Abstract
Clinical decision support (CDS) knowledge, embedded over time in mature medical systems, presents an interesting and complex opportunity for information organization, maintenance, and reuse. To have a holistic view of all decision support requires an in-depth understanding of each clinical system as well as expert knowledge of the latest evidence. This approach to clinical decision support presents an opportunity to unify and externalize the knowledge within rules-based decision support. Driven by an institutional need to prioritize decision support content for migration to new clinical systems, the Center for Knowledge Management and Health Information Technology teams applied their unique expertise to extract content from individual systems, organize it through a single extensible schema, and present it for discovery and reuse through a newly created Clinical Support Knowledge Acquisition and Archival Tool (CS-KAAT). CS-KAAT can build and maintain the underlying knowledge infrastructure needed by clinical systems.
Introduction
Vanderbilt University Medical Center’s (VUMC) long history of innovation in biomedical informatics has led to the creation of unique and increasingly complex clinical decision support (CDS) over multiple decades. These CDS resources are embedded in the many information systems within the institution at the point of care. This paper presents VUMC’s efforts to externalize this varied content in a systematized manner, leading to the creation of a comprehensive CDS schema and knowledge acquisition and archival tool.
At VUMC, CDS is tightly integrated with the system in which it is implemented. Horizon Expert Orders (HEO), the computerized physician order entry (CPOE) system originally developed at Vanderbilt as WizOrder,1 presents CDS ranging from allergy checking and drug-drug interactions to discharge instructions and complex protocols. Within HEO, CDS is written in a locally-developed scripting language called Vanderbilt Generalizable Rules (VGR) that allows user screens to be overridden with simple display templates and HTML files.2–4 Within StarPanel,5 Vanderbilt’s electronic health record (EHR) system, built-in CDS visually presents relevant information derived from the patient’s data. These display resources are called Indicators6 because they rapidly indicate critical information about the patient. Indicators are created by professional developers in the high-level programming language Perl. Other systems such as the outpatient order entry system, VOOM, similarly integrate unique CDS directly into the system with which providers interact. This approach has led to the implementation of CDS through diverse languages and mechanisms.
Support for Horizon Clinicals, the suite of clinical systems7 used by VUMC which includes HEO, HED (the inpatient nurse documentation system), AdminRx (the barcoded medications administration system), and HMM (the pharmacy management system) will be discontinued. In light of this, VUMC leadership created the Clinical Systems 2.0 initiative8 to replace the clinical systems with a new suite of products. The part of this institution-wide effort presented in this paper is focused on transitioning select CDS content knowledge from the current systems and implementations to representations that retain clinical significance and intent and can be implemented in the new clinical systems. The Center for Knowledge Management (CKM) team was approached directly by the Chief Informatics Officer and asked to apply its unique set of skills towards this project in March 2015.
The CKM team at VUMC aids in supporting the information-strata of the organization through innovative processes of knowledge acquisition and organization for the purposes of archiving and reuse. As such, the CKM team is well situated to work on this project given our close linkage to biomedical informatics and our expertise with metadata schema development, long standing provision of actionable evidence in the clinical setting, relational database and interface creation, content curation, and extraction and identification of linkages between disparate information sources.9,10
Preliminary meetings with leadership, CDS content creators, and stakeholders further demonstrated both the scope of the institution’s need and that CKM’s skills could easily, and immediately, be directed towards managing the existing content. For example, as evidence has been a critical part of creating and updating CDS resources, the CKM team recognized the opportunity to extend and update this evidence alongside the executable content. Further, production databases hold both content that is currently used and content that had been intentionally deactivated, presenting an opportunity for archiving and future retrieval. Additionally, the meetings further aided CKM in fostering collaboration with HealthIT’s Knowledge Engineering team at VUMC. The knowledge engineers, using their relationship with clinicians and end-users of CDS content, helped shape the approach the CKM team used to refine tool development, and eventually, content validation. The meeting outcomes, along with CKM’s long history of expert evidence provision in complex environments,11 made ongoing content maintenance another key goal for the project.
Implementation Background and Methods
Synthesizing the institutional need, the preliminary data we gathered from meeting with experts and stakeholders, and our own skills, we arrived at the following goals for a unified approach to managing knowledge around clinical decision support: extract content and medical context from application-specific representations, preserve complexity, identify and update evidence to support current practice, enable cross-system search for the discovery of previously obscured linkages, validate content, and aid in the process of making decisions on active versus archival retention. These goals would be tested over the entirety of active content from two types of CDS: VGRs and Indicators. The rationale behind this implementation was dictated by our goals and the relevant literature.
Our first and most critical goal was to extract content directly from the multiple applications in which it was created and represent it in a unified way. As this initiative was started well in advance of new clinical systems being implemented, VUMC did not have a replacement suite of systems selected, and it became a major design goal to keep the extracted knowledge system-agnostic. We therefore focused first on standardization efforts such as Arden Syntax, Knowledge Artifacts, and GELLO that show the value of representing CDS in a generic way using a consistent schema. In Arden Syntax, basic descriptions of content are standardized, while data to support the logic are kept entirely application-specific, which means the decision support must be mapped to each application to be reused.12 Knowledge Artifacts solve this challenge through an XML representation of logic using the Virtual Medical Record (VMR) domain analysis model of the HL7’s Reference Information Model (RIM) to promote interchange and interoperability.13,14 GELLO likewise uses the RIM to enumerate all possible data types and type extensions.15 The CKM team conducted a thorough literature search of CDS standards, schemas, taxonomies, ontologies, and development methods and considered the aforementioned standards as ways of creating implementation-independent representations of the CDS resources at scale, while allowing for extensibility to meet evolving needs.
Another goal of the CKM team’s effort was to facilitate unambiguous human understanding of complex rules while ensuring ease of use. Local CDS that implements a protocol or guideline is complicated, and to allow users to truly understand that protocol we had to correctly capture all of its intricacies. The Clinical Decision Support Consortium created a model for representing guidelines that uses multiple layers of increasing complexity from text to semistructured, structured, and executable,16 similar in nature to the multiple representation formats of DeGeL.17 This allows consumers of the representation to pick the level that best suits their needs, potentially increasing use.16 CKM similarly chose to create a tiered approach that allows both complexity and simplicity depending on user preference.
A chief concern of the CKM team was enabling discoverability of resources in as many ways as possible. As we were starting from a collection of software artifacts, some understanding of methods from software reuse were beneficial to our efforts. Software reuse is a software engineering concept that starts from a piece of code that provides an important function and creates a searchable abstraction. The goal of this process is that useful snippets of code can be reused as much as possible, saving future development time. While not quite the same as our goal of recreating rather than reusing these resources, software reuse provides four key components of reusability: abstraction, concise description, specificity enough to distinguish between descriptions, and a clear understanding of relationships to other artifacts.18 Further, software reuse literature describes eight classical methods of abstraction that can fulfill these needs,19 whether for reuse or adaption.
Other work in biomedical informatics has further described the content and context of decision support beyond standards. Decision support taxonomies in the literature describe the functions decision support can provide, based on empirical observation.3,20–24 Greenes presented a general schema based on purpose and design.25 This body of work provided context for thinking about discoverability specific to CDS.
It was important to the team to automatically extract as much as possible from the existing disparate systems and present the resulting information in a single repository for users to find and view CDS. Sittig et al. defined four tools needed to effectively manage clinical knowledge: an external repository of clinical content, a collaborative space to develop content, a terminology control tool, and a mechanism for end users to provide feedback.26 Our focus was on creating this external repository through iterative feature development based on stakeholder requirements.
Finally, it was critical to identify clinical owners, validate our work, and prioritize the CDS content to move into the new systems. Validation and decision-making on CDS prioritization was provided by the Knowledge Engineering team in partnership with clinical content owners. Further, the first step in validation was ensuring the schema was robust through testing across a varied set of highly used and logically separate resources as a representative sample. Reference documentation was used to aid in application of the schema to each clinical system. Knowledge Engineering team members were also encouraged to contribute to populating any content fields that their expertise could provide.
This intervention took place within the clinical systems of VUMC’s tertiary-care teaching hospitals, which serve middle Tennessee and the surrounding areas, as well as inpatient and outpatient facilities, all of which use StarPanel as the EHR.
Results
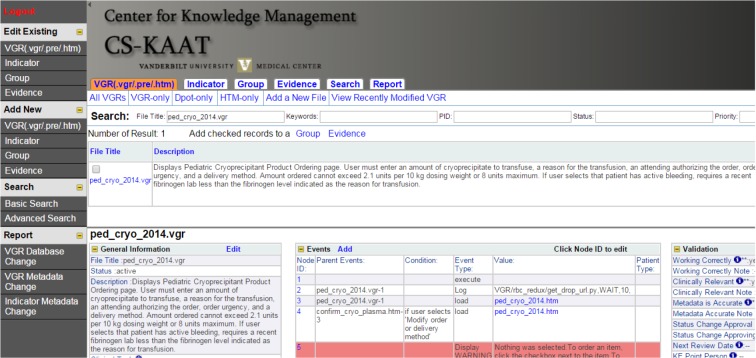
In all, the CKM team extracted 1024 Indicators (965 active; 59 inactive), 3147 VGRs (1671;1476), 1875 display templates (945;930), and 1951 HTML pages (705;1246) for representation in our in-house created web-based tool. This tool (Figure 1), named the Clinical Support Knowledge Acquisition and Archival Tool (CS-KAAT), allows all the above knowledge to be viewed and edited. CS-KAAT uses a MySQL database with a PHP front end. Access is restricted by login, and user role enables various functions from access to editing specific fields. Codified information is collected automatically via scripting and validated by human agents. If the automatically extracted data needs to be modified, the new values for that field are clearly marked as edited, allowing end users to differentiate which party to trust for the accuracy of the information. Changes to production databases are captured periodically and automated content extraction and manual reconciliation are performed on all new or modified resources to ensure that content in CS-KAAT remains current with production systems.
Figure 1.
Clinical Support Knowledge Acquisition and Archival Tool (CS-KAAT).
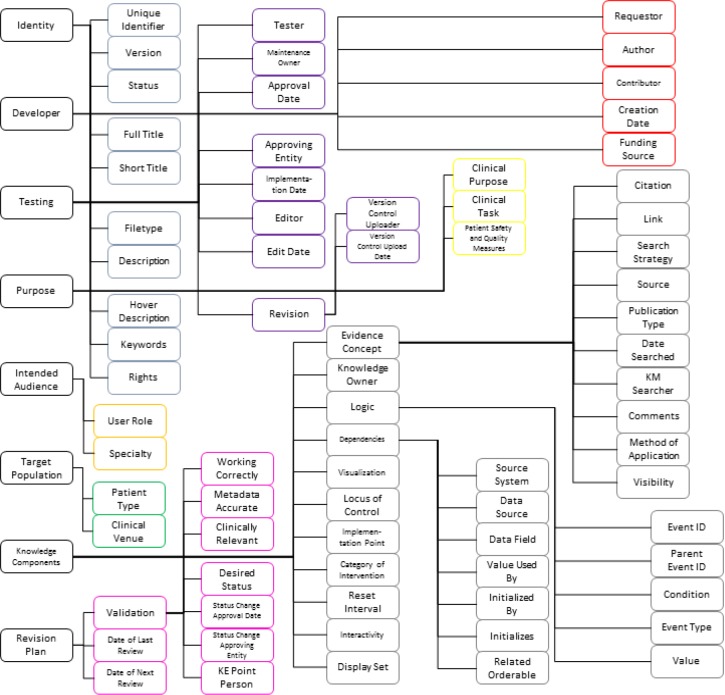
Our schema provides 70 elements informed by multiple literature sources and modified to fully describe CDS content at VUMC. Content is described along central facets with controlled vocabularies that enable comparison. This method of software abstraction allows high precision and recall, while providing transparent and easily understood access to users.19 The schema driving CS-KAAT can be divided into important components - for instance, divisions taken from the high-level concepts of the GEM model27 - to present the elements (Figure 2). Identity elements uniquely identify a resource. Developer elements are the names and roles of people involved in the creation of a resource, and Testing elements identify editorial oversight and modifications. Purpose elements define the reasons a resource was implemented. Intended Audience elements identify the clinicians impacted by a resource, while Target Population elements delineate patients affected by the resource. Knowledge Components elements store the knowledge from applicable evidence, local practice, and implemented logic and display. Finally, Revision Plan elements ensure currency of the resources.
Figure 2.
Schema elements. Adapted from elements presented in references [3,12,13,21–25,27,31]
The schema applies to decision support of any type by mapping the features of that particular type of content to the appropriate schema elements. Mapping to each system allowed us to see that the schema element ‘Category of Intervention’ would receive the same value for all Indicators, but individual VGRs could have values for this element different from each other. From this realization, schema elements are mapped to the most appropriate of four levels to fully describe the CDS resource: system, group, resource, or resource part.
System-level elements apply equally to each resource from the same system. For example, the ‘Locus of Control’ is “system” and ‘Rights’ are “Copyright VUMC” for every VGR, and are therefore applied once to the entire system.
Within CS-KAAT, all users are able to create a generic group and can include any resource as members of their newly created group. This allows users to collocate resources for specific interests or needs, without restriction, and retrieve those resources easily (e.g., all resources associated with a clinical intervention or a workflow process). Group elements enable the user to describe the reasons for collocation and create multiple levels of specificity for this description. Elements that apply to these groups are ‘Description,’ ‘Date of Next Review,’ and ‘Visualization’.
The resource level denotes each distinctly named CDS resource. Elements around creation and maintenance are naturally applied here.
Critical points of interest, called events, were identified and parsed from each resource to create the resource part level. Events currently used are listed below (Table 1). ‘Logic’ and ‘Patient Type’ elements are captured around these events.
Table 1.
Events within clinical decision support rules.
| Control | orderables |
| Order | |
| Modify | |
| Discontinue | |
| Control program flow | Execute |
| Load | |
| Require specified exit checks | |
| Exit without ordering | |
| External notification | Send email |
| Send page | |
| Log | |
| User interaction | Display information/warning |
| Radio button | |
| Checkbox | |
| Dropdown | |
| Button | |
| Textbox |
Each event is assigned a human-readable explanation of the logical condition that must be true for the event to execute. Additionally, each event has a unique identifier that is used to link events together. Finally, the value of the event is captured; for example, the specific file being loaded or the parameters of an order being modified are extracted from the source code.
Events are not tied to logical blocks, but are the points at which a resource affects the environment outside of itself, such as executing a program or interacting with a user or a different clinical system. It is common that several events will happen due to the same condition being true, or may be alternatives where a user will only be affected by one event from a set. For example, selecting to order Esmolol Infusion in a cardiac step-down unit requires electrophysiologist approval. Providing this approval places the order in the queue to be signed, logs the approving physician’s name, and emails a pharmacist. Each of these actions takes point in a different block of code: in fact, in two different files. Our interest in these operationally interconnected events is that they were all triggered by one action and that we may decide they should all happen together in the future system regardless of how many steps it currently takes to achieve that function. The loose structure of events therefore allows us to extract the clinical content and medical context of each event from the application’s execution requirements. In the VGR system, each CDS resource has several events, ranging from 3 to over 300, due to the needs of the HEO system for presenting alternatives to the clinician and providing real-time response without the delay of querying underlying databases after each selection. By defining clinically and operationally significant events rather than the blocks of code that exist now, future CDS implementations are not bound by past technological requirements.
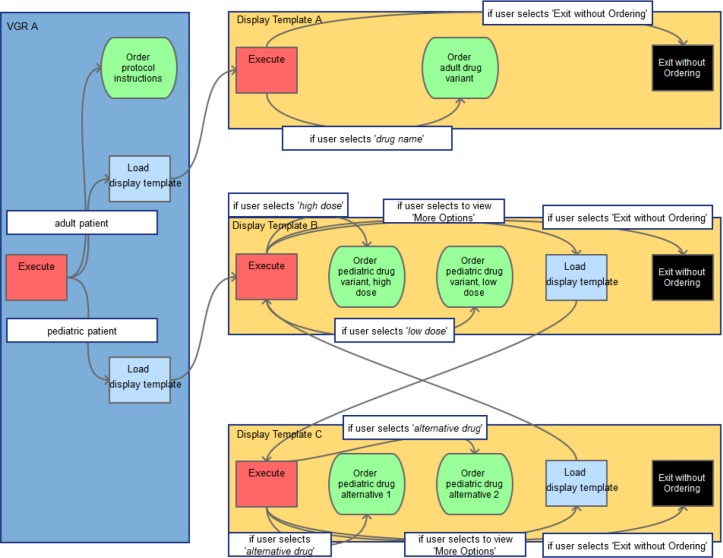
In linking specific events to one another, only sequences have been preserved explicitly. Sequence is captured by human coders by expanding on all possible sets of events that generate all the data within that event’s condition and value. Alternatives in this sequence are allowed through a simple description logic that supports unions and intersections. Alternatives have been captured only implicitly, allowing future representations to reorder content or take pieces from resources if desired. Events are linked through backward chaining sequences, allowing the user to understand what prior data had to exist to make the current event possible. Together, all these individual data points allow for robust visualization of highly connected networks that create a comprehensive picture (Figure 3) from initial states to ultimate possible outcomes.
Figure 3.
Generic protocol visualization. Red, blue, green, and black boxes represent events. Arrows indicate the direction of relationships. White boxes note conditions that must be fulfilled.
Controlled vocabularies come from taxonomies around decision support if possible, and local taxonomies if necessary. Specifically, ‘Category of Intervention,’ ‘Locus of Control,’ ‘Clinical Task,’ and ‘Method of Application’ come from taxonomies. For ‘Clinical Venue’ and ‘Patient Type,’ we had to create our own local taxonomy based on the values used in the local systems. Location data are inherently administrative and not able to be standardized across institutions, and grouping patients into similar types is also based entirely on local needs. To fully capture these elements, therefore, we started from the data definitions within our systems rather than an external source. This allows us to understand whether the current functionality can be exactly recreated or must be modified to work in a new system.
In any system allowing multiple users to create displays for different needs, there will be variability in interface design. Systematically capturing these techniques involved manual effort in both cultivating a controlled vocabulary and applying it. Display techniques captured in the ‘Category of Intervention’ element illustrate the evolution of a local controlled vocabulary. Each HTML page and display template overrides the common HEO interface and is manually coded. This has led to many different displays for clinicians that employ various tactics to present information effectively. The front-end tools taxonomy of Wright et al.24 provided a foundation, as front-end effects are often displayed in addition to executing - displaying a medication order sentence before placing that order, for example. Focusing only on information display, we removed several elements of the taxonomy that were purely functional and added new vocabulary elements for display techniques not presented in the taxonomy, such as ‘drug name/brand name display’. We also created controlled terms of general design techniques for successfully engaging users with clinical systems from the human-computer interaction review of Horsky et al.28 By applying taxonomies of CDS design, we enable users to analyze different designs in a rigorous manner and reuse successful features, an idea presented as a potential benefit of taxonomies in Greenes.25 The exact moment in the clinical workflow that CDS is triggered is one aspect (‘Implementation Point’) captured in a taxonomy that allows users to compare differences between content initiated when an orderable is requested and content initiated after all orders are placed.
Scripting allows for the automation of some of the above tasks. Events were parsed from source code by exploiting the syntax of the underlying programming language and using regular expressions to extract and identify each event type. Based on the events present in a resource and the relationships to other resources found, the ‘Initializes,’ ‘Initialized By,’ ‘Related Orderable,’ and ‘Implementation Point’ elements were automatically inferred. System- level fields were automatically populated and passed to all resources within that system. Finally, elements that could be factually stated without ambiguity were also automated - ‘Revision,’ ‘Full Title,’ ‘Short Title,’ ‘Hover Description,’ ‘Filetype,’ and ‘Dependencies’.
Evidence is preserved in a separate workflow from the CDS it informs, as evidence around a concept can affect more than one resource. This conceptual grouping is an extension of the generic grouping feature. For example, clopidogrel metabolization is affected by genetic variation in the CYP2C19 gene. Furthermore, the evidence supporting differentiation in clopidogrel dosing by CYP2C19 variation is not simply one resource, but a collection of external resources, local evaluation of evidence quality and impact, and potentially even local practice constraints. Therefore, this concept of evidence-based practice regarding clopidogrel metabolization is the logical unit at which to maintain the evidence and local decisions on usefulness of the evidence. Attaching this concept to each resource that requires it (five resources in the current systems to perform additional decision support recommending alternative treatment for intermediate and hypo-metabolizers, and flag patients’ charts a predetermined color for those who have a recorded drug-genome interaction) allows us to periodically evaluate whether the evidence supporting this concept has changed and whether all our current decision support involving this concept is ideal.
Validity of all resources is ensured in three steps. First, the metadata for each resource is checked for accuracy by automated and manual processes. An automated check requires valid data for controlled fields, and accepts the links between specific events only if they are structurally possible between the two resources. Two manual checks, one by a CKM information scientist and one by a knowledge engineer, ensure that the human-readable logic and other event and resource-level fields are correct. Second, the resource is thoroughly tested in the production environment to determine it is still working correctly. Third, in order to determine what content is important to carry forward, a team of knowledge engineers is continuing to assess the clinical relevance of each resource by reviewing usage data, working closely with subject matter experts and system developers. At each of these points, documentation and resolution of any errors becomes part of ongoing content maintenance tasks. A companion document was created for each clinical system to provide the team with up to date information and examples of how each element should be correctly applied.
Discussion
Despite the many benefits of having CDS tightly coupled with the executing applications, this approach does require “the author to be familiar with not only the clinical domain but also with the syntax and features of the programming language.”29 For an outsider to a system to understand its decision support content required additional training in that system, even with clinical expertise and experience creating other decision support content. The VGRs were designed as a way to allow clinical experts to learn a simplified functional language rather than the programming language of HEO,4 but this benefit only applies to the single system. Ordersets are generated through a user-friendly tool that masks the programming complexities that enable each to work in the HEO application,30 but again this tool only assists in orderset creation and maintenance. We have successfully described content from two very distinct systems in a common format. Our project has shown it is possible to represent all decision support content in a consistent manner, within one single system. Each type of decision support being system-specific does require authors and editors to learn those respective tools, but content can be considered outside of the narrow bounds of the application through CS-KAAT.
In the guidelines modeling literature,16,17 work is conducted from a textual representation to an executable one to allow different interested parties access points to the model. The CKM team worked in the opposite direction, from executables to semi-structured representation, to similarly create access points and enable flexibility to transition decision support to any number of potential systems. Multiple layers of abstraction allow end users to view content as complexly or simply as they desire. At the highest level, there is a basic description or a visual grouping. Below this, each resource and the key events that it comprises are described fully. Finally, data inputs and data flow are presented as structured data, the layer of representation closest to the functional source code.
In each of the above aspects, existing standards did not fully meet our needs. Our literature review and focus on discoverability highlighted several important descriptive facets that are not included in existing standards. Therefore, to fully meet our goals, it was important to adapt our schema with taxonomies extending standards. The guiding process for adding elements to the schema was to present a use case for each element to provide information that could not come from any existing element. This focus on institutional need led us to view each source as a potential contributor to the schema. With different needs, these extensions into literature-derived elements and local vocabularies will be different. For any institution interested in establishing a similar process, this has averaged 6.5 FTE equivalent effort. Given the diversity and complexity of the work, completing knowledge extraction and transformation for one named CDS resource has ranged from ten minutes to two days.
Separating the review processes for evidence and content allows flexibility in involving the appropriate parties for each step in the maintenance process. Knowledge management information scientists, who have extensive experience providing evidence, are well positioned to ensure that the latest evidence is provided to the content specialist and developers creating CDS resources. Then, clinicians can provide local context on best evidence to determine to create, modify, or retire CDS resources. This is anticipated to relieve some of the burden of engaging with CDS content for maintenance decisions. Consistent, externalized evidence provision also provides the institution the ability to more effectively integrate evidence with CDS at the point of care by making each relevant citation a focal point of the model, one that is purposefully saved and maintained. Finally, CS-KAAT provides the institution with a robust citation repository that is up to date, expertly curated, and has institution-wide scope.
CS-KAAT has made data dependencies traceable across systems. To illustrate the revelations that our tool has provided, we can follow an orderable from order creation in HEO to patient evaluation in StarPanel. A user initiates an order which is flagged to trigger a VGR when it is requested. From there, a series of VGRs eventually ends with the correct order being placed. After passing through an interface that links the different operational systems from the CPOE to the pharmacy system, data about the successful filling of this order are again sent through this generic interface engine to the medical record. An intermediate database duplicates information about the order of interest and distributes that information to the patient’s chart via an Indicator. The VGR and Indicator in this example are clearly related, but this information was obscured by the complex relationship between the two CDS resources. Because our schema has a facet for related orderables, the relationship is clearly seen at the semi-structured level and directly searchable. Additionally, the application of UMLS keywords to each of the records allows for files to be linked by concept, something that was also previously unavailable to users. Similarly, each facet gives users of CS-KAAT the powerful capability to understand connections and associations that previously only a few developers had.
Validation efforts enforced adherence to the schema. In each area where scripting was used for content extraction, users were able to manually override the harvested value. Periodic review of these overrides led to editing the scripts, making data collection more authoritative. We found that the compiler for the VGR local scripting language allowed slight variations to syntax, for example, and adjusted our extraction scripts to capture these. While these scripts are only useful in parsing knowledge from the current versions of local systems, this method both saved time for our team members and provides trusted content to end users. For data that could not be extracted automatically, validation by at least two people other than the documenter provides another level of assurance to end users that the content of CS-KAAT is trusted.
This was a significant effort that required many people to contribute different skill sets and to approach the work with a great deal of collaboration. The work at Partners and Intermountain Healthcare demonstrate that this is an extensive process, fraught with difficulty.31,32 It has been successful at VUMC through two key human features of our work: flexibility and communication. Our schema went through many early iterations, and as workflows changed, both the team and the CS-KAAT adapted to evolving needs. The CKM team provided support for each other and interchanged roles to assist in completing different portions of the work. Communication between teams and with stakeholders has allowed progress to be measured and created buy-in. For example, the user interface has been improved after first time users from Knowledge Engineering provided usability feedback.
The separation of clinical content from application is critical to the long term usefulness and maintenance of CDS.33 While there are knowledge editors for certain CDS types,30 these useful and highly used tools do not have institutionwide scope, a situation described as typical of most institutions today.31 CS-KAAT demonstrates an effective means for separating clinical content from native applications, and further makes important steps towards an institutionwide knowledge editor through a unified acquisition and archival tool.
Future Work
The content within CS-KAAT will continue to be used to prioritize CDS for the new clinical systems, and as the new technical requirements of continued clinical needs are defined will be remixed to meet those requirements. The CKM team is focused on continued enhancements to the tool to enrich lifecycle support and enable retention of collaborative documentation for future use. As CS-KAAT has proven to be effective in the realization of our stated goals, the team will continue collaboration with Knowledge Engineering and others within the HealthIT group to incorporate CDS resources from other systems with the ultimate goal of representing all CDS content that affects care at VUMC. Further, the team is exploring various tactics for displaying evidence underlying CDS to clinicians at the point of care to enhance usefulness of decision support systems while retaining clinical user satisfaction.
Conclusion
VUMC has externalized decision support content from multiple sources in a consistent schema. Descriptive facets from decision support standards, as well as empirical taxonomies and controlled vocabularies, create many new ways for users to find and evaluate content. This understanding is supported through a focus on human-readable descriptions and multiple levels of representational complexity. In all, this flexible structure positions the medical center to effectively maintain the entirety of its decision support at the institutional level and continue to develop new methods of effectively supporting clinical care.
Acknowledgments
The authors would like to acknowledge the dedication of the CKM team members who have made this project a success: Mallory Blasingame and Jing Su. Additionally, we would like to thank all of the members of the Knowledge Engineering team, including Tina French, Shari Just, Sylinda Littlejohn, Janos Mathe, Michael McLemore, Lorraine Patterson, Debbie Preston, Audra Rosenbury, and Charlie Valdez, for their collaboration and contributions.
References
- 1.Geissbuhler A, Miller RA. A new approach to the implementation of direct care-provider order entry.. Proceedings of the AMIA Annual Fall Symposium; American Medical Informatics Association; 1996. [PMC free article] [PubMed] [Google Scholar]
- 2.Heusinkveld J, Geissbuhler A, Sheshelidze D, Miller R. A programmable rules engine to provide clinical decision support using HTML forms. Proceedings of the AMIA Symposium; American Medical Informatics Association; 1999. [PMC free article] [PubMed] [Google Scholar]
- 3.Miller RA, Waitman LR, Chen S, Rosenbloom ST. The anatomy of decision support during inpatient care provider order entry (CPOE): empirical observations from a decade of CPOE experience at Vanderbilt. J BiomedInform. 2005;38(6):469–85. doi: 10.1016/j.jbi.2005.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Starmer JM, Talbert DA, Miller RA. Experience using a programmable rules engine to implement a complex medical protocol during order entry; Proceedings of the AMIA Symposium; American Medical Informatics Association; 2000. [PMC free article] [PubMed] [Google Scholar]
- 5.Giuse DA. Supporting communication in an integrated patient record system; AMIA Annual Symposium Proceedings; American Medical Informatics Association; 2003. [PMC free article] [PubMed] [Google Scholar]
- 6.Starmer J, Giuse DA. A real-time ventilator management dashboard: toward hardwiring compliance with evidence-based guidelines; AMIA Annual Symposium Proceedings; American Medical Informatics Association; 2008. [PMC free article] [PubMed] [Google Scholar]
- 7.Raths D. Vanderbilt University Medical Center begins Epic journey. Healthcare Informatics. 2015 [Google Scholar]
- 8.Govern P. Nashville, TN: VUMC News and Communications; 2015. Clinical information systems in line for major upgrade. [cited 2016 March 3]; Available from: http://news.vanderbilt.edu/2015/04/clinical- information-svstems-in-line-for-maior-upgrade/. [Google Scholar]
- 9.Koonce TY, Giuse DA, Beauregard JM, Giuse NB. Toward a more informed patient: bridging health care information through an interactive communication portal. J Med Libr Assoc. 2007;95(1):77–81. [PMC free article] [PubMed] [Google Scholar]
- 10.Giuse NB, Kusnoor SV, Koonce TY, et al. Strategically aligning a mandala of competencies to advance a transformative vision. J Med Libr Assoc. 2013;101(4):261–67. doi: 10.3163/1536-5050.101.4.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giuse DA, Giuse NB, Miller RA. Evaluation of long-term maintenance of a large medical knowledge base. J Am Med Inform Assoc. 1995;2(5):297–306. doi: 10.1136/jamia.1995.96073832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Health Level Seven. Health Level Seven Arden Syntax for medical logic systems, version 2.10. HL7. 2014.
- 13.Health Level Seven. Clinical Decision Support Knowledge Artifact specification, release 1 DSTU release 1.3. HL7. 2015.
- 14.Health Level Seven. Reference Information Model, release 6. HL7. 2014.
- 15.Health Level Seven. GELLO: a common expression language, release 2. HL7. 2010.
- 16.Boxwala AA, Rocha BH, Maviglia S, et al. A multi-layered framework for disseminating knowledge for computer-based decision support. J Am Med Inform Assoc. 2011;18(Suppl 1):i132–i9. doi: 10.1136/amiajnl-2011-000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shahar Y, Young O, Shalom E, et al. A framework for a distributed, hybrid, multiple-ontology clinical- guideline library, and automated guideline-support tools. J Biomed Inform. 2004;37(5):325–44. doi: 10.1016/j.jbi.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Krueger CW. Software reuse. ACM Comput Surv. 1992;24(2):131–83. [Google Scholar]
- 19.Mili A, Mili R, Mittermeir RT. A survey of software reuse libraries. Ann Softw Eng. 1998;5:349–414. [Google Scholar]
- 20.Wang JK, Shabot MM, Duncan RG, Polaschek JX, Jones DT. A clinical rules taxonomy for the implementation of a computerized physician order entry (CPOE) system; Proceedings of the AMIA Symposium; American Medical Informatics Association; 2002. [PMC free article] [PubMed] [Google Scholar]
- 21.Sim I, Berlin A. A framework for classifying decision support systems; AMIA Annual Symposium Proceedings; American Medical Informatics Association; 2003. [PMC free article] [PubMed] [Google Scholar]
- 22.Berlin A, Sorani M, Sim I. A taxonomic description of computer-based clinical decision support systems. J Biomed Inform. 2006;39(6):656–67. doi: 10.1016/j.jbi.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Wright A, Goldberg H, Hongsermeier T, Middleton B. A description and functional taxonomy of rule- based decision support content at a large integrated delivery network. J Am Med Inform Assoc. 2007;14(4):489–96. doi: 10.1197/jamia.M2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright A, Sittig DF, Ash JS, et al. Development and evaluation of a comprehensive clinical decision support taxonomy: comparison of front-end tools in commercial and internally developed electronic health record systems. J Am Med Inform Assoc. 2011;18(3):232–42. doi: 10.1136/amiajnl-2011-000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenes RA. In: Greenes RA, editor. Clinical decision support: the road to broad adoption. 2 ed. New York, NY: Elsevier; 2014. Features of computer-based clinical decision support; pp. 111–144. [Google Scholar]
- 26.Sittig DF, Wright A, Simonaitis L, et al. The state of the art in clinical knowledge management: an inventory of tools and techniques. Int J Med Inform. 2010;79(1):44–57. doi: 10.1016/j.ijmedinf.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shiffman RN, Karras BT, Agrawal A, Chen R, Marenco L, Nath S. GEM: a proposal for a more comprehensive guideline document model using XML. J Am Med Inform Assoc. 2000;7(5):488–98. doi: 10.1136/jamia.2000.0070488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horsky J, Schiff GD, Johnston D, Mercincavage L, Bell D, Middleton B. Interface design principles for usable decision support: a targeted review of best practices for clinical prescribing interventions. J Biomed Inform. 2012;45(6):1202–16. doi: 10.1016/j.jbi.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Jenders RA. Greenes RA Clinical decision support: the road to broad adoption. 2 ed. New York, NY: Elsevier; 2014. Decision rules and expressions; pp. 417–434. [Google Scholar]
- 30.Geissbuhler A, Miller RA. Distributing knowledge maintenance for clinical decision-support systems: the“ knowledge library” model; Proceedings of the AMIA symposium; American Medical Informatics Association; 1999. [PMC free article] [PubMed] [Google Scholar]
- 31.Rocha RA, Maviglia SM, Sordo M, Rocha BH. Greenes RA Clinical decision support: the road to broad adoption. 2 ed. New York, NY: Elsevier; 2014. A clinical knowledge management program; pp. 773–818. [Google Scholar]
- 32.Hulse NC, Galland J, Borsato EP. Evolution in clinical knowledge management strategy at Intermountain Healthcare; AMIA Annual Symposium Proceedings; American Medical Informatics Association; 2012. [PMC free article] [PubMed] [Google Scholar]
- 33.Greenes RA. Greenes RA Clinical decision support: the road to broad adoption. 2 ed. New York, NY: Elsevier; 2014. Definition, scope, and challenges; pp. 3–47. [Google Scholar]