Abstract
Clinical trial coordinators refer to both structured and unstructured sources of data when evaluating a subject for eligibility. While some eligibility criteria can be resolved using structured data, some require manual review of clinical notes. An important step in automating the trial screening process is to be able to identify the right data source for resolving each criterion. In this work, we discuss the creation of an eligibility criteria dataset for clinical trials for patients with two disparate diseases, annotated with the preferred data source for each criterion (i.e., structured or unstructured) by annotators with medical training. The dataset includes 50 heart-failure trials with a total of 766 eligibility criteria and 50 trials for chronic lymphocytic leukemia (CLL) with 677 criteria. Further, we developed machine learning models to predict the preferred data source: kernel methods outperform simpler learning models when used with a combination of lexical, syntactic, semantic, and surface features. Evaluation of these models indicates that the performance is consistent across data from both diagnoses, indicating generalizability of our method. Our findings are an important step towards ongoing efforts for automation of clinical trial screening.
Introduction
With the amount of data collected during patient care growing, researchers have proposed a number of scenarios in which knowledge extracted from clinical data can be leveraged effectively1. Information found in the Electronic Health Record (EHR) can have an important secondary use in identifying patients eligible for clinical trials2. Identifying the patient cohort involves understanding the exact information sought by the eligibility criteria for trials and searching for it in the subject’s EHR. However, this manual process of eligibility determination is challenging and time-consuming3. Therefore, a number of initiatives have been undertaken to introduce automation into this process4.
The EHR captures patient information using both structured data, stored in the form of tables or flowsheets, and unstructured data, stored in the form of clinical notes or free-text reports. While structured data are comprised of laboratory values, demographics, diagnosis codes, etc., unstructured clinical notes are written by physicians, nurses and other health professionals to document a patient’s condition, prognosis, response to various clinical interventions, and future plans for treatment. Eligibility criteria based on data found in structured data fields can be processed easily using structured queries over databases (e.g., “At least 18 years of age at the time of enrollment.”). Although structured data enables quick and easy retrieval of data, unstructured text is a preferred means of documentation for physicians5. Thus, the information need of eligibility criteria that involve clinical nuances (e.g., “Patients with a known severe symptomatic primary pulmonary disease.”) can only be answered using unstructured clinical notes or discussion with the treating physician.
Gaps between data found in the EHR and the desired data for trial recruitment have been studied6. Köpcke et al.7 found that there was a significant gap (65%) between the structured data documented for patient care and the data required for eligibility assessment. Li et al.8 studied the strengths and weaknesses of using structured or textual clinical data for clinical trials eligibility screening. They concluded that insights obtained from natural language processing of patient reports supplement important information for eligibility screening and should be used in combination with structured data. Therefore, researchers have devised methods9,10 using NLP to identify relevant text in clinical notes and resolve eligibility criteria to screen patients into clinical trials. However, using NLP techniques on clinical data is computationally expensive. Moreover, certain criteria (e.g., “Patients between the ages of 18 and 90.”) can be answered accurately and quickly using structured data alone. An ideal system for identifying patients would maximize the use of structured data, and only use inference over clinical narratives when necessary. Thus, it would be desirable to know whether eligibility can be concluded for a criterion using structured data, or if it requires an NLP pipeline for its resolution. Such knowledge would not only result in identifying patient cohorts with better accuracy but also optimize the computational efficiency of the task.
Related Work
Eligibility criteria for clinical trials are specified in natural language and hence they are not amenable to computational processing. In their comprehensive review of formal representations of eligibility criteria, Weng et al.11 discuss the variety of intermediate representations used by researchers to overcome this challenge. The authors discuss classification of eligibility criteria along three dimensions: content, use in eligibility determination, and complexities of semantic patterns. The first dimension deals with the information needed to answer eligibility queries. Multiple studies12,13,14 have been published characterizing this dimension of eligibility criteria. The TrialBank project14 classifies criteria into three large categories: age-gender, ethnicity-language, and clinical. Tu et al.15 classified criteria from a clinical perspective: stable, variable, controllable, subjective, and special. Metz et al.16 classified criteria for cancer trials as demographics, contact information, personal medical history, cancer diagnosis, and treatment to date. The majority of these classifications are very specific to the downstream task of these studies. Moreover, since most of these are annotated manually, they are also time-consuming and expensive.
Very few studies have explored automatic classification of eligibility criteria. Luo et al.17 classified eligibility criteria into 27 semantic categories. They show that machine learning classifiers using semantic types from the Unified Medical Language System (UMLS) as features perform better than those using traditional bag of words features; with J48 decision trees performed better than Bayesian models. Bhattacharya and Cantor18 present a study where they automatically map trial protocol documents from the pharmaceutical industry to openly available trial specifications. They compare the eligibility criteria from both sources and propose the creation of template criteria for standardization. Levy-Fix et al.19 propose automatic mapping of eligibility criteria to the widely adopted Common Data Model using a clustering method that uses UMLS semantic type based features. Hong et al.20 define “diagnostic criteria” as a combination of signs, symptoms, and test results used by clinicians to determine the correct diagnosis, and propose mapping of such criteria to the Quality Data Model.
In this paper, we target a very specific problem of automatically classifying whether an eligibility criterion should be resolved using structured data or by using NLP techniques. This is an application driven problem that is common to a number of approaches undertaken by the biomedical informatics community for the secondary use of EHR data. We present a detailed evaluation of our approach using data from two disparate diagnoses. The paper is organized as follows: in order to evaluate an approach that can automatically classify eligibility criteria, we first describe the creation of a gold standard dataset. This is followed by a description of the features used for the machine-learning based classification task and our results. We then examine errors made by our system, followed by a discussion and conclusion of our work.
Dataset Creation
The National Library of Medicine and the National Institutes of Health maintain the websitewww.clinicaltrials.gov, a registry and a database of publicly and privately supported clinical studies across the globe. Almost all of the studies described above, which analyze eligibility criteria of clinical trials, use this registry as their data source. We searched the registry with the query “heart failure” and randomly sampled 50 trials from the result. This was repeated for the query “chronic lymphocytic leukemia.” We then extracted all eligibility criteria from these 100 trials and split them into individual criterion sentences using a sentence chunking module from Lingpipe21 that was trained on MEDLINE articles, along with custom rules for refinement. The goal was to annotate each criterion with the preferred data source for resolution, either “Structured’ or “Unstructured.” We selected trials from two disparate diseases to test the generalization capabilities of our method. The goal was to verify if preferences for resolving these criteria would remain consistent across trials of different diagnoses.
Our annotation team consisted of two senior undergraduate nursing students, a medical student (KR), and a physician (CH). The first author (CS) developed detailed guidelines for the annotators in consultation with the physician on the team. While creating the annotation guidelines, we assumed that the criteria were being resolved by searching for patient information in the EHR software from Epic Systems (Verona, WI) at our institution, The Ohio State University’s Wexner Medical Center. In order to ensure that the annotation guidelines were complete, we conducted three training rounds with ten trials each. This was followed by a discussion period, and disagreements were resolved by the physician. The annotation guidelines were updated after every training round to reduce differences among annotators. We ensured that trials used in the training rounds were not included in the final set of trials used for the study. The training rounds consisted of 30 trials for annotation; criteria texts and annotations from these trials were then used for feature development.
During the discussions, our annotators argued that although the EHR might have a structured field for a criterion, the field may or may not be consistently used by providers for documentation. We addressed this by assuming that for a particular criterion, if the EHR had an available structured field for documentation, it would be classified as structured. In cases of criteria where evidence from both structured and unstructured sources was required, we treated the annotation as unstructured, since criterion resolution could not proceed unless a clinical note was referred to. The end-goal of the classifier being developed is to separate eligibility criteria that can be resolved exclusively by structured data from others, in order to speed up screening. Thus, if a criterion can be partially resolved using structured data but mandates reading of notes for its full resolution, it cannot be processed exclusively using structured data and hence does not contribute to speedup. We therefore do not consider a separate category for “Structured AND Unstructured.”
Another observation made by our annotation team was the presence of eligibility criteria in trials that could not be resolved by information in the EHR at all. These included criteria that would need to be resolved in person or after the start of the study, such as informed consent, willingness to comply with certain restrictions, and criteria associated with post-recruitment details such as randomization constraints, etc. Therefore, in addition to the two categories of structured and unstructured data, we included a third category “Neither” for our annotations. We also observed that the automatic sentence segmentation was not accurate; these were hand-corrected by our annotators. The final kappa among annotators on the set of 50 heart failure trials was κ = 0.83, with κ = 0.81 for the CLL trials; these indicate excellent inter-annotator agreement. The dataset consists of 766 criteria across 50 heart failure trials and 677 criteria across 50 CLL trials. Table 1 summarizes the distribution of annotations across the three categories, which is fairly consistent across the two diagnoses. Figure 1 shows sample criteria across each of the three categories.
Table 1.
Overview of annotations in the dataset
| Number of criteria | Percentage | |||||
|---|---|---|---|---|---|---|
| Annotation | HF | CLL | Combined | HF | CLL | Combined |
| Unstructured | 491 | 368 | 859 | 64.1 | 54.4 | 59.5 |
| Structured | 194 | 193 | 387 | 25.4 | 28.5 | 26.8 |
| Neither | 81 | 116 | 197 | 10.5 | 17.1 | 13.7 |
Figure 1.
Sample eligibility criteria across the three classification categories.
Unstructured
Major heart event or heart procedure within the 6 weeks prior to study entry.
Stable, chronic, mild to severe heart failure as defined as subjects with symptoms of heart failure who do not require IV diuretics, inotropes, or vasodilators or those that require support with a left ventricular assist device.
Progressive lymphocytosis with an increase of > 50% over a 2 month period, or an anticipated doubling time of < 6 months.
Structured
porphyria
Individuals who are morbidly obese
Has significant, moderate-severe renal dysfunction or disease, confirmed by serum creatinine of greater than 2mg/dl (180 micromol/L)
Neither
Patients must be able to provide informed consent.
Concomitant administration of oral ACE inhibitor medication from 2 hours before the start of the study drug until 30 minutes after the start of the study drug.
Patient is physically capable and willing to perform repeated physically demanding tests associated with the study.
Feature Representation
We extracted four sets of features for creating a model to classify the eligibility criteria statements. As discussed earlier, these were designed by examining the criteria from the training rounds. We used MetaMap22 for all UMLS related features and the Stanford Parser23 for other shallow linguistic features. We explain each feature and the rationale for its design below:
Surface Features
M1. Number of words (excluding stop words): Short criteria can be often inferred from structured data.
M2. Number of numerical quantities: Presence of numerical quantities reduces ambiguity and can thus be inferred from structured data.
M3. Number of arithmetic symbols: Presence of arithmetic symbols such as ‘<‘, ‘>‘ or ‘=‘ typically indicates a criterion that can be easily converted into a structured query.
M4. Number of delimiters: Protocol writers often combine multiple sub-criteria using a semi-colon or comma. Multiple information sources may be required to resolve such a criterion.
M5. Number of measurement units: These are often lab values and can be reliably inferred from structured data.
M6. Number of UMLS concepts: This has the same intuition as M1. Criteria with multiple concepts are likely to be complex and thus would mandate reading of notes.
M7. Number of illustrative terms: A generic eligibility criterion is often explained using examples. Illustrative terms used for such a construction are: e.g., example, such as, defined by.
Syntactic Features
SY1. Number of conjunctions: A single criterion often has multiple sub-criteria in it separated by conjunctions (such as and, or). A criterion with multiple sub-criteria is likely to be resolved by reading notes.
SY2. Number of adjectives: Adjectives are used to describe qualitative judgments, possibly making the criterion ambiguous from a computational perspective. It is hard to conclude about such criteria using structured data.
Semantic Features
SM. Semantic types of concepts: Semantic types from UMLS provide a level of abstraction that is intuitive for criteria classification. For example, it is generally accepted that criteria involving laboratory values and medications are resolved using structured data. These correspond to well-defined UMLS semantic types “Laboratory or Test Result” and “Pharmacological Substance” respectively.
Lexical Features
L1. Bag of Words: As discussed earlier, all criteria are from trials of specific diagnoses. Thus, similar words will be commonly used for defining criteria from each of the three categories. These features will capture this pattern.
L2. Bag of UMLS Concepts: This has the same rationale as L1 with one difference: UMLS concepts are often multi-word expressions, and a concept level mapping helps to mitigate this phenomenon.
Results
We evaluated the contributions of different features for the criteria classification task using machine learning classifiers. The evaluation of these tasks was carried out using accuracy as a metric. The distribution of the three categories in our data shows that the simplest classifier, which always predicts the category “Unstructured” for every instance, can achieve an accuracy of 64% on heart failure and 54.4% on CLL. This “Majority” classifier is the simplest baseline.
We chose two different machine learning algorithms for our experiments, namely Naïve Bayes and Support Vector Machines (SVMs). While Naïve Bayes is a simple classifier, it can deliver quick results. SVMs are state of the art machine learning algorithms for classification tasks and work well even with datasets that are not linearly separable. Although it can be time consuming to tune SVM parameters on large datasets, the relatively small dataset in our work meant tuning was not an issue. Following Luo et al.,17 we also tried J48 decision trees, but did not observe any significant performance gain in comparison with the Naïve Bayes model.
We set up the classification task in two settings. First, we carried out a leave-one-out validation on a per-trial basis. Of the 50 trials in our dataset, we trained a classifier on eligibility criteria across 49 trials and tested it on the remaining trial. This was repeated 50 times, such that each trial appeared in the test set once, to calculate the reported accuracy. We refer to this as the trial-dependent setup. Second, we carried out criteria classification independent of their association with a trial. Each criterion statement was considered as a separate instance and evaluation was performed using ten-fold cross validation. While such a setup may induce some knowledge into the model since criteria from the same trial may appear in the training set, it is a more practical setup. When such a system will be deployed for practical use, it would be trained on criteria from different trials. The results of these two evaluations are summarized in Table 2. We found that SVMs outperformed Naïve Bayes significantly; interestingly, the trial-independent setup gave better results with both models than the trial-dependent one.
Table 2.
Comparison of classification accuracy in different setups.
| Classification | Naïve Bayes | SVM | ||||
|---|---|---|---|---|---|---|
| Setup | HF | CLL | Combined | HF | CLL | Combined |
| Trial dependent | 70.4 | 71.7 | 69.3 | 75.2 | 77.7 | 78.1 |
| Trial independent | 71.3 | 74.2 | 72.6 | 79.8 | 82.1 | 80.6 |
Feature Analysis
The Naïve Bayes classifier assumes that each of the features it uses is conditionally independent of the others, given some class label. SVMs do not make this assumption. They transform a given set of data points into a high dimensional space and construct a hyperplane for classification. We hypothesized that the Naïve Bayes model does not perform well because it does not consider the interaction between different features. In order to verify this, we evaluated the contribution of our various features towards the performance of the two models, using the trial independent setup. We compare feature contributions for both the Naïve Bayes and SVM classifiers. The following sections discuss experiments for these evaluations.
Naïve Bayes
We created simple classification models using the Naïve Bayes classifier in combination with the L1, L2, and SM feature sets individually. A fourth model was also created combining all the morphological and syntactic features. All models were created using the Weka24 toolkit. The results of these individual models are summarized in Table 3.
Table 3.
Accuracy using Naïve Bayes.
| Feature set | Constituents | Numberof features | Accuracy | ||||
|---|---|---|---|---|---|---|---|
| HF | CLL | Combined | HF | CLL | Combined | ||
| Majority | - | - | - | - | 64.1 | 54.4 | 59.1 |
| Bag of Words | L1 | 1874 | 1616 | 2769 | 68.8 | 72.0 | 70.4 |
| Bag of Concepts | L2 | 2246 | 1815 | 3255 | 65.1 | 70.4 | 68.7 |
| Bag of Semantic Types | SM | 101 | 102 | 110 | 67.9 | 69.1 | 65.6 |
| Morphology and Syntax | M1-M5, SY1-SY2 | 9 | 9 | 9 | 50.3 | 40.7 | 45.0 |
| All | M, L, SY, SM | 4230 | 3542 | 6143 | 71.3 | 74.2 | 72.6 |
Support Vector Machines
SVMs belong to a class of algorithms known as kernel methods, which have been shown to be very effective for classification. This is achieved through a kernel function, which is a similarity function K that maps two inputs x and y from a given domain into a similarity score that is a real number25. Formally, it is a function K(x,y) =< Ø(x), Ø(y) > → R, where Ø(x) is some feature function over instance x. For a function to be a valid kernel, it should be symmetric and positive semi-definite.
To analyze the contribution of different features, we constructed individual linear kernels (regularization parameter C=0.25) using each of the four feature sets described earlier: words, concepts, semantic types, and morphology and syntax features. This involves creating a kernel matrix (or gram matrix) comprised of similarity values between all pairs of instances in the dataset, obtained using the kernel function K. All models were implemented using the LibSVM26 library. Joachims et al.27 also showed that given two kernels K1 and K2, the composite kernel K(x,y) = K1(x,y) + K2(x,y) is also a valid kernel. This can be achieved by a simple addition of two kernel matrices. Thus, we combined different kernels to test the contribution of individual feature sets towards classification. The results are summarized in Table 4.
Table 4.
Accuracy using different feature kernels.
| Kernel | Features | Accuracy | ||
|---|---|---|---|---|
| HF | CLL | Combined | ||
| Majority | - | 64.0 | 54.4 | 59.1 |
| Words | L1 | 76.2 | 76.8 | 74.5 |
| Concepts | L2 | 75.0 | 76.8 | 75.7 |
| Semantic Types | SM | 74.1 | 70.0 | 69.6 |
| Morphology and Syntax | M1-M5, SY1-SY2 | 66.1 | 55.0 | 61.2 |
| All | L, SM, M, SY | 79.8 | 82.1 | 80.6 |
Comparing results from Table 3 and Table 4, we observe that SVMs perform better at the classification task by modeling interactions between different features. In, the following section we discuss the performance of this classifier across each of the class labels: “Structured”, “Unstructured” and ‘“Neither”. We also discuss common errors made by the classifier in this task.
Error Analysis
As discussed in the section on data creation, the corpus is skewed, with the majority of the eligibility criteria labeled as resolved from unstructured data. Table 5 presents the confusion matrix of the SVM classifier (using the “All” kernel combination from Table 4) on the combined data. These errors are representative of the individual datasets as well. Most of the errors stem from the class “Structured,” while the “Unstructured’ class has the least.
Table 5.
Confusion matrix and class-wise accuracy of the best performing classifier.
| Prediction | |||||
|---|---|---|---|---|---|
| Label | Unstructured | Structured | Neither | Number of instances | Accuracy |
| Unstructured | 768 | 72 | 19 | 859 | 89.4 |
| Structured | 129 | 251 | 7 | 387 | 64.8 |
| Neither | 46 | 7 | 144 | 197 | 73.1 |
We examined the criteria for these errors. Many criteria are simply too brief and the classifier has very little context to predict the right label. For example, the criteria “Any etiology” or “Diabetes Mellitus” are labeled as “Structured’ but misclassified as “Unstructured.” Similarly, “Previous exposure to KW-3902” or “End stage cancer” are labeled as “Unstructured’ but predicted as “Structured.”
Interestingly, some errors also stemmed from information that would be classified as “semi-structured” data by humans. These were discussed by the annotators during our training rounds for consensus. For example, one of the guidelines was to label criteria associated with numerical fields found in ECG and EKG data as “Unstructured,” since they are not present as discrete values in our EHR. The classifier made errors for statements such as “EF < 45%” or “QRS > 120 msec.” Similar observations can also be made for criteria such as “Folstein MMSE score less than 20” or “Walk < 450 meters during 6 minute hall walk test.”
Although certain criteria statements had a single class label assigned to them, there was a possibility of multiple labels being associated with them. For example, consider the criterion statement: “Adult patients age >= 18 admitted with CHF exacerbation with NYHA Class III-IV symptoms at screening.” The annotators marked this criterion as “Unstructured,” whereas our model classified it as “Structured”. One can observe that this statement has two subcriteria: one related to the patient being adult, and the other one about her CHF. While the age related part can be resolved using structured data, the CHF part requires reading clinical notes. A similar claim can be made for “WBC > 1.5; ANC >500; Plt >50,000 unless documented as due to disease” which was marked “Unstructured’ by our annotators (because of the judgment required to determine if these laboratory abnormalities were related to the disease) and predicted “Structured’ by our model.
Some kinds of criteria were quite rare and hence the model was not able to predict the correct label. For example, the criterion “Currently a prisoner” was labeled as “Structured’ but predicted as “Unstructured.” This is the sole criterion about prisoners in our dataset. Similarly, “Inability to lie flat for MR study” is labeled as “Neither’“ and misclassified as “Unstructured.” There were some obvious generalization errors as well. For example, “Current treatment on any Class I or III antiarrhythmic, except amiodarone.” which was labeled as “Structured’ but incorrectly predicted as “Unstructured.” In the trials for heart failure, mentions of “Class” are associated with New York Heart Association (NYHA) Class I, II, III or IV and are labeled as “Unstructured’ multiple times in the dataset. Similarly, for the criterion “Patients who are unable to refrain from taking acetaminophen” the phrase “unable to refrain” makes it “Unstructured,” however the remaining is a medication that can be looked up in the structured medication list, so our model classified it as “Structured.” A larger labeled dataset or lexical features with longer n-grams could potentially avoid such errors.
Some errors could not be explained. For example, “Patients having undergone revascularization procedures within 6 months” or “Current treatment of calcium channel blockers except for long acting dihydropyridines” were labeled as “Structured’ and misclassified as “Unstructured.” Similarly, “End-stage renal disease on dialysis or imminent” was labeled as “Unstructured’ (would not be able to determine “imminent” without reading a note) and misclassified as “Structured.
Discussion
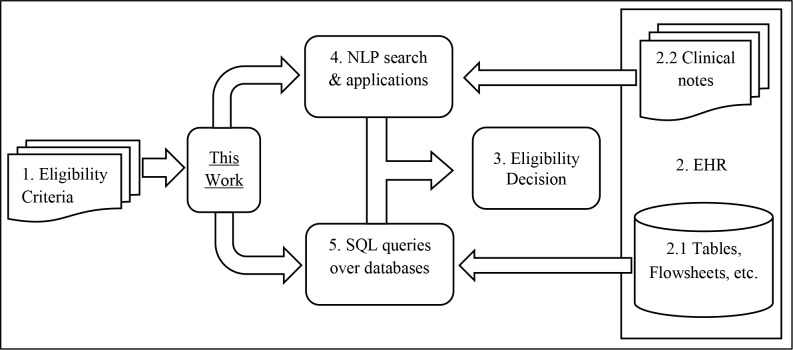
Figure 2 outlines the role of our work in an automated clinical trial screening workflow. A number of clinical research studies mandate selecting patients as subjects based on stringent eligibility criteria. Clinical trials are a particular type of such a research study. These eligibility criteria are specified in natural language text (block 1), and trial coordinators examine these criteria to locate information in the EHR (block 2) that can be used to resolve the criteria. This information is present in the form of structured content such as tables and flowsheets (block 2.1), or in the form of clinical notes and reports (block 2.2). Eligibility decisions (block 3) are made based on evidence from these sources.
Figure 2.
Role of this work in an automated clinical trial screening workflow
There has been a lot of research focused around introducing automation into the process of cohort identification, with various efforts in the research community addressing different stages in the screening process. Recent studies in NLP focus on the development of techniques that can be used to automate cohort identification9,28. These can assist the human effort involved in reading the unstructured content from the EHR for eligibility determination (block 4). A number of studies in the past have proved that structured information such as billing information29 can be used effectively for cohort identification. These are stored as structured content in the EHR in the form of databases. Simple SQL queries can be used to extract this information (block 5). There has been a large body of work to translate eligibility criteria into such queries11. However, to the best of our knowledge, there has been no work in the past to triage an eligibility criterion along one of these two routes. This work is an effort towards introducing automation in this step of the workflow. This work has utility in various applications such as clinical question-answering30 and decision support31 where the same problem of choosing a preferred data source for evidence gathering is encountered.
Limitations
This is an initial investigation for a potentially larger problem and is not without limitations. As discussed in the section on dataset creation, we made some assumptions for the task of annotation. Our most crucial assumption was the availability of data in a structured field; in practice, presence of a structured field in the EHR does not imply availability and reliability of the values in that field. Retrospectively, we realized that although the annotation team had high agreement, some decisions were subjective based on the annotators’ knowledge of our EHR, and could differ at other institutions. In some cases, the criterion may be best resolved by looking at both structured and unstructured data which cannot be sped up despite the classification step using our model. Finally, annotations obtained for this study were based only on observing and reading the criteria texts. A retrospective study examining agreement for our annotations with actual screening undertaken by clinical trial coordinators would confirm our findings.
Conclusion
We introduced the problem of identifying the right data source type for resolving an eligibility criterion in clinical trial screening. A gold standard dataset for automating the task using 100 trials (50 for heart failure, and 50 for CLL) was created. We focused on the language used in the criterion text and designed a variety of intuitive features for determining which data source to use to best resolve a particular criterion. These include morphological, lexical, syntactic and semantic features. An empirical comparison of two popular machine learning algorithms shows that a combination of these features results in optimal performance using SVM as a classifier. We note that most of our remaining errors stem from structured criteria being predicted as unstructured. Our system’s current performance shows promise, but has room for improvement. Our work is a step towards efficient automation of clinical trial recruitment by triaging each eligibility criterion to the right data source for its resolution.
Acknowledgements
We would like to thank Jennifer Fox and Christy Starr for their help with the annotation effort. We would also like to thank Denis Griffis for his help in preparing in this paper. Research reported in this publication was supported by the National Library of Medicine of the National Institutes of Health under award number R01LM011n6, the Intramural Research Program of the National Institutes of Health, Clinical Research Center, and through an Inter-Agency Agreement with the US Social Security Administration. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Prokosch HU, Ganslandt T. Perspectives for medical informatics. Reusing the electronic medical record for clinical research. Methods Inf Med. 2009 Jan.48(1):38–44. [PubMed] [Google Scholar]
- 2.Kopcke F, Prokosch H-U. Employing Computers for the Recruitment into Clinical Trials: A Comprehensive Systematic Review. J Med Internet Res. 2014 Jan.16(7):e161. doi: 10.2196/jmir.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Penberthy LT, Dahman BA, Petkov VI, DeShazo JP. Effort required in eligibility screening for clinical trials. J Oncol Pract. 2012 Nov.8(6):365–70. doi: 10.1200/JOP.2012.000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shivade C, Raghavan P, Fosler-Lussier E, Embi PJ, Elhadad N, Johnson SB, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc. 2014 Mar.21(2):221–30. doi: 10.1136/amiajnl-2013-001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenbloom ST, Denny JC, Xu H, Lorenzi N, Stead WW, Johnson KB. Data from clinical notes: a perspective on the tension between structure and flexible documentation. J Am Med Inform Assoc. 2011;18(2):181–6. doi: 10.1136/jamia.2010.007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raghavan P, Chen JL, Fosler-lussier E, Lai AM. How essential are unstructured clinical narratives and information fusion to clinical trial recruitment?. Proceedings of the 2014 AMIA Clinical Research Informatics Conference; 2014; pp. 218–23. [PMC free article] [PubMed] [Google Scholar]
- 7.Köpcke F, Trinczek B, Majeed RW, Schreiweis B, Wenk J, Leusch T, et al. Evaluation of data completeness in the electronic health record for the purpose of patient recruitment into clinical trials: a retrospective analysis of element presence. BMC Med Inform Decis Mak. 2013 Jan.:37. doi: 10.1186/1472-6947-13-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L, Chase HS, Patel CO, Friedman C, Weng C. Comparing ICD9-encoded diagnoses and NLP-processed discharge summaries for clinical trials pre-screening: a case study. Proceedings of the AMIA Annual Symposium. 2008:404–8. [PMC free article] [PubMed] [Google Scholar]
- 9.Shivade C, Hebert C, Loptegui M, de Marneffe MC, Fosler-Lussier E, Lai AM. Textual inference for eligibility criteria resolution in clinical trials. J Biomed Inform. 2015 doi: 10.1016/j.jbi.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ni Y, Kennebeck S, Dexheimer JW, McAneney CM, Tang H, Lingren T, et al. Automated clinical trial eligibility prescreening: increasing the efficiency of patient identification for clinical trials in the emergency department. J Am Med Inform Assoc. 2014 Jul. doi: 10.1136/amiajnl-2014-002887. amiajnl – 2014–002887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weng C, Tu SW, Sim I, Richesson R. Formal representation of eligibility criteria: a literature review. J Biomed Inform. 2010 Jun.43(3):451–67. doi: 10.1016/j.jbi.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross J, Tu S, Carini S, Sim I. Analysis of eligibility criteria complexity in clinical trials. AMIA Summits Transl Sci Proc. 2010 Jan.2010:46–50. [PMC free article] [PubMed] [Google Scholar]
- 13.Van Spall HGC, Toren A, Kiss A, Fowler RA. Eligibility criteria of randomized controlled trials published in high-impact general medical journals: a systematic sampling review. JAMA. 2007 Mar.297(11):1233–40. doi: 10.1001/jama.297.11.1233. [DOI] [PubMed] [Google Scholar]
- 14.Sim I, Olasov B, Carini S. An ontology of randomized controlled trials for evidence-based practice: content specification and evaluation using the competency decomposition method. J Biomed Inform. 2004 Apr.37(2):108–19. doi: 10.1016/j.jbi.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Tu SW, Kemper CA, Lane NM, Carlson RW, Musen MA. A methodology for determining patients’ eligibility for clinical trials. Methods Inf Med. 1993 Aug.32(4):317–25. [PubMed] [Google Scholar]
- 16.Metz JM, Coyle C, Hudson C, Hampshire M. An Internet-based cancer clinical trials matching resource. J Med Internet Res. 2005 Jul 1;7(3) doi: 10.2196/jmir.7.3.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo Z, Yetisgen-Yildiz M, Weng C. Dynamic categorization of clinical research eligibility criteria by hierarchical clustering. J Biomed Inform. 2011 Dec.44(6):927–35. doi: 10.1016/j.jbi.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacharya S, Cantor MN. Analysis of eligibility criteria representation in industry-standard clinical trial protocols. J Biomed Inform. 2013 Oct.46(5):805–13. doi: 10.1016/j.jbi.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 19.Levy-Fix G, Yaman A, Weng C. Structuring Clinical Trial Eligibility Criteria with the Common Data Model. Joint Summits on Clinical Research Informatics. 2015.
- 20.Hong N, Li D, Yu Y, Liu H, Chute CG, Jiang G. Representing Clinical Diagnostic Criteria in Quality Data Model Using Natural Language Processing. Workshop on Biomedical Natural Language Processing. 2015. pp. 177–81.
- 21.LingPipe 4.1.0. 2008. Available from: http://alias-i.com/lingpipe/
- 22.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proceedings of the Annual AMIA Symposium. 2001. pp. 17–21. [PMC free article] [PubMed]
- 23.Klein D, Manning CD. Accurate unlexicalized parsing. Proceedings of the 41st Annual Meeting on Association for Computational Linguistics-Volume 1; 2003; pp. 423–30. [Google Scholar]
- 24.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software. ACM SIGKDD Explor Newsl. ACM. 2009 Nov.11(1):10. [Google Scholar]
- 25.Hofmann T, Schölkopf B, Smola AJ. Kernel methods in machine learning. Ann Stat. Institute of Mathematical Statistics. 2008 Jun.36(3):1171–220. [Google Scholar]
- 26.Chang C-C, Lin C-J. LIBSVM: A library for support vector machines. ACM Trans Intell Syst Technol. ACM. 2011;2(3):27. [Google Scholar]
- 27.Joachims T, Cristianini N, Shawe-Taylor J. Morgan Kaufmann Publishers Inc; 2001. Composite Kernels for Hypertext Categorisation. Proceedings of the Eighteenth International Conference on Machine Learning; pp. 250–7. [Google Scholar]
- 28.Ni Y, Kennebeck S, Dexheimer JW, McAneney CM, Tang H, Lingren T, et al. J Am Med Inform Assoc. 1. Vol. 22. The Oxford University Press: 2015. Jan. Automated clinical trial eligibility prescreening: increasing the efficiency of patient identification for clinical trials in the emergency department; pp. 166–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birman-Deych E, Waterman AD, Yan Y, Nilasena DS, Radford MJ, Gage BF. Accuracy of ICD-9-CM codes for identifying cardiovascular and stroke risk factors. Med Care. 2005 May;43(5):480–5. doi: 10.1097/01.mlr.0000160417.39497.a9. [DOI] [PubMed] [Google Scholar]
- 30.Demner-Fushman D, Lin J. Comput Linguist. 1. Vol. 33. MIT Press: 2007. Mar. Answering Clinical Questions with Knowledge-Based and Statistical Techniques; pp. 63–103. [Google Scholar]
- 31.Sim I, Gorman P, Greenes RA, Haynes RB, Kaplan B, Lehmann H, et al. J Am Med Informatics Assoc. 6. Vol. 8. The Oxford University Press: 2001. Nov. Clinical Decision Support Systems for the Practice of Evidence-based Medicine; pp. 527–34. [DOI] [PMC free article] [PubMed] [Google Scholar]