability to Electronically Query Patient Health Information from Outside Their Organization and System. Office of the National Coordinator for Health Information Technology. 2014.
- 7.IHIE. Indiana Health Information Exchange. www.ihie.org 2016
- 8.KLAS. Patient Portals: The Path of Least Resistance. www.KLASresearch.com 2012
- 9.NATE. PHR Ignite Pilot. National Association for Trusted Exchange . 2014.
- 10.ONC. About Blue Button. Office of the National Coordinator for Health Information Technology. 2015.
- 11.Microsoft. HealthVault. Microsoft . 2016.
- 12.Lewis N. Consumers Slow To Adopt Electronic Personal Health Records.; Information Week; 2011. [Google Scholar]
- 13.Lohr S. Google Offers Personal Health Records on the Web. New York Times. 2008 [Google Scholar]
- 14.Moore J. Indivo Health: Further Details on Dossia Agreement.; Chilmark Research; 2007. [Google Scholar]
- 15.Yoon PW, Scheuner MT, Jorgensen C, Khoury MJ. Developing Family Healthware, a family history screening tool to prevent common chronic diseases. Prev. Chronic Dis 6 1 A33 2009. [PMC free article] [PubMed] [Google Scholar]
- 16.Cohn W, Ropka M, Pelletier S, et al. Health Heritage, a web-based tool for the collection and assessment of family health history: initial user experience and analytic validity. Pub Health Gen. 2010;13(7-8):477–491. doi: 10.1159/000294415. [DOI] [PubMed] [Google Scholar]
- 17.Dunbrack L. IDC Health Insights Survey. IDC; 2011. [Google Scholar]
- 18.Tsai J, Rosenheck RA. Use of the internet and an online personal health record system by US veterans: comparison of Veterans Affairs mental health service users and other veterans nationally. J Am Med Inform Assoc. 2012;19(6):1089–1094. doi: 10.1136/amiajnl-2012-000971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beranek L, Horan TA. Prospective Personal Health Record Use Among Different User Groups: Results of a Multi-wave Study.; Proceedings of the 41st Hawaii International Conference on System Sciences; 2008. [Google Scholar]
- 20.Nazi KM. The Personal Health Record Paradox: Health Care Professionals’ Perspectives and the Information Ecology of Personal Health Record Systems in Organizational and Clinical Settings. J Med Internet Res. 2013;15(4):e70. doi: 10.2196/jmir.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gartrell K, Trinkoff AM, Storr CL, Wilson ML, Gurses AP. Testing the Electronic Personal Health Record Acceptance Model by Nurses for Managing Their Own Health: A Cross-sectional Survey.; Appl Clin Inform.; 2015. Apr. pp. 224–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansen MA, Henriksen E. The evolution of personal health records and their role for self-management: a literature review.; Stud Health Technol Inform ; 2014. pp. 458–62. [PubMed] [Google Scholar]
- 23.CDC, National Data, Center for Disease Control, 2013.
- 24.Jiaquan X, Murphy SL,, Kochanek KD, Bastian BA. Deaths: Final Data for 2013. National Vital Statistics Reports. 2016;64(2) [PubMed] [Google Scholar]
- 25.HealthBoards. Health Message Board healthboards.com
- 26.PatientForum. The Patient Forum. Thepatientforum.com
- 27.Giraud A. The Next Frontier - User-Friendly Applications in the Business DSAG Helps Its Members Prepare for Easier and More Intuitive Business Applications. SAPinsider. 2014;15(3) [Google Scholar]
- 28.MongoDB. Mongo DB for Giant Idea. Mongodb.com
- 29.UMLS. Unified Medical Language System. U.S. National Library of Medicine.
- 30.Splice Machine. First RDBMS Powered by Hadoop and Spark. splicemachine.com



