Abstract
Abbreviation disambiguation in clinical texts is a problem handled well by fully supervised machine learning methods. Acquiring training data, however, is expensive and would be impractical for large numbers of abbreviations in specialized corpora. An alternative is a semi-supervised approach, in which training data are automatically generated by substituting long forms in natural text with their corresponding abbreviations. Most prior implementations of this method either focus on very few abbreviations or do not test on real-world data. We present a realistic use case by testing several semi-supervised classification algorithms on a large hand-annotated medical record of occurrences of 74 ambiguous abbreviations. Despite notable differences between training and test corpora, classifiers achieve up to 90% accuracy. Our tests demonstrate that semi-supervised abbreviation disambiguation is a viable and extensible option for medical NLP systems.
Introduction
The frequent use of abbreviations in clinical texts is a major challenge for natural language processing (NLP) systems. Medical abbreviations are especially challenging because they tend to be highly ambiguous: a 2001 survey reports that nearly a third of shorter abbreviations catalogued in the Unified Medical Language System (UMLS) are ambiguous;1 a later study reveals that even the UMLS sense inventory provides spotty coverage of all possible abbreviation senses.2 The problem is even worse in clinical notes created for patient care, as opposed to other biomedical texts mostly derived from peer-reviewed literature, due to issues ranging from higher word ambiguity3 to mistakes in spelling and dictation.4 Even detecting which words are abbreviations is not a trivial task, and modern clinical NLP systems have much room for improvement in this area.5
Resolving ambiguities in clinical texts is a major concern for improving medical information retrieval outcomes.6,7 As such, an active area of research in medical NLP is in the normalization of abbreviations.8 For ambiguous abbreviations, this is generally treated as a special case of the word sense disambiguation (WSD) problem: determining the sense of a single string that may have multiple distinct semantic interpretations.9,10 Supervised machine learning approaches to abbreviation disambiguation based on WSD techniques have been generally successful.11,12 However, most prior studies in this domain have evaluated only a handful of abbreviations; the generalizability of these results to more realistic use cases, with hundreds or thousands of abbreviations, is limited.
Fully supervised methods are also subject to the limitation that they require training data that is slow and expensive to obtain. Furthermore, to keep up with the rapid and ongoing proliferation of clinical abbreviations, such data would require constant maintenance. To obviate the need for labeled data, some researchers have proposed unsupervised13 or knowledge-based14 strategies to the disambiguation problem. Still, published research in this area generally focuses on fewer than 20 abbreviations.
Another possibility is a semi-supervised (or “distantly supervised”) approach, which requires some attention to data collection but not nearly the commitment necessary for fully hand-annotated data. Normalizing abbreviations differs from other WSD problems in that the senses (i.e., long forms) have distinct string realizations. This property can be exploited to generate examples of virtual occurrences of an abbreviation by targeting its various long forms in a search of unlabeled text. This general approach has been developed for biomedical texts15 and clinical notes,10,16 with reported accuracy approaching 90%. Stevenson et al. apply a similar corpus generation process to Medline abstracts that have abbreviations co-occurring locally with their long forms.11 (Note that these convenient cooccurrences are rare for clinical notes.) They employ a battery of features to achieve 99% accuracy for 20 abbreviations.
The main objective of the present study was to conduct a large-scale test of the semi-supervised method just described. Ours differs from prior research in two important ways. First, training and test data are drawn from different corpora. This point is key, as training and testing on homogeneous data is not a realistic evaluation of an NLP application for many cases—versatile clinical NLP systems should be useful for a wide variety of texts, not just those similar to data on which its models were trained. Other studies on semi-supervised biomedical acronym normalization which train and test on different corpora are by Pakhomov et al., who disambiguate 8 acronyms with up to 67.8% accuracy,16 and Xu et al., who disambiguate 13 abbreviations from typed hospital admission notes with up to 87.5% accuracy.12
Second, we test a wider range of abbreviations than has been considered previously: 74 ambiguous abbreviations frequent in clinical texts but not specific to any particular field of medicine. The only other published studies that test a comparable number all focus on the same abbreviations that we do (or a subset of them).17–19 These studies, however, rely on fully supervised cross-validation on a single corpus rather than separately generated training and test sets. By applying a semi-supervised approach to a wide scope of abbreviations in a more realistic test case, we present the most extensible method to date for disambiguating abbreviations in clinical texts. (The test set we use actually comprises 74 initialisms, commonly referred to in the literature as “acronyms,” which are a sub-type of abbreviation that involves the concatenation of initial letters of words in a phrase. We continue to use the term “abbreviation” because our methods should be equally applicable to acronyms and to other abbreviations.)
The semi-supervised method is appealing because it should maintain the high accuracy and efficiency of fully supervised approaches without requiring expensive human tagging of corpora. It does depend, however, upon an assumption that the distributions of an abbreviation and of its long forms are similar. This assumption seems intuitively well supported, but at the same time it is easy to imagine reasons why distributions might differ— abbreviated forms may tend to occur more often in notes more reliant on abbreviations, or a writer may consciously avoid abbreviations in contexts where they would be too opaque or unusual.
That said, it is not evident a priori that these weaknesses in the assumption lead to unsatisfactory results. The present study is an empirical evaluation of the semi-supervised method, simulating a real-world use case: training a normalization system on machine-annotated data and testing it on natural occurrences of a wide range of abbreviations.
Methods
Corpora and sense inventory
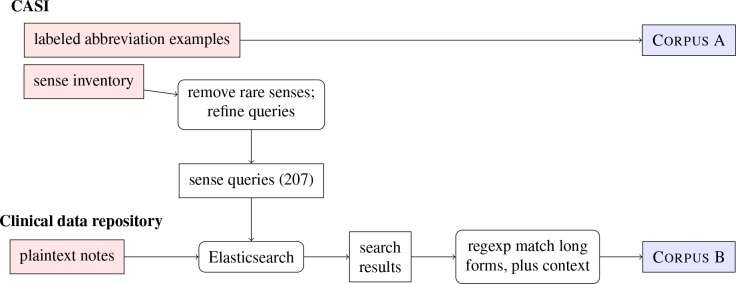
Two corpora were used in this study. The first was taken from the Clinical Abbreviation Sense Inventory.20 This publicly available data set (“CASI”) lists 440 common clinical abbreviations along with their long forms, mappings to medical concepts, and other information. Most of these abbreviations were found to have a dominant sense accounting for 95% or more of 500 randomly sampled occurrences; for those 74 that do not, CASI provides anonymized plaintext data and manually annotated senses for 500 samples of each (37,000 total). These samples, minus the 223 marked as having ‘unsure sense’, constitute Corpus A.
Corpus B was built by querying a large clinical data repository in the Fairview Health Services system (about 90 million notes) for long forms of these abbreviations using Elasticsearch. (All notes in the health record were searched, so this corpus contains results from many different types of documents across numerous specialties. We used definitions of notes as based off of the HL7-LOINC document ontology and as catalogued in our data repository.) In most cases, unmodified long forms themselves could serve as queries. A few, however, required slight adjustments. For example, searching for the long form ‘computed tomographic angiography’ returned very few results; the slightly modified (case-insensitive) query ‘ct angiography’ returned many more, while still being unambiguous. In another case, the expansion ‘gutta’ (for ‘GT’) was rare in the corpus (and ambiguous with a physician’s name), while the semantically equivalent ‘gtts’ was more common. Overall, we kept any modification of the searches to a minimum; queries were only altered if an informal examination of the search results showed them to be scant, overly repetitive, or obviously incorrect.
827,647 total notes were returned by queries across 207 different senses, with a maximum of 5,000 notes per sense. Samples were taken from the body text of these notes by identifying the long form in them and matching (greedily) any appearances of that form both preceded and followed by 40 to 100 characters. This step effectively excluded any examples with too little context, such as those from very short notes. Up to 8,000 samples were generated for each sense (allowing for multiple tokens of a sense per sample). In all, a total of 857,724 annotated virtual abbreviations were generated.
The sense inventory used to collect documents for Corpus B was built from CASI. 357 total senses are represented in the data set, although 150 are exceptionally rare (occurring as fewer than 5 of its abbreviation’s 500 samples) and were excluded; in all, these account for 0.6% of the total data. (Note that examples of these rare senses were retained in the test set and thus contributed to the errors of a model not trained on them.) The procedure for generating both corpora is visualized in Figure 1. Histograms showing the number of senses per abbreviation and the number of training samples per sense in Corpus B are given in Figure 2a and 2b, respectively.
Figure 1.
The procedure for generating the two corpora used in this study (far right) from existing data (far left). Rounded boxes represent operations; angular boxes represent data.
Figure 2a.
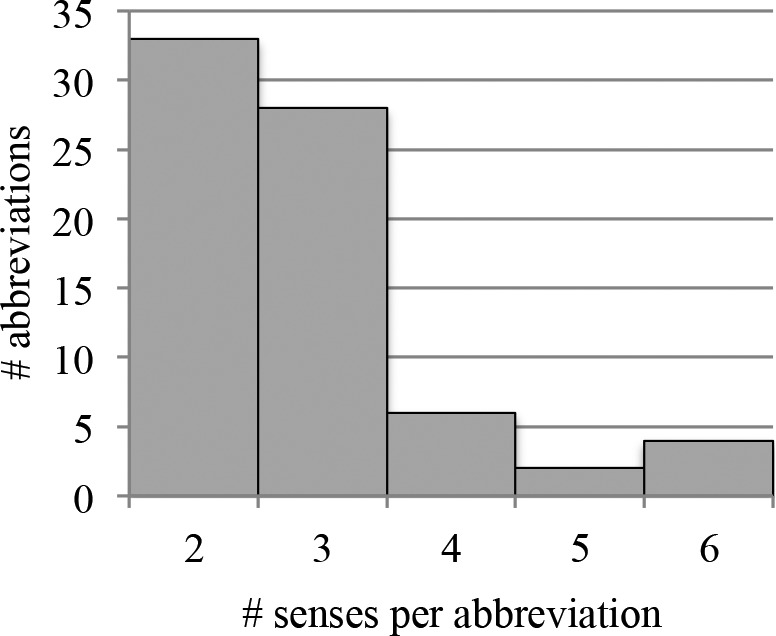
Distribution of number of senses per abbreviation, excluding very rare senses.
Figure 2b.
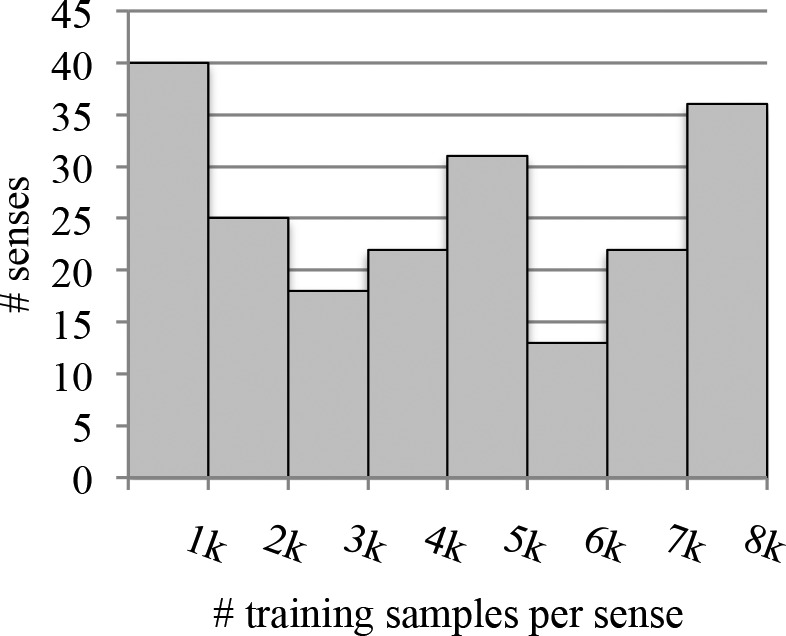
Distribution of number of virtual abbreviation tokens generated per sense (Corpus B).
Word vectors
Feature vectors for each instance of an abbreviation were calculated as simple co-occurrence counts within a window of text, a “bag-of-words” representation. Tokens were lemmatized, case was ignored, and numerals were collapsed into broad categories: single-digit integers, multi-digit integers, and decimals. By these criteria, Corpus A has 13,877 unique words, and Corpus B 72,514. (Note that Corpus A has been anonymized with generic strings whereas B has not, so names, addresses, etc. contribute to the latter’s word count.)
Although word order was not taken into account explicitly, counts were weighted to increase the contribution of words in closer context. Weights were computed by a sigmoid function W(d) that decreases with the context token’s (always positive) distance d in words from the token of interest:
where r is the number of words away at which weight drops to half and α controls the steepness of the curve. Various parameters for the rate and point of falloff were tested; good values were found to be around r = 9 with a shallow falloff of α = 0.3. Words weighted at less than 0.25 (greater than 12 words away for these values) were not counted at all. This weighting maintains a minor effect of word order and was found to slightly improve classification accuracy overall. Beyond about 9 or 10 words, a larger window does not generally improve accuracy for this kind of task.17
Researchers have suggested various other features to enhance supervised classification—salient N-grams,11 knowledge sources,14 neural word embeddings.19 The central question here, however, is not how best to tailor features to maximize performance; rather, we are concerned with whether the machine-annotated data can effectively serve as a training corpus given minimal assumptions and adjustments. Bag-of-words features are simple, general, and quite reliable, and they should give a fair indication of whether it is worth the effort to further tune models and features for these data.
Machine learning algorithms
Several well-known classification algorithms were implemented, all using the features described above:
-
-
Naïve Bayes (NB): A single Gaussian was fitted for each feature across all examples of a given class (sense). Class priors were not considered, as these would favor more frequent classes in the training data, and the relative frequencies for long forms may not be representative of sense frequencies for abbreviations—recall the earlier discussion that some senses are more likely to be abbreviated than others. Hypotheses at test time were chosen by maximizing likelihood, with consideration only of the non-zero entries in the test vector. Given the data’s high dimensionality and natural-language origins, many probabilities at test time come out at or near zero, so natural log probabilities were kept to a minimum of -100 to prevent a single unusual or unseen word from dooming a candidate altogether. A separate model was trained for each of the 74 acronyms.
-
-
Multinomial logistic regression (LR): A classifier was trained for every abbreviation. The intercept term typical of logistic regression was not used, for the same reason class priors were not used for NB. Models were trained by 100 iterations of gradient descent, which usually converged in 3 to 10 iterations. Regularization generally did not improve results and was not used in the reported tests.
-
-
Support vector machine (SVM): This classifier was identical to LR but with a standard hinge loss function (ε = 1) rather than log loss.
-
-
Cosine similarity (COS): Test items were classified by maximizing the cosine of the angle between the test context vector and a normalized vector representing the centroid of all training items of each sense. Two other adjustments were made to these centroid sense vectors before normalization. First, all values were compressed via a square root, which effectively favors less common words and therefore diverse lexical contexts (note that this would have little effect for other classifiers, which do not average training examples). Second, words in training and testing context vectors were weighted by their inverse document frequency (IDF). The cosine similarity metric is not a discriminative or maximum-likelihood strategy, so it is essential to reduce the impact of words with low predictive power.
Other features: hyperdimensional indexing
Another method that has received some recent attention has been the use of high-dimensional vectors to represent words. Rather than unitary representation as a single dimension in a context vector, word vectors themselves are “hyperdimensional,” having on the order of a few thousand dimensions. (Note that word vectors are actually of lower dimensionality than the “one-hot” vectors used for a traditional word vector space, but that these representations depend on encoding several of these dimensions, whereas the one-hot vectors are predictable from the vocabulary.) Context vectors are represented by “bundling” word vectors, typically through a sum or similar operation. Word vectors are not orthogonal as unitary word vectors in a traditional vector space are, but are all nearly orthogonal due to their high dimensionality. Context vectors are equidimensional with word vectors, so they are actually shorter (but less sparse) than the context vectors considered for the other classifiers. Two hyperdimensional approaches were tested:
-
-
Random indexing (RI): Each unique word is represented as a ternary vector of 1,800 elements, with a random four values set to 1, another random four to −1, and the rest to 0. These are the same parameters described by Kanerva et al.21 Other hyperparameter values were explored but did not significantly improve classification. Context vectors are created by summing word vectors within the context window (9 tokens on either side) subject to the same IDF weighting used for the vector space model. Senses were chosen by the same similarity criterion as the COS classifier described above. In the clinical domain, RI has been applied effectively towards capturing distributional similarity between long forms and their abbreviations.22
-
-
Binary spatter code (BSC): Each unique word is represented by a 10,000-dimensional binary vector, with all values randomly assigned. During training, context vectors are calculated not for the senses themselves, but for all words within 9 tokens of an abbreviation, by summing together vectors that are the “product” (bitwise XOR) of an abbreviation and its labeled sense. Word frequency is accounted for in these sums by entropy weighting, and the final vector chosen by voting on the weighted counts of ones and zeros in each dimension. For testing, contexts are “divided” (XOR again) by the abbreviation vector to recover a context vector correlated with the most likely sense. The implementation hews very closely to the description given by Berster et al.;23 see also Kanerva24 for a theoretical discussion and Moon et al. for its suitability to abbreviation normalization.
Test conditions
Recall that this study considers two corpora derived from clinical texts—Corpus A, smaller and hand annotated; and Corpus B, larger and automatically annotated. We performed tests with each as the training set and each as the test set. Whenever a single corpus was used for both training and test, 10-fold cross-validation was performed.
The original aim of the study was to evaluate an abbreviation normalizer that is not dependent on fully supervised data. Corpus B can be created cheaply, while A contains actual occurrences of abbreviations; thus, training on B and testing on A is the intended use case and gives the best indication of performance on real-world clinical data.
To mitigate problems related to overdeveloping to this use case, we halved Corpus A into development and validation sets. Examples were split by stratified random sampling: for every sense of every abbreviation, half of the examples selected at random went to each set, with the validation set receiving the remainder for odd counts. All refinements of the models were performed according to performance on the development set (18,278 samples total), and all statistics reported in this paper are from the validation set (18,499 samples total). For conditions other than training on B and testing on A, this split was ignored and Corpus A was used in its entirety.
We have made code for the experiments in this study available online (https://github.com/gpfinley/towards_comprehensive). Corpus A is freely available; however, Corpus B contains protected health information and cannot be publicly shared.
Results & discussion
Classification accuracy
The results for classifiers tested on Corpus A are reported in Table 1. Scores reported in the left column are results of 10-fold cross-validation, applied in a classically supervised manner to Corpus A. The right column of the table represents the intended use case of the semi-supervised method.
Table 1.
Classification accuracy for classifiers tested on the hand-annotated Corpus A. Scores in the left column are averages of 10-fold cross-validation. Baseline is the average majority sense score for all abbreviations.
| CV on A | Train on B, test on A | |
|---|---|---|
| NB | .949 | .854 |
| LR | .966 | .880 |
| SVM | .964 | .887 |
| COS | .961 | .900 |
| RI | .945 | .817 |
| BSC | .935 | .761 |
| baseline | .735 | |
| chance | .398 | |
Baseline accuracy for tests on Corpus A is defined as the majority sense—the score obtained by simply guessing the most common sense for each abbreviation. Recall that no classifiers tested in this study explicitly take sense frequency into account.
The results of cross-validation on Corpus A confirm that traditionally supervised methods are highly accurate. Hyperdimensional approaches were slightly less accurate but still reasonable. For the semi-supervised case, however, all classifiers suffered somewhat. The drop in performance, as compared to the fully supervised crossvalidation results, was modest for classifiers operating on word count vectors, with COS losing only 6.1% accuracy. Recall the assumption that abbreviations and their long forms share the same distribution in text. Given that this assumption is certainly not guaranteed, it can be said that LR, SVM, and especially COS perform fairly well to any variation introduced by training on long forms and testing on short forms. Corpus B was built with very light supervision, so other sources of noise probably also contribute to the drop in accuracy.
The hyperdimensional classifiers, on the other hand, showed a sharper drop in performance, up to 17.4%. Lexical differences between the two corpora may be responsible: high-dimensional word vectors are not perfectly orthogonal, causing some overlap in context vectors that would be difficult to account for if some words are present in one corpus but not another. Alternatively, it may be that hyperdimensional context vectors are especially sensitive to other differences between corpora that are unimportant for abbreviation disambiguation.
Whatever the explanation, it is clear that the RI and BSC algorithms suffer more from using machine-annotated training data than those classifiers based on the vector space model. Hyperdimensional approaches certainly have some advantages for representing lexical semantics in NLP, such as compactness and flexibility,21,25 but they appear ill suited to the task here (especially because maintaining context vectors for abbreviation senses requires less storage than for an entire vocabulary). This result is especially noteworthy given the excellent results reported for BSC in fully supervised abbreviation disambiguation by Moon et al.18
The same classifiers were also evaluated using Corpus B as a test set, with results shown in Table 2. Note that this is a highly artificial case, as the notes used to derive Corpus B contain the long forms rather than ambiguous abbreviations. Note also that a majority-sense baseline is less meaningful for Corpus B (it would be 54.6%), which targeted the long forms directly rather than sampling actual occurrences of the abbreviation, and thus is not thought to be representative of actual sense frequency.
Table 2.
Classification accuracy for classifiers tested on the hand-annotated Corpus B. Scores in the right column are averages of 10-fold cross-validation.
| Train on A, test on B | CV on B | |
|---|---|---|
| NB | .717 | .959 |
| LR | .742 | .990 |
| SVM | .748 | .988 |
| COS | .804 | .982 |
| RI | .762 | .934 |
| BSC | .610 | .948 |
| chance | .349 | |
Fully supervised cross-validation was even better for Corpus B than Corpus A, a fact which may be due to the sheer size of Corpus B, the frequent use of boilerplate language in the source notes, or even altogether redundant notes, which were not screened out when searching the database.
In the reverse case, training on Corpus A and testing on B, no classifier performed exceptionally well (the COS classifier achieves the highest accuracy with 80.4%). Such a condition—normalizing virtual abbreviations in texts originally lacking them—is unlikely to arise in practical use. However, it may speak to fundamental differences between the corpora. While it is true that Corpus B is much larger, it is unlikely that this discrepancy alone leads to poor performance, given the finding by Moon et al.17 that a training set a fraction of the size of Corpus A can still give decent results for abbreviation normalization. A more likely explanation is that Corpus A is simply less general and diversified than B: data in Corpus A are primarily verbally dictated and transcribed, whereas Corpus B contains notes created through dictation as well as a variety of combinations of preformed templates, macros, typing, and voice recognition software. Variation inherent to Corpus A is present in B, but not vice-versa. (Note also that the impact of domain transfer may be overstated in these results because we have not controlled for the differences between hand- and machine-labeled annotations; i.e., instances of abbreviations in Corpus A are not found using semi-supervised methods.)
In terms of practical application, the performance discrepancy between the two experiments emphasizes the importance of using well-diversified data to train robust WSD systems. The semi-supervised approach, in conjunction with access to a large database of clinical notes, handles this with ease.
Error analysis
We also investigated several of the abbreviations that were responsible for the accuracy lost between the fully supervised and semi-supervised conditions (i.e., the two columns of Table 1). Our focus is specifically on the COS classifier, as it had the best performance. All abbreviations with an accuracy differential between the two tasks of at least ten percentage points are shown in Table 3.
Table 3.
Differences in the accuracy of the COS classifier for the semi- and fully supervised cases. Only abbreviations with a performance differential of at least 10 percentage points are shown. Sense counts exclude very rare senses.
| abbrev. | senses | B/A acc. | A/A acc. | diff. |
|---|---|---|---|---|
| MOM | 2 | .425 | .998 | .573 |
| DT | 3 | .673 | .956 | .283 |
| T1 | 3 | .672 | .944 | .272 |
| PAC | 4 | .726 | .962 | .236 |
| PCP | 4 | .713 | .944 | .231 |
| AB | 3 | .760 | .958 | .198 |
| NAD | 2 | .745 | .942 | .197 |
| RT | 3 | .773 | .962 | .189 |
| PA | 4 | .777 | .948 | .171 |
| DC | 4 | .755 | .904 | .146 |
| IT | 6 | .780 | .905 | .125 |
| ER | 3 | .856 | .974 | .118 |
| GT | 3 | .825 | .940 | .115 |
| CD4 | 3 | .865 | .972 | .107 |
| RA | 3 | .855 | .962 | .107 |
| BMP | 3 | .761 | .866 | .105 |
For most of these abbreviations, it is not immediately evident why the semi-supervised classifier performs poorly. For a handful, though, some educated guesses can be made:
-
-
MOM: Nearly all of the errors on this abbreviation were misidentifying the sense ‘multiples of median’ as ‘milk of magnesia’. The former long form was difficult to target in the notes corpus, as it is usually abbreviated, so there were few training examples for what happened to be the most common sense of ‘MOM’.
-
-
T1: ‘thoracic (level) 1’ was frequently misidentified as ‘T1 (MRI)’, which was difficult to target as a long form. We ultimately designed the query for the latter to match ‘T1’ in close context with ‘MRI’, and it appears to have erroneously matched several uses that refer to the first thoracic vertebra in an MRI context.
-
-
PAC: ‘physician assistant certification’ was often missed. This sense almost always follows a name, and a key difference between Corpus A and Corpus B is that the former has had all names collapsed as part of deidentification.
-
-
IT: The pronoun ‘it’ was frequently identified as an abbreviation. The contexts for ‘it’ are exceptionally diverse, so it may have been difficult to motivate ‘it’ over other hypotheses. These errors could easily be sidestepped if tokens were tagged for part of speech, as most medical NLP systems do, by assuming that pronouns are not abbreviations.
It is fair to say that certain senses of these abbreviations are represented in Corpus B differently from in A. Some differences arise from terms that rarely appear as long forms (e.g., ‘multiples of median’), whereas others may reflect difficulties inherent to the semi-supervised approach (‘T1’, ‘PAC’).
Some errors, particularly in this latter category, could be substantially reduced with more attention paid to the searches used to retrieve data. The intent for this project was specifically not to do so; the goal was to determine how well minimally managed data collection would work to train machine learning models. The results obtained here for the semi-supervised case should be considered a conservative estimate of performance, and more systematic attention to queries will likely increase accuracy further.
Extending to other abbreviations
Our test set encompasses hundreds of senses from 74 unique abbreviations. If a system were required to only account for these 74, then a fully supervised approach might be the best recommendation. Of course, this is but a fraction of known medical abbreviations. Though our coverage is still incomplete, the semi-supervised methods that we validate here are extensible to other abbreviations and their known senses. More comprehensive sense inventories could be built either from existing medical knowledge sources, such as the SPECIALIST Lexicon,26 or by using unsupervised machine learning methods, such as the clustering approach proposed by Xu et al.13
Evaluating the accuracy beyond these 74 is difficult without acquiring more hand-annotated test data. Nevertheless, there is no reason to believe that other abbreviations should be less suitable for the semi-supervised approach. Our tests are also rather conservative, in both the minimal treatment of database queries and the use of very general machine learning methods.
Generally speaking, better performance on primary NLP tasks such as WSD should improve outcomes in secondary tasks. While an in-depth discussion of WSD applications in clinical NLP is beyond the scope of the present paper, we would like to note briefly that the ability to identify context-appropriate senses of ambiguous medical terms, abbreviations in particular, is fundamental to several higher-level NLP and information retrieval tasks, including accurate concept mapping to standardized vocabularies and conceptual indexing of electronic health records. Fully supervised systems are feasible only for a small and static set of ambiguous terms; however, clinical notes are “living” documents whose authors often do not follow conventions and abbreviate medical terms ad libitum for convenience and speed of documentation. Unsupervised or semi-supervised systems of the kind described in this paper offer the potential to keep up with these new ambiguous patterns dynamically introduced into clinical documentation. It may even be of interest to have models configured to text in specific clinical domains, especially if there are idiosyncrasies in documentation style between providers or specialties.
Comparison to other published results
A subset of the data from Corpus A was also used for training and testing by Moon et al.17 and Moon et al.;18 the former tested Na¨ve Bayes and SVM classifiers, the latter a hyperdimensional binary spatter approach. Figures reported previously in the current study are on all 74 abbreviations of the published data set. When testing on only the 50 abbreviations considered in the other two papers, our accuracy results are extremely similar: marginally lower for BSC (our 92.9% versus their 93.5%) and higher for NB and SVM (our 94.2% and 96.2% versus their 93.7% and 93.9%).
Note that the above comparison is only for fully supervised methods. Comparisons to other tests of semi-supervised methods are difficult to make because most other studies draw training and test items from the same corpus, which simplifies the problem considerably. The closest comparison is with Xu et al.,12 who investigate 13 abbreviations that have at least one disease sense. They draw training data from dictated discharge summaries and test data from typed hospital admission notes. Their methods for generating the corpora are nearly identical to ours: training samples are built by targeting long forms directly in a text search, while test samples are manually annotated. As such, some performance comparisons can be reasonably made. Our peak performance at 90.0%, using only bag-of- words features (with some positional weighting), slightly exceeds theirs at 87.5%, which uses a combination of bag- of-words features, positional features, section headings, and sense frequency information. Xu et al.’s reliance on sense frequency in particular may introduce bias because frequencies are calculated off of the test corpus itself, albeit with unsupervised methods. (We use sense frequency only initially to determine if any senses are vanishingly rare, and not as a feature for classification.) Xu et al. report accuracy of 79.2% when not using sense frequency.
Limitations
There are a few limitations to this study that should be considered. First, we do rely somewhat on sense frequency in Corpus A to exclude rare senses (less than 1% of a given abbreviation’s occurrences) from consideration. Including these senses would have required the acquisition of much more training data and would probably have had some negative impact on accuracy. Some degree of medical expertise would be helpful to determine which senses are very rare.
Second, several of the database queries that we perform do require some attention and modification. The amount of labor is significantly less than that required for fully supervised labeling of examples, and it does not scale with the size of the data set. Nevertheless, queries optimized for one database and search protocol might not transfer well to others.
Finally, implementing the semi-supervised method with clinical notes as training data requires access to an appropriately large database, which many members of the larger research community do not have. Sharing models trained on clinical note data, such as the ones in this study, is difficult due to confidentiality concerns and would require removal of potentially identifying information.
Future work
Though our results show some success with semi-supervised methods, a more thorough investigation should be conducted to quantify the actual distributional similarity between abbreviations and their long forms and to understand how the assumption of similarity affects the results. Our own preliminary investigation has found that this assumption generally holds but is much more valid for some abbreviations than for others. Distributional similarity may depend on a number of factors—such as text domain, writing style, and accidental homonymy with other terms—and may interact with NLP task accuracy in interesting ways.
Conclusion
In this study we demonstrated the suitability of a semi-supervised approach for disambiguating abbreviations in clinical text on a large scale. Our tests covered more data and more distinct abbreviations than have been tested previously by this method, and our test conditions are a better representation of real-world problems. Generating machine-annotated training data requires little human supervision and can be easily extended to other abbreviations. Applying this method has the potential to significantly improve information retrieval outcomes for clinical text.
Acknowledgments
The National Institutes of Health through the National Library of Medicine (R01LM011364 and R01GM102282), Clinical and Translational Science Award (8UL1TR000114-02), UMN Academic Health Center Faculty Development Award, and Fairview Health Services supported this work. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health.
References
- 1.Liu H, Lussier YA, Friedman C. A study of abbreviations in the UMLS; AMIA Annu Symp Proc; 2001. pp. 393–397. [PMC free article] [PubMed] [Google Scholar]
- 2.Xu H, Stetson PD, Friedman C. A study of abbreviations in clinical notes; AMIA Annu Symp Proc; 2007. pp. 821–825. [PMC free article] [PubMed] [Google Scholar]
- 3.Savova GK, Coden AR, Sominsky IL, Johnson R, Ogren PV, de Groen PC, Chute CG. Word sense disambiguation across two domains: Biomedical literature and clinical notes. J Biomed Inform. 2008;41(6):1088–1100. doi: 10.1016/j.jbi.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Moon S, McInnes B, Melton GB. Challenges and practical approaches with word sense disambiguation of acronyms and abbreviations in the clinical domain. Healthc Inform Res. 2015;21(1):35–42. doi: 10.4258/hir.2015.21.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Y, Denny JC, Rosenbloom ST, Miller RA, Giuse DA, Xu H. A comparative study of current clinical natural language processing systems on handling abbreviations in discharge summaries; AMIA Annu Symp Proc; 2012. pp. 997–1003. [PMC free article] [PubMed] [Google Scholar]
- 6.Kuhn IF. Abbreviations and acronyms in healthcare: when shorter isn’t sweeter. Pediatr Nurs. 2007;33(5):392–399. [PubMed] [Google Scholar]
- 7.Walsh KE, Gurwitz JH. Medical abbreviations: writing little and communicating less. Arch Dis Child. 2008;93(10):816–817. doi: 10.1136/adc.2008.141473. [DOI] [PubMed] [Google Scholar]
- 8.Cohen AM, Hersh WR. A survey of current work in biomedical text mining. Brief Bioinform. 2005;6(1):57–71. doi: 10.1093/bib/6.1.57. [DOI] [PubMed] [Google Scholar]
- 9.Schuemie MJ, Kors JA, Mons B. Word sense disambiguation in the biomedical domain: An overview. J Comput Biol. 2005;12(5):554–565. doi: 10.1089/cmb.2005.12.554. [DOI] [PubMed] [Google Scholar]
- 10.Pakhomov S. Semi-supervised maximum entropy based approach to acronym and abbreviation normalization in medical texts; Proc 40th Annu Meet Assoc Comput Ling; 2002. pp. 160–167. [Google Scholar]
- 11.Stevenson M, Guo Y, Amri AA, Gaizauskas R. Disambiguation of biomedical abbreviations; Proc Workshop Curr Trends Biomed Nat Lang Process; 2009. pp. 71–79. [Google Scholar]
- 12.Xu H, Stetson PD, Friedman C. Combining corpus-derived sense profiles with estimated frequency information to disambiguate clinical abbreviations; AMIA Annu Symp Proc; 2012. pp. 1004–1013. [PMC free article] [PubMed] [Google Scholar]
- 13.Xu H, Stetson PD, Friedman C. Methods for building sense inventories of abbreviations in clinical notes. J Am Med Inform Assoc. 2009;16(1):103–108. doi: 10.1197/jamia.M2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McInnes BT, Pedersen T, Liu Y, Pakhomov SV, Melton GB. Using second-order vectors in a knowledge-based method for acronym disambiguation; Proc 15th Confe Comput Nat Lang Learn; 2011. pp. 145–153. [Google Scholar]
- 15.Liu H, Lussier YA, Friedman C. Disambiguating ambiguous biomedical terms in biomedical narrative text: An unsupervised method. J Biomed Inform. 2001;34(4):249–261. doi: 10.1006/jbin.2001.1023. [DOI] [PubMed] [Google Scholar]
- 16.Pakhomov S, Pedersen T, Chute CG. Abbreviation and acronym disambiguation in clinical discourse; AMIA Annu Symp Proc; 2005. pp. 589–593. [PMC free article] [PubMed] [Google Scholar]
- 17.Moon S, Serguei Pakhomov S, Melton GB. Automated disambiguation of acronyms and abbreviations in clinical texts: Window and training size considerations; AMIA Annu Symp Proc; 2012. pp. 1310–1319. [PMC free article] [PubMed] [Google Scholar]
- 18.Moon S, Berster BT, Xu H, Cohen T. Word sense disambiguation of clinical abbreviations with hyperdimensional computing; AMIA Annu Symp Proc; 2013. pp. 1007–1016. [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Y, Xu J, Zhang Y, Xu H. Clinical abbreviation disambiguation using neural word embeddings; Proc Workshop Biomed Nat Lang Process; 2015. pp. 171–176. [Google Scholar]
- 20.Moon S, Pakhomov S, Liu N, Ryan JO, Melton GB. A sense inventory for clinical abbreviations and acronyms created using clinical notes and medical dictionary resources. J Am Med Inform Assoc. 2014;21(2):299–307. doi: 10.1136/amiajnl-2012-001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanerva P, Kristoferson J, Holst A. Random indexing of text samples for latent semantic analysis; Proc 22nd Annu Conf Cogn Sci Soc; 2000. p. 1036. [Google Scholar]
- 22.Henriksson A, Moen H, Skeppstedt M, Daudaravičius V, Duneld M. Synonym extraction and abbreviation expansion with ensembles of semantic spaces. J Biomed Sem. 2014;5(6):1–25. doi: 10.1186/2041-1480-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berster BT, Goodwin JC, Cohen T. Hyperdimensional computing approach to word sense disambiguation; AMIA Annu Symp Proc; 2012. pp. 1129–1138. [PMC free article] [PubMed] [Google Scholar]
- 24.Kanerva P. Hyperdimensional computing: An introduction to computing in distributed representation with highdimensional random vectors. Cogn Comput. 2009;1(2):139–159. [Google Scholar]
- 25.Sahlgren M. Method Appl Semant Index Workshop 7th Int Conf Terminol Knowl Eng. TKE; 2005. An introduction to random indexing; p. 5. [Google Scholar]
- 26.Humphreys BL, Lindberg DAB, Schoolman HM, Barnett GO. The unified medical language system: An informatics research collaboration. J Am Med Inform Assoc. 1998;5(1):1–11. doi: 10.1136/jamia.1998.0050001. [DOI] [PMC free article] [PubMed] [Google Scholar]