Abstract
We explored the feasibility of representing nursing research data with the Observational Medical Outcomes Partners (OMOP) Common Data Model (CDM) to understand the challenges and opportunities in representing various types of health data not limited to diseases and drug treatments. We collected 1,431 unique data items from 256 nursing articles and mapped them to the OMOP CDM. A deeper level of mapping was explored by simulating 10 data search use cases. Although the majority of the data could be represented in the OMOP CDM, potential information loss was identified in contents related to patient reported outcomes, socio-economic information, and locally developed nursing intervention protocols. These areas will be further investigated in a follow up study. We will use lessons learned in this study to inform the metadata development efforts for data discovery.
Introduction and Background
Wide adoption of Electronic Health Record (EHR) systems and the development of secure cloud-based infrastructure to store large amounts of clinical and health research data have been opening new opportunities for data-driven discovery and validation of knowledge. Observational and pragmatic studies are gaining traction as alternatives to Randomized Controlled Trials (RCT) [1–3]. Although the highest quality of clinical evidence is still produced via RCTs [4] it is infeasible to perform an RCT for every clinical problem of interest for every possible patient situation. Pharmaco-surveillance [5,6], various comparative effectiveness studies [7,8] and cohort discoveries [9,10] are a few example areas that have benefited by the increasing availability of health data. However, these data also pose non-trivial challenges such as the handling of missing and/or noisy data and the integration of data generated from various sources, each with its own idiosyncratic representation. The Big Data to Knowledge (BD2K) initiative by NIH aims to address such challenges and to promote the utilization a large amount of biomedical data (i.e., “big data”) by improving discoverability, accessibility, interoperatbility, and reusability of data and advanced data analytics [11,12].
bioCADDIE – indexing for big data discovery
bioCADDIE (biomedical and healthCAre Data Discovery Index Ecosystem) is a BD2K consortium dedicated to establishing a user friendly and robust means to discover and index biomedical data [13,14]. At its core are the bioCADDIE metadata, a minimum set of information about a dataset that needs to be made available to data seekers to facilitate search for data sets. The bioCADDIE metadata specification was developed by incorporating existing major metadata schemas in the biomedical domain (i.e., top-down) and high-priority dataset search use cases identified by the bioCADDIE leadership group and the user community. The version 1 metadata specification was released in August of 2015 and is currently under revision based on the feedback obtained from the bioCADDIE community [15]. Some of the referenced existing metadata and use cases reflect clinical and/or healthcare domains to a certain extent (mostly diagnoses, laboratory tests, and medications). However, overall, clinical and healthcare research domains are relatively under-represented in bioCADDIE metadata when compared to omics research domains. As a first step to augmenting the clinical and healthcare domains, the bioCADDIE team has started exploring the interoperability between the bioCADDIE metadata and the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM), a widely accepted, standardized model for healthcare data [16].
OMOP CDM – a common data model for healthcare research data
OMOP was formed as a partnership between public and private sectors to establish the safe and effective use of observational healthcare data to study the effectiveness of medical products [17]. Observational Health Data Sciences and Informatics (OHDSI) is an international collaboration that aims to promote data reusability of the observational healthcare data produced from various venues by achieving data interoperability among them [18]. The OMOP CDM is adopted and now continuously revised by OHDSI [19]. Patient Centered Outcomes Research network (PCORnet), a national network funded by PCORI, is another major effort that aims at utilizing large scale clinical and healthcare data research networks for patient-centered outcomes research. A CDM plays a critical role in achieving data interoperability [20–22]. PCORnet CDM [23] and OMOP have recently been cross mapped by the OHDSI research team [24].
Gaps in representing nursing data for big data science
Many studies explored the use of the OMOP CDM to conduct large scale drug surveillance studies as well as to integrate clinical data repositories [22,25–29]. However, few studies reported the experience on standardizing and integrating data generated from clinical or healthcare research with OMOP CDM, except for studies that involved drugs or medical devices.
The recent Precision Medicine Initiatives (PMI) calls for collecting and utilizing a full spectrum of biomedical data to improve patient care: from microbiology to physical exams, laboratory tests, behavioral and environmental exposures [30]. In addition, the National Academy of Medicine (NAM), formerly Institutes of Medicine (IOM), recommended the incorporation of social and behavioral domains of data in EHRs in such a way that would support the reuse of those data in patient care and research [31]. Therefore, integrating different types of data in a way that supports secondary analyses has become critical. Nursing research targets a wide range of nursing interventions and problems, which often may not involve specific drugs, medical devices, or medical diagnoses. It often includes topics such as patients’ functional status, education, satisfaction toward the provided care and quality of life. This might indicate that nursing and nursing research data present a unique opportunity to understand the challenges and special considerations in integrating data generated in variety of health domains. However, few studies investigated the feasibility of representing various types of nursing and nursing research data with a CDM except for a very specific nursing domain such as pressure ulcer risk assessment and prevention [32,33].
The purpose of this study was to test the feasibility of representing nursing research data with the OMOP CDM using data reported in published articles and subsequently indexing them with the bioCADDIE tools. Specifically, we evaluated (1) the content coverage of the OMOP CDM (version 5) toward the data content reported in 256 published nursing research articles and (2) the feasibility of conducting data searches using the use cases extracted from the 10 randomly selected research articles.
Materials and Methods
Data preparation
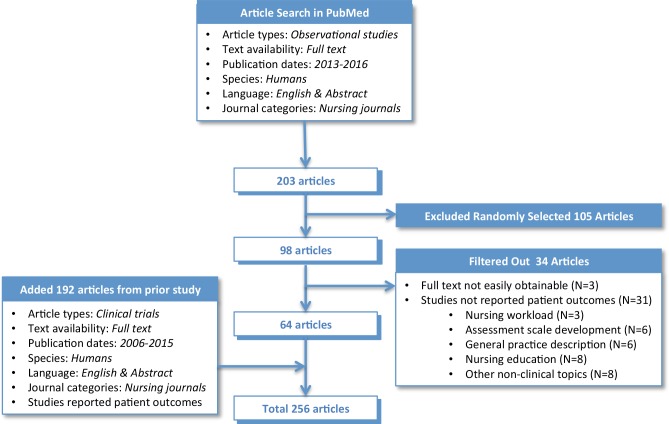
In a prior separate study, we reviewed 192 articles on clinical trials published in nursing journals between 2006-2015. From this review, we annotated the articles with a list of metadata such as study types, target research problems, key interventions, comparison groups, sample demographics, data items used/generated, data sources, funding supports, data access information, and locations and settings of the studies. To augment this collection with observational studies we conducted additional literature review. Using PubMed and its structured search filters, we retrieved articles classified as Observational Studies published in Nursing Journals for the past 3 years (i.e., 2013 ~ February of 2016). This search yielded 203 articles. We randomly selected about half of the retrieved articles (N=98). We then read the full text articles and annotated each article with the same set of metadata.
During the full text review we excluded 34 articles for which full text papers were not easily obtainable (N=3) or the articles that did not investigate patient problems from nursing perspectives thus consequently no patient outcomes were reported (N=31). For example, behavioral issues among nursing students [34], assessment scale development [35], observing hand washing behavior among nurses without reporting associated patient safety outcomes [36] and nursing management topics such as nursing shifts and workload estimation [37,38] were excluded from this review. With this additional review, we generated metadata for 64 articles on nursing observational studies. Therefore, combining the new articles with the previously tagged ones, we included a total of 256 articles to this study. The article selection process is summarized in Figure 1. We also classified the data items reported in 256 articles based on the modes of data collection – i.e., routinely collected clinical data, Patient Reported Outcomes (PRO), and data collected through mobile devices.
Figure 1.
Article selection process.
Mapping to the OMOP CDM
A total of 2,438 data items were extracted from the 256 articles. After removing duplicate entries we classified 1,431 unique data items into concept classes of the OMOP CDM to establish a high (i.e., general) level of mapping. This mapping was largely guided by the definition and usage description provided for each concept class in OMOP CDM. However, we also checked the attributes of concept classes to ensure a given data item had a relevant attribute that could be used to hold the information about the data item. For example, one might expect that various socio-economic, and demographic items such as marital status and education level would belong to the PERSON class as done in many healthcare databases. However OMOP’s PERSON class does not provide a place to hold those items as it was designed to hold permanent information about a person such as birth year and race. Two of the authors (HK, JC) collaboratively conducted this high level mapping. We also aimed at identifying data items that did not fit into any of the OMOP CDM classes during this high level mapping activity.
Next we conducted a small scale, deeper level mapping and investigated to what extent the semantics of the content items presented in the selected articles were represented in the OMOP CDM. We fully utilized the attributes and the constraints associated with the concept classes for this mapping. We first created 10 data search use cases from 10 articles randomly selected from the 256 articles pool by asking ourselves “how do you query a clinical data repository to replicate the study reported in this article?” These use cases incorporated the eligibility criteria of the studies and thus provided an opportunity to evaluate additional content items not reflected in the data items at the deeper level (i.e., value level). Next we simulated the dataset search use cases by converting them into a data query form similar to the SQL code style (i.e., pseudo SQL codes). We used the mapping established between the OMOP CDM and the data items as a database schema of a hypothetical data repository. A use case and its data query form are presented as an example in Figure 2.
Figure 2.
Data search use case and data query form translation
To simplify the conversion we omitted certain details and employed a few of our own conventions: we did not specify the primary key and foreign key relations to simplify the conversion. We used standardized concept id attributes (e.g., gender_concept_id, observation_concept_id, etc.) to represent data item names and non-numeric data item values. Instead of specifying mapped standardized concept codes, we put data item or value names in brackets to imply the values are the concept codes of the concept names presented in the brackets. For example, the code observation_concept_id [] = [marital status] in Figure 2 implies that the observation_concept_id attribute takes the concept code of an OMOP-recognized standardized vocabulary of choice, for instance “125680007” when the Systematized Nomenclature of Medicine- Clinical Term (SNOMED-CT) is used to encode values.
Results
The majority of the data items identified from the 256 articles were mapped to the concept classes in the OMOP CDM, except for 16 data items. A few examples of unmapped items were the socio-economic and/or demographic information of health care providers (e.g., “number of years working as a nurse”) and the items that potentially embedded multiple content items, each of which would need to have been mapped to different concept classes. For example, “operating room nurses’ task list” and “advice provided by healthcare provider” might need to be mapped to different classes depending on what values they take.
MEASUREMENT was the concept class that most data items were related to, followed by OBSERVATION. The data items mapped to each concept class in the OMOP CDM is summarized in examples in Table 1.
Table 1.
Number of data items mapped to each concept class
| Mapped OMOP CDM Class | Count (N=1,431) | Examples |
|---|---|---|
| Measurement | 529 | Charlson comorbidity index score, Ejection fraction, Vital signs |
| Observation | 319 | Treatment history, Smoking status, Marital status, Catheter leakage |
| Condition_Occurrence | 223 | Problem list, Admitting diagnoses |
| Procedure_Occurrence | 109 | Renal transplant, Chemotherapy, Detoxification protocol, Relaxation |
| Person | 94 | Age, Race, Gender, Ethnicity |
| Drug_Exposure | 48 | Opioid dose, Type of opioid used, Pain medication |
| Device_Exposure | 25 | Mechanical ventilator use, Traction device type, CPAP use |
| Visit_Occurrence | 27 | Admission date, Discharge date |
| Care_Site | 15 | Type of hospital, Admitting service |
| Provider | 3 | APN specialty, Service provider type |
| Payer_Plan_period | 6 | Payer, Health insurance type, Insurance status |
| Visit_Cost | 5 | Average cost per patient per patient day, Average cost per patient |
| Drug_Era | 4 | Period of hormone therapy, Period of chemotherapy |
| Death | 4 | Hospital death, Death |
| Procedure_Cost | 3 | Wound dressing cost, Overhead |
| UNMAPPED | 17 | Influence of ward condition, Counselor’s age, Hygiene compliance |
PRO items collected using a questionnaire (both standardized or custom developed) were reported in 82 articles and data generated through mobile devices were reported in 2 articles.
Our attempt to simulate data search from the use cases was not completely successful in all 10 cases. We observed potential information loss in 2 cases where (1) the study outcomes were assessed by comparing dates and times of a surgical procedure and (2) a device study that required dates and times of certain clinical findings. The PROCEDURE and DEVICE classes of the OMOP CDM stored only date information, thus precise comparison was not deemed possible. The metadata annotated for the 256 articles, the full results of CDM mapping, and data search use case simulations are available to download from https://idash-data.ucsd.edu/download/folder/4825/AMIA2016.zip.
Discussion
We attempted to represent the data contents reported in 256 nursing articles on health studies using the OMOP CDM version 5. Our goal was to start understanding the scope of the representation supported by the OMOP CDM, a major information model standard, with regards to content areas not related to conventional comparative effectiveness research, using the data from nursing research. It is promising that the majority of the clinical contents used in nursing research were well represented in the OMOP CDM, especially through the Standardized Clinical Data domain. However, it is our conclusion that the OBSERVATION and the MEASUREMENT classes had to be somewhat abused in this mapping as a place to hold many non-clinical and/or unusual clinical findings and assessment items.
Representing socio-economic and demographic information of patients, family members, and healthcare providers
PERSON and PROVIDER are two classes of the OMOP CDM that hold information on human subjects in the healthcare domain. The former is dedicated to describing patients, and the latter describes healthcare providers. Family dynamics is expected to have a significant impact on patient outcomes therefore it is an important research topic in nursing. Many articles reviewed in our study reported information about family members, especially those who play the role of a main care taker. We mapped those items into the OBSERVATION class, although this class was designed to capture the clinical facts not represented in other concept classes in the OMOP CDM. Similarly, many socio-economic items (e.g., marital status, education level, employment status, etc.) about patients had to be mapped to the OBSERVATION class since attributes in the PERSON class are limited to key “unchangeable” demographics such as age, gender, race, and ethnicity. Similar challenges were observed with the socio-demographic information about healthcare provider. We did not map these items to OBSERVATION, as the latter is to capture “any clinical facts about a patient obtained in the context of examination, questioning, or a procedure”[17]. These additional socio-demographic items on healthcare providers seem to be potential areas for expansion to fully support observational studies in nursing.
Representing patient reported outcomes
A third of the articles reviewed in this study reported assessment of patient status using standardized and/or custom-built assessment scales and survey questionnaires. Some of these assessment tools are completed by patients and some are by healthcare providers. Differentiating the source of information is important as it provides additional context to consider when studying patient outcomes. Of note, the CONDITION table of the PCORnet CDM captures this information through the condition_source attribute, whose structured value set contains patient reported medical history. It might be possible to infer the source of information for the outcomes measured with standardized scales that are widely used and well documented, although this practice still increases the risk for information loss by requiring additional steps for obtaining and associating the information. However, the source of information for outcomes measured with custom-built survey questionnaire is still highly likely be lost.
Representing nursing specific intervention concepts
Many articles we reviewed reported the effectiveness of specific nursing interventions, which often include new care protocols and complementary therapies. As recognized by the leaders in nursing informatics as a high priority task in nursing informatics, having nursing specific data sufficiently represented in “big data” sets is important to promote the incorporation of nursing’s holistic approaches to patient care into data-driven knowledge generation [39,40]. Despite the relative small scale of concept mapping for nursing interventions pursued in this study, we found that the majority of complementary therapies are covered by SNOMED-CT. However, locally developed specific care protocols are challenging to represent, since they are built from a collection of nursing actions. Consequently, a care protocol name used to describe an intervention concept might be considered less informative than the listing of nursing actions. Although it seems somewhat convoluted, this challenge can be addressed by establishing a member relation between a set of nursing interventions and a specific care protocol through the FACT_RELATIONSHIP table. We will continue investigating this issue in future studies.
Nursing problems cover wide areas of health domains: from disease-oriented matters to emotional wellbeing. Nursing research thus also includes concepts there are not frequently used in other clinical studies. However, we found that the OMOP CDM provides sufficient representation capability, even for unusual nursing problem concepts through less restrictive concept classes like OBSERVATION and MEASUREMENT, along with CONDITION_OCCURRENCE.
Limitations and future directions
This study used data items reported in a small sample of nursing research articles. Therefore, its findings may not be generalized to the entire content domains of nursing research data. In addition, the OMOP CDM strives for a high level of standardization and hence all key concepts in the CDM classes are required to be encoded with a standardized vocabulary. This means that the class and attribute levels of association does not always result in successful representation of nursing research concepts, as information loss can occur due to the lack of a standardized concept code. Using the OMOP CDM as a hypothetical database model to simulate data search use cases is another limitation. Information loss can happen during the transformation of a local database model to the OMOP CDM, and our simulation approach might not have been robust enough to capture this type of loss.
Recognizing these limitations, along with potential over-use of certain concept classes as previously described, we plan to continue this work by expanding the scope and revisiting identifiably challenging cases. We also plan to seek collaboration with OHDSI to discuss and substantiate lessons learned in this study.
Conclusion
Nursing research targets wide ranges of topic areas that are not limited to diseases or medical treatments. Therefore, investigating the standardized representation of the data contents in published nursing research provides a unique opportunity to understand challenges and opportunities in representing various types of health data so that they can be found and reused. In this study, we explored the feasibility of representing nursing research data with the OMOP CDM, using the data items reported in 256 nursing articles. The OMOP CDM provided a good representation for the majority of data items but we also observed potential gaps that might lead to information loss. These gaps will be further investigated in a follow-up study. Lessons learned in this study will help inform metadata development efforts for the Data Discovery Index.
Acknowledgment
This project was supported in part by the grant 1U24AI117966 (NIH/BD2K).
References
- 1.Moses C, Celi LA, Marshall J. Pharmacovigilance: an active surveillance system to proactively identify risks for adverse events. Population health management [Internet] 2013 Jun.16(3):147–9. doi: 10.1089/pop.2012.0100. [cited 2016 Mar 6] Available from: http://www.ncbi.nlm.nih.gov/pubmed/23530466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badawi O, Brennan T, Celi LA, Feng M, Ghassemi M, Ippolito A, et al. Making big data useful for health care: a summary of the inaugural mit critical data conference. JMIR medical informatics [Internet] 2014 Jan.2(2):e22. doi: 10.2196/medinform.3447. [cited 2016 Mar 6] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4288071&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Safran C, Bloomrosen M, Hammond WE, Labkoff S, Markel-Fox S, Tang PC, et al. Toward a national framework for the secondary use of health data: an American Medical Informatics Association White Paper. Journal of the American Medical Informatics Association: JAMIA [Internet] Jan.14(1):1–9. doi: 10.1197/jamia.M2273. [cited 2016 Mar 6] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2329823&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns PB, Rohrich RJ, Chung KC. The levels of evidence and their role in evidence-based medicine. Plastic and reconstructive surgery [Internet] 2011 Jul.128(1):305–10. doi: 10.1097/PRS.0b013e318219c171. [cited 2015 Oct 15] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3124652&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Low YS, Caster O, Bergvall T, Fourches D, Zang X, Norén GN, et al. Cheminformatics-aided pharmacovigilance: application to Stevens-Johnson Syndrome. Journal of the American Medical Informatics Association: JAMIA [Internet]. The Oxford University Press. 2015 Oct. doi: 10.1093/jamia/ocv127. [cited 2016 Mar 5] ocv127 Available from: http://jamia.oxfordjournals.org/content/early/2015/10/23/jamia.ocv127.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCoy AB, Cox ZL, Neal EB, Waitman LR, Peterson NB, Bhave G, et al. Real-time pharmacy surveillance and clinical decision support to reduce adverse drug events in acute kidney injury: a randomized, controlled trial. Applied clinical informatics [Internet] 2012 Jan.3(2):221–38. doi: 10.4338/ACI-2012-03-RA-0009. [cited 2016 Mar 5] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3377180&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lund JL, Sturmer T, Sanoff HK. Comparative effectiveness of postoperative chemotherapy among older patients with non-metastatic rectal cancer treated with preoperative chemoradiotherapy. Journal of geriatric oncology [Internet] 2016 Feb. doi: 10.1016/j.jgo.2016.01.011. [cited 2016 Mar 5] Available from: http://www.ncbi.nlm.nih.gov/pubmed/26926829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Widdifield J, Moura CS, Wang Y, Abrahamowicz M, Paterson JM, Huang A, et al. The Longterm Effect of Early Intensive Treatment of Seniors with Rheumatoid Arthritis: A Comparison of 2 Population-based Cohort Studies on Time to Joint Replacement Surgery. The Journal of rheumatology [Internet] 2016 Feb. doi: 10.3899/jrheum.151156. [cited 2016 Mar 5] Available from: http://www.ncbi.nlm.nih.gov/pubmed/26879353. [DOI] [PubMed] [Google Scholar]
- 9.Anderson N, Abend A, Mandel A, Geraghty E, Gabriel D, Wynden R, et al. Implementation of a deidentified federated data network for population-based cohort discovery. Journal of the American Medical Informatics Association: JAMIA [Internet] 2012 Jun.19(e1):e60–7. doi: 10.1136/amiajnl-2011-000133. [cited 2016 Mar 6] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3392860&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Namjou B, Marsolo K, Lingren T, Ritchie MD, Verma SS, Cobb BL, et al. A GWAS Study on Liver Function Test Using eMERGE Network Participants. PloS one [Internet] 2015 Jan.10(9):e0138677. doi: 10.1371/journal.pone.0138677. [cited 2016 Mar 6] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4586138&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.BD2K Home Page | Data Science at NIH [Internet] [cited 2015 Sep 10]Available from: https://datascience.nih.gov/bd2k.
- 12.Margolis R, Derr L, Dunn M, Huerta M, Larkin J, Sheehan J, et al. The National Institutes of Health’s Big Data to Knowledge (BD2K) initiative: capitalizing on biomedical big data. Journal of the American Medical Informatics Association: JAMIA [Internet] Jan.21(6):957–8. doi: 10.1136/amiajnl-2014-002974. [cited 2015 Dec 9] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4215061&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.bioCADDIE | biomedical and healthCAre Data Discovery and Indexing Ecosystem [Internet] [cited 2015 May 31] Available from: https://biocaddie.org/
- 14.Ohno-machado L, Alter G, Fore I, Martone M, Sansone S-A, Xu H. bioCADDIE White Paper; 2015. [Google Scholar]
- 15.Sansone S-A, Rocca-Serra P, Gonzalez-Beltran A, Vardigan M. Members bioCADDIE W. Working Group 3: Metadata Specifications bioCADDIE [Internet]; 2015. [cited 2015 Sep 10] Available from: https://biocaddie.org/group/working-group/working-group-3-metadata-specifications. [Google Scholar]
- 16.Kim H, Marmor R, Singh S, Ohno-Machado L. Exploring clinical research data network metdata for phenotype dataset discovery; Workshop on Data Science, Learning and Applications to Biomedical and Health Sciences (DSLA-BHS2016); 2016. pp. 7–11. [Google Scholar]
- 17.Observational Medical Outcomes Partnership [Internet] [cited 2015 Nov 29] Available from: http://omop.org/
- 18.OHDSI | Observational Health Data Sciences and Informatics [Internet] [cited 2015 Nov 29] Available from: http://www.ohdsi.org/
- 19.Hripcsak G, Duke JD, Shah NH, Reich CG, Huser V, Schuemie MJ, et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. Studies in health technology and informatics [Internet] 2015 Jan.216:574–8. [cited 2015 Nov 4] Available from: http://www.ncbi.nlm.nih.gov/pubmed/26262116. [PMC free article] [PubMed] [Google Scholar]
- 20.Collins FS, Hudson KL, Briggs JP, Lauer MS. PCORnet: turning a dream into reality. Journal of the American Medical Informatics Association: JAMIA [Internet] Jan;21(4):576–7. doi: 10.1136/amiajnl-2014-002864. [cited 2015 Nov 29] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4078299&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.PCORnet, the National Patient-Centered Clinical Research Network - PCORnet [Internet] [cited 2015 Nov 29] Available from: http://www.pcornet.org/
- 22.Ohno-Machado L, Agha Z, Bell DS, Dahm L, Day ME, Doctor JN, et al. pSCANNER: patient-centered Scalable National Network for Effectiveness Research. Journal of the American Medical Informatics Association: JAMIA [Internet] Jan;21(4):621–6. doi: 10.1136/amiajnl-2014-002751. [cited 2015 May 31] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4078293&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The National Patient-Centered Clinical Research Network. PCORnet Common Data Model (CDM) Specification. 2015. Version 3.0.
- 24.Belenkaya R, Mirhaji P, Khayter M, Torok D, Khare R, Ong T, et al. Establishing Interoperability Standards between OMOP CDM v4, v5, and PCORnet CDM v1; OHDSI workshop; 2015. [Google Scholar]
- 25.Rijnbeek PR. Converting to a common data model: what is lost in translation?: Commentary on “fidelity assessment of a clinical practice research datalink conversion to the OMOP common data model.”. Drug safety [Internet] 2014 Nov;37(11):893–6. doi: 10.1007/s40264-014-0221-4. [cited 2015 Nov 30] Available from: http://www.ncbi.nlm.nih.gov/pubmed/25187018. [DOI] [PubMed] [Google Scholar]
- 26.Matcho A, Ryan P, Fife D, Reich C. Fidelity assessment of a clinical practice research datalink conversion to the OMOP common data model. Drug safety [Internet] 2014 Nov.37(11):945–59. doi: 10.1007/s40264-014-0214-3. [cited 2015 Nov 22] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4206771&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makadia R, Ryan PB. Transforming the Premier Perspective Hospital Database into the Observational Medical Outcomes Partnership (OMOP) Common Data Model; EGEMS; (Washington, DC) [Internet]. 2014. Jan. 1110 pp. [cited 2016 Feb 16] Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4371500&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogunyemi OI, Meeker D, Kim H-E, Ashish N, Farzaneh S, Boxwala A. Identifying appropriate reference data models for comparative effectiveness research (CER) studies based on data from clinical information systems. Medical care [Internet] 2013 Aug.51(8) 3:S45–52. doi: 10.1097/MLR.0b013e31829b1e0b. [cited 2015 Nov 30] Available from: http://www.ncbi.nlm.nih.gov/pubmed/23774519. [DOI] [PubMed] [Google Scholar]
- 29.Voss EA, Makadia R, Matcho A, Ma Q, Knoll C, Schuemie M, et al. Feasibility and utility of applications of the common data model to multiple, disparate observational health databases. Journal of the American Medical Informatics Association: JAMIA [Internet] 2015 May;22(3):553–64. doi: 10.1093/jamia/ocu023. [cited 2015 Nov 3] Available from: http://www.ncbi.nlm.nih.gov/pubmed/25670757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collins FS, Varmus H. A New Initiative on Precision Medicine. The New England journal of medicine [Internet] 2015 Jan.372(9):793–5. doi: 10.1056/NEJMp1500523. [cited 2015 Feb 6] Available from: http://www.ncbi.nlm.nih.gov/pubmed/25635347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hripcsak G, Forrest CB, Brennan PF, Stead WW. Informatics to support the IOM social and behavioral domains and measures. Journal of the American Medical Informatics Association: JAMIA [Internet] 2015 Jul.22(4):921–4. doi: 10.1093/jamia/ocv035. [cited 2016 Feb 16] Available from: http://www.ncbi.nlm.nih.gov/pubmed/25914098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vittorini P, Tarquinio A, di Orio F. XML technologies for the Omaha System: a data model, a Java tool and several case studies supporting home healthcare. Computer methods and programs in biomedicine [Internet] 2009 Mar.93(3):297–312. doi: 10.1016/j.cmpb.2008.10.009. [cited 2016 Mar 6] http://www.ncbi.nlm.nih.gov/pubmed/19070934. [DOI] [PubMed] [Google Scholar]
- 33.Harris MR, Langford LH, Miller H, Hook M, Dykes PC, Matney SA. Harmonizing and extending standards from a domain-specific and bottom-up approach: an example from development through use in clinical applications. Journal of the American Medical Informatics Association: JAMIA [Internet] 2015 May;22(3):545–52. doi: 10.1093/jamia/ocu020. [cited 2016 Mar 6] Available from: http://www.ncbi.nlm.nih.gov/pubmed/25670750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rabanales Sotos J, López Gonzalez Á, Párraga Martínez I, Campos Rosa M, Simarro Herraez MJ, López-Torres Hidalgo J. Prevalence of hazardous drinking among nursing students. Journal of advanced nursing [Internet] 2015 Mar.71(3):581–90. doi: 10.1111/jan.12548. [cited 2016 Mar 4] Available from: http://www.ncbi.nlm.nih.gov/pubmed/25363658. [DOI] [PubMed] [Google Scholar]
- 35.Lucchini A, De Felippis C, Elli S, Schifano L, Rolla F, Pegoraro F, et al. Nursing Activities Score (NAS): 5 years of experience in the intensive care units of an Italian University hospital. Intensive & critical care nursing: the official journal of the British Association of Critical Care Nurses [Internet] 2014 Jun.30(3):152–8. doi: 10.1016/j.iccn.2013.10.004. [cited 2016 Mar 4] Available from: http://www.ncbi.nlm.nih.gov/pubmed/24370275. [DOI] [PubMed] [Google Scholar]
- 36.Fisher DA, Seetoh T, Oh May-Lin H, Viswanathan S, Toh Y, Yin WC, et al. Automated measures of hand hygiene compliance among healthcare workers using ultrasound: validation and a randomized controlled trial; Infection control and hospital epidemiology [Internet]; 2013. Sep. pp. 919–28. [cited 2016 Mar 4] Available from: http://www.ncbi.nlm.nih.gov/pubmed/23917905. [DOI] [PubMed] [Google Scholar]
- 37.Calhoun AW, Boone MC, Dauer AK, Campbell DR, Montgomery VL. Using simulation to investigate the impact of hours worked on task performance in an intensive care unit. American journal of critical care: an official publication, American Association of Critical-Care Nurses [Internet] 2014 Sep.23(5):387–95. doi: 10.4037/ajcc2014756. [cited 2016 Mar 4] Available from: http://www.ncbi.nlm.nih.gov/pubmed/25179034. [DOI] [PubMed] [Google Scholar]
- 38.Moreno-Casbas MT, Ruzafa-Martinez M, Rol MA, Madrid JA, Serrano Pinto A, González-María E, et al. Sleepiness in Spanish nursing staff--influence of chronotype and care unit in circadian rhythm impairment: research protocol. Journal of advanced nursing [Internet] 2014 Jan;70(1):211–9. doi: 10.1111/jan.12200. [cited 2016 Mar 4] Available from: http://www.ncbi.nlm.nih.gov/pubmed/23834526. [DOI] [PubMed] [Google Scholar]
- 39.Brennan PF, Bakken S. Nursing Needs Big Data and Big Data Needs Nursing. Journal of nursing scholarship: an official publication of Sigma Theta Tau International Honor Society of Nursing / Sigma Theta Tau [Internet] 2015 Aug.47(5):477–84. doi: 10.1111/jnu.12159. [cited 2015 Sep 1] http://www.ncbi.nlm.nih.gov/pubmed/26287646. [DOI] [PubMed] [Google Scholar]
- 40.Westra BL, Clancy TR, Sensmeier J, Warren JJ, Weaver C, Delaney CW. Nursing Knowledge: Big Data Science-Implications for Nurse Leaders. Nursing administration quarterly [Internet] Jan.39(4):304–10. doi: 10.1097/NAQ.0000000000000130. [cited 2015 Sep 9] Available from: http://www.ncbi.nlm.nih.gov/pubmed/26340241. [DOI] [PubMed] [Google Scholar]