Abstract
Viral testing for pediatric inpatients with respiratory symptoms is common, with considerable associated charges. In an attempt to reduce testing volumes, we studied whether data available at the time of admission could aid in identifying children with low likelihood of having a particular viral origin of their symptoms, and thus safely forgo broad viral testing. We collected clinical data for 1,685 pediatric inpatients receiving respiratory virus testing from 2010-2012. Machine-learning on the data allowed us to construct pre-test models predicting whether a patient would test positive for a particular virus. Text mining improved the predictions for one viral test. Cost-sensitive models optimized for test sensitivity showed reasonable test specificities and an ability to reduce test volume by up to 46% for single viral tests. We conclude that diverse forms of data in the electronic medical record can be used productively to build models that help physicians reduce testing volumes.
Introduction
Upper respiratory infections comprise one of the most common emergency department (ED) diagnoses in the pediatric population accounting for up to 25% of visits during influenza seasons1,2. In the majority of children, the respiratory viral infection is mild and considered self-limiting with adequate supportive care3–5. A large variety of viruses may cause respiratory symptoms and routine testing for key viruses is now available for children visiting the ED6–11.
The clinical utility of routine virus testing has been demonstrated previously, with children testing positive receiving less antibiotics and incurring fewer overall charges12,13. Also, children with positive results that are admitted after their ED visit may be properly isolated to prevent the spread of hospital-acquired infections14. However, with most children testing negative for the majority of viruses on the routine panels, the high billable charges of these panels, and the self-limiting nature of most virus infections, there are open questions about the overall utility of routine viral testing for ED visits.
There has been growing interest in the secondary use of EMR data to aid in clinical decision making and test ordering. Recognizing the prevalence of overtesting in the ICU setting, one group used fuzzy modeling of readily available data, like vital signs, urine collection, and transfused products, to identify tests that likely would not contribute to information gain in patients with gastrointestinal bleeding15. Other studies have looked to clinical documentation as a source of variables for developing classifiers that screen for disease. For example, emergency department free-text reports were used as the source of data for machine learning classifiers that outperformed expert Bayesian classifiers in the detection of influenza16. Another study has also employed the use of natural language processing of clinical notes to identify patients who may have Kawasaki disease, a diagnosis that is notoriously difficult to make17.
Our goal was to develop a practice-based workflow for the evaluation and optimization of respiratory virus test utilization in the pediatric population. The workflow would use data that is available at the time of the ED visit. The workflow would be helpful in identifying children with low likelihood of having a particular viral origin of their symptoms, who could safely forgo broad viral testing. The overall goal would be the design of smaller, custom viral test panels that test only those viruses that are most likely present given the pre-test patient data.
Using a retrospective study design, we collected clinical, seasonal, and demographic data for patients that underwent viral testing, and identified key characteristics that are indicative for patients with subsequent negative test results. Using machine learning, we thus optimized a classifier for the robust identification of patients with negative test results based on pre-test data. Based on current clinical knowledge of pediatric respiratory virus infections, we hypothesized that age and season would be the strongest predictors of test result when used in machine learning classifiers. We also hypothesized that the inclusion of clinical concepts in admission notes as predictive variables would enhance classifier performance.
Methods
The study population consisted of general pediatric inpatients who received respiratory virus laboratory testing between March 2010 and March 2012. The following inclusion criteria were employed: adequate nasopharyngeal swab sample, 18 years of age and younger, general inpatient admission, and test ordering within two days of admission. The following data were collected and de-identified for each patient: age, gender, season of test ordering, test results, and history and physical admission notes. Patients were tested with DFA and/or PCR panels. The DFA panel consisted of tests for adenovirus, influenza A/B, parainfluenza 1-3, and RSV. The PCR panel tested for the same viruses in addition to human metapneumovirus and rhinovirus. The study was performed using protocols reviewed and approved by the Yale University Institutional Review Board.
Of the clinical natural language processing (NLP) tools available, MetaMap 2013 was used to identify medical concepts in the History of Present Illness of each note. MetaMap 2013 is a program made available by the National Library of Medicine that allows users to map biomedical text to the Unified Medical Language System Metathesaurus18,19. While clinical NLP frameworks exist that are arguably more powerful, like cTAKES, HITEX, and Sophia, MetaMap was chosen for its ease of implementation. The following options were used in MetaMap: restriction of terms to the SNOMED CT terminology, limitation of semantic types to “signs and symptoms” and “diseases and syndromes”, utilization of all derivational variants of a word, allowance for concept gaps, expansion of acronyms and abbreviations, and identification of negated concepts. The presence or absence of each mapped concept was added as additional variables for model development. To determine the precision of the MetaMap software when run on the corpus of clinical notes, concepts identified by MetaMap were compared to the original text. For 100 randomly selected notes, the identified concepts were reviewed and compared to the original statements by the physicians. Partial-match precision was calculated, as previously published20. Recall was not calculated because we were primarily interested in the concepts that MetaMap was actually able to identify and not terms outside of its matching capabilities.
Based on these variables, we developed models using machine learning algorithms to predict the likelihood that a given patient would test positive or negative for a specific virus. For each viral diagnosis, three models were built based on: 1) billing data alone, 2) MetaMap concepts, and 3) billing data in addition to MetaMap concepts. For this task, we used Weka 3.7.9, a freely available Java based implementation that implements numerous machine learning algorithms, as well as common tools for data mining, such as data pre-processing and attribute selection21. Using Weka, we performed attribute selection on the variables to determine which variables provided the greatest gain of information, given the specific viral test to be modeled22,23. Attribute selection reduced the number of variables for inclusion in each model from over 400 to less than 20, depending on the test. A cost sensitive classifier was applied on top of machine learning algorithms to weigh against false negative model predictions with varying cost thresholds24. The decision tree learning algorithm, J48 (the Java implementation of C4.5), was chosen for use in the development and evaluation of each model25. To reduce the likelihood of overfitting, we employed ten-fold cross-validation. Hyperparameter optimization was not explored in this study.
Using the sensitivities and specificities resulting from the numerous cost thresholds, we constructed a specialized receiver operating characteristic (ROC) curve, also referred to as an ROC instance-varying transformation26. We calculated the area under the curve (AUC) for each model using the trapezoidal rule. The standard error for each model’s ROC was calculated, as previously published27. To simulate a scenario in which tests can be selected individually for a customized panel (e.g. in house PCR vs. multiplex PCR), we created three different types of customized panels and modeled the performance of the panels on the study sample. For each type of customized panel, a given patient would be serially tested until either a test returned positive or the patient had received all of the recommended testing. We created a control panel in which patients were assigned a panel with a random order. A second panel was composed of a fixed order based on the prevalence of diagnoses. The third and experimental panel took into consideration the predictions generated by the decision tree models. Patients would be tested for viruses, based on the recommendation of the model and then ordered from highest to lowest prevalence, and testing would cease after a positive result or all negative results for the recommended tests.
Results
In total, 1,972 orders for respiratory virus testing were placed during 1,848 visits. For each patient, the test consisted of a panel of 6 viruses - adenovirus, influenza, parainfluenza, RSV, hMPV, and rhinovirus. Each hospital visit was associated with 1 or more orders for respiratory virus testing. Table 1 summarizes the age, gender, and season of the study population by etiology. Negative test results accounted for 69.5% of all tests ordered during the study period. Males comprised around half of each viral diagnosis except for adenovirus diagnoses of which males comprised 68.8%. The mean age for all viral diagnoses was less than 5 years of age, except for positive cases of influenza, where the mean age of diagnosis was 8.24.
Table 1.
Summary basic clinical variables of general admission pediatric inpatients by etiology (2010-2012)
| Cases | Adenovirus (n = 32) | Influenza (n = 40) | Parainfluenza (n = 93) | RSV (n = 234) | hMPV (n = 29) | Rhinovirus (n = 180) | Multiple viruses (n = 57) | Negative (n = 1519) |
|---|---|---|---|---|---|---|---|---|
| Males, No. (%) | 22 (68.8) | 21 (52.5) | 47 (50.5) | 125 (53.4) | 15 (51.7) | 91 (50.6) | 31 (54.4) | 858 (56.5) |
| Age, mean ± SD | 3.75 ± 3.78 | 8.24 ± 6.53 | 2.99 ± 4.10 | 2.14 ± 3.20 | 4.89 ± 5.00 | 4.40 ± 4.74 | 2.39 ± 3.39 | 4.88 ± 5.29 |
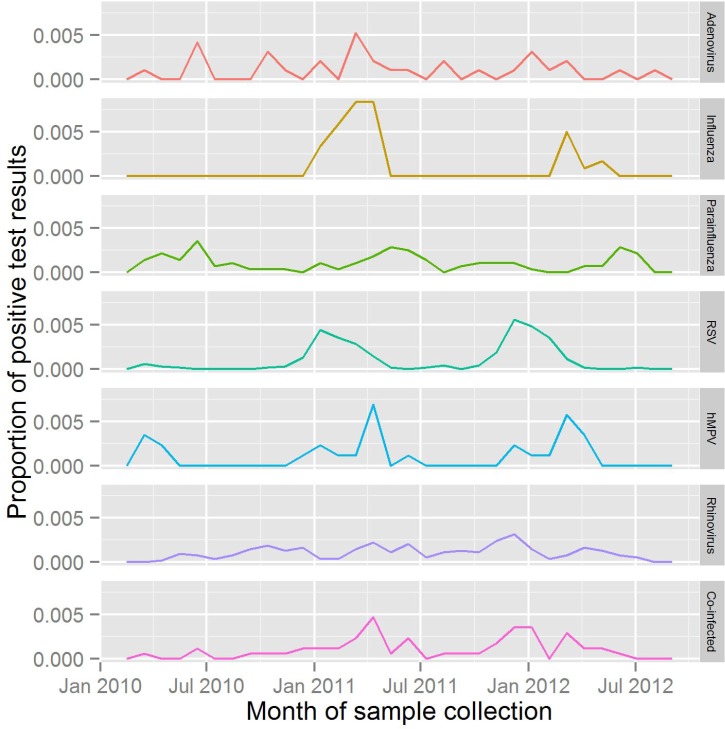
Both DFA and PCR panels are ordered throughout the year and show similar patterns in peak months of test ordering, during the winter seasons. DFA panels (n = 2152) were ordered 3.9 times more often than PCR panels (n = 550). Figure 1 shows the distribution of positive test results aggregated by month during the study duration. Adenovirus and rhinovirus are detected in all months with no clear pattern. Cases of co-infection, influenza, hMPV, parainfluenza, and RSV demonstrated regular seasonal fluctuations. The distribution of co-infected cases were present at low rates during the entire year, although demonstrated peaks in the winter months of each year. No cases of influenza occurred outside the winter or spring months in the study population. Test results for hMPV had a similar distribution to influenza infections. Cases of RSV arose primarily during the winter months, although the onset of the RSV season differed between the two years.
Figure 1.
Positive test results as a proportion of all test results for each detectable virus, by month.
In order to reduce viral testing in patients, we evaluated models for their predictive ability, based on pre-test data, to identify patients with low likelihood of a particular virus. Some models consumed data from text mining clinical notes. We evaluated the performance of the MetaMap program in identifying concepts by calculating the partial-match precision. The partial-match precision was calculated to be 0.724 across a random sample of 100 notes.
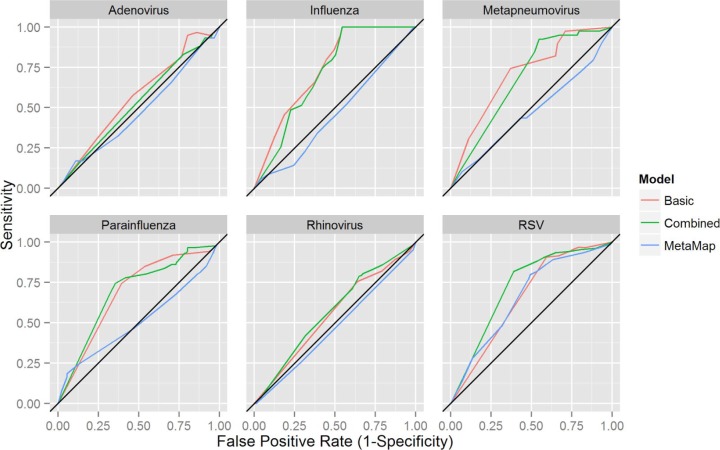
Using Weka, models based on the decision tree algorithm, J48, were derived. By varying the type of data used to derive the models, three types of models were developed for the detection of each virus. A basic model was derived using age, gender, and season. A text-mining model was derived using MetaMap concepts as predictive variables. A combined model was derived using age, gender, season, and MetaMap concepts. The ROC curves are depicted in Figure 2 and the corresponding AUC values are presented in Table 2. Basic models showed some predictive ability (AUC > 0.65) for four of the six viruses under study: influenza, parainfluenza, RSV, and hMPV. The text-mining models showed no predictive ability, except for RSV, where the text-mining model performed similarly to the basic model (AUC: 0.661 vs. 0.658, respectively). The combined models showed similar discriminative ability to the basic models for all viruses, except for RSV, where the combined model outperformed both the basic and text-mining.
Figure 2.
J48 decision tree classifier models predicting the outcome of laboratory tests for individual viruses.
Table 2.
Receiver operator characteristic curve characteristics
| ROC curve AUC (SE) | |||
|---|---|---|---|
| Basic model | MetaMap model | Combined model | |
| Adenovirus | 0.568 (0.114) | 0.480 (0.099) | 0.532 (0.108) |
| Influenza | 0.743 (0.126) | 0.451 (0.084) | 0.715 (0.122) |
| Parainfluenza | 0.686 (0.078) | 0.510 (0.061) | 0.694 (0.078) |
| RSV | 0.658 (0.048) | 0.661 (0.048) | 0.722 (0.051) |
| hMPV | 0.713 (0.143) | 0.474 (0.103) | 0.682 (0.138) |
| Rhinovirus | 0.549 (0.047) | 0.471 (0.041) | 0.570 (0.048) |
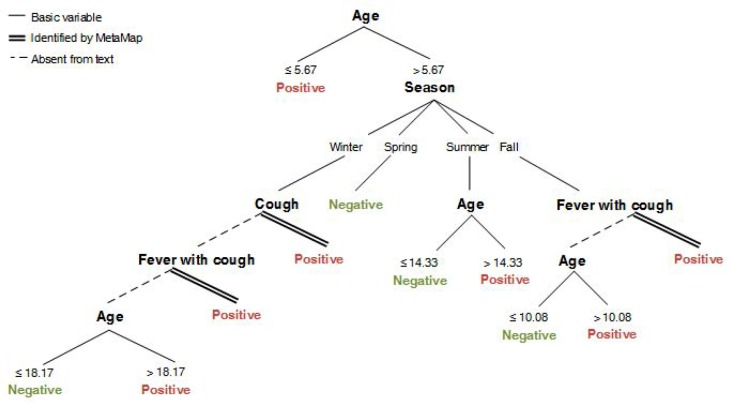
To determine how the MetaMap terms factor into the combined model for RSV, we mapped out the decision tree of the combined model (Figure 3). Age was the first branching point followed by branching points based on a combination of season, MetaMap concepts, and other age ranges.
Figure 3.
The graphical representation of the J48 decision tree classifier for RSV when based on both billing data and MetaMap concepts. Thresholds for age and season were determined by the J48 algorithm based on the information gain provided by the attribute splits. These are denoted by the single lines. The presence or absence of clinical terms in the HPIs are shown as branching nodes, followed double lines and dashed lines, respectively.
To assess how the models would perform as “in silico” screening tests prior to actual diagnostic testing, we computed the test characteristics for the combined models. Unlike the general model presented above, we would assess models with specific performance thresholds. At a target sensitivity of > 95% for the combined models for each virus, we calculated the sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) (Table 3). The target of ≥ 95% sensitivity (i.e. no more than 5% of cases with a respiratory infection are missed) was achieved by using a cost sensitive classifier that was applied on top of machine learning algorithms to weigh against false negative predictions. As detailed in Table 3, specificity ranged from 3.7% to 45.5% for all of the models at a sensitivity ~95%. A cost-sensitive classifier was thus able to safely sort patients into those that will eventually test positive for a virus (with at least 95% sensitivity), while robustly identifying patients that will not test positive (no viral infection). Some models were able to identify up to 45.5% of patients with no viral infection, and thus would lead to a sizeable reduction in test volume, as no testing is warranted. Models for adenovirus and rhinovirus showed the lowest specificities. While the PPV ranged from 50.1% to 64.7% for all models, the NPV ranged from 52.1% to 100%. For influenza, parainfluenza, RSV, and hMPV, the NPV was above 80.7%.
Table 3.
Test characteristics of combined model for each of the viruses
| Virus | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|
| Adenovirus | 0.966 | 0.037 | 0.501 | 0.521 |
| Influenza | 1.00 | 0.455 | 0.647 | 1.00 |
| Parainfluenza | 0.965 | 0.198 | 0.546 | 0.850 |
| RSV | 0.953 | 0.196 | 0.542 | 0.807 |
| hMPV | 0.974 | 0.205 | 0.551 | 0.887 |
| Rhinovirus | 0.943 | 0.076 | 0.505 | 0.571 |
*A minimum threshold sensitivity of 95% was set for each of the combined models (sensitivity of Rhinovirus was slightly below the set threshold)
In order to assess how model predictions might be useful in clinical practice, where patients are routinely tested for all 6 viruses, we simulated the effects of “customized panels” on the average number of tests that a given patient in the study population would need to undergo. In an environment where tests for individual viruses could be serially performed without being prohibitive in regard to time, we developed three types of customized panels that might be employed, as described in the methods section: 1) random order, 2) order based on the prevalence of a virus, 3) order based on model predictions. Each panel would test a patient serially until either a positive test result was returned or all tests on the panel had been performed. For the model based on random order, an average of 4.93 tests (SD = 1.69) would be performed; for the model based on prevalence, 4.52 tests (SD = 2.03; p < 0.001); and for the model based on the predictive models, 3.93 tests (SD = 1.64; p < 0.001). For the customized panel based on the predictive models, 22 patients would have been untested, who would have otherwise tested positive for a virus and 57 patients would have been missed for an additional viral co-infection.
Discussion
This retrospective study evaluated respiratory virus testing in the general pediatric population at a tertiary academic children’s hospital. Of the tests that were ordered, the vast majority resulted in a negative finding. Our objective was to develop clinical models using information available at the time of test ordering that might reduce test volume, while ensuring that patients with detectable infections are still tested, given the previously published benefits7, 12, 14, 28–33. This study found that predictive models built on clinical variables were able to discriminate patients who should receive testing better than chance. For 4 out of 6 viruses included in our study, age, season, and gender could be used to build models with fair predictive ability. We also hypothesized that concepts contained in the history of present illness portion of the clinician’s admission documentation could improve the predictive ability of our models. In the case of RSV, a model based only on concepts in the HPI had the same predictive power as the model based on billing data. Combining these two sources of data improved prediction of RSV. The precision of our concept matches via MetaMap were in line with previously published results34–36. Our findings suggest a useful role for admission notes, aside from documentation.
Effective implementation of these predictive models as screening tests requires practical understanding of the test characteristics as they relate to an institution’s clinical goals. By limiting testing of patients who would otherwise test negative, the population receiving diagnostic testing would be enriched for positive cases. A practical, cost-saving clinical model would thus strive for the highest possible sensitivity that is associated with a reasonable specificity. That is, high test sensitivity (almost no patient missed that will test positive) may be achieved while still achieving above zero specificity (patients with low likelihood of testing positive). We show, for example, that for influenza testing, we could achieve 100% sensitivity at 46% specificity. In other words, we can build a pre-test model that safely identifies all patients that are going to test positive for influenza, while identifying many patients that will not test positive, and for which no testing needs to be carried out. The model, as seen in Figure 1, is mostly determined by assessing the season of the patient visit.
It is important to note that the presented predictive models are not designed to predict positive cases with high accuracy. While we sought models that resulted in the highest sensitivities, these models often had very high false positive rates. Thus, in our models, the predictive model should be considered to predict “high risk for positive result” and “low risk for positive test result.” The distinction is important to the proposed function of the models, as the models were intended to aid the practitioners in reducing the number of overall tests that they order.
From a practical point of view, predictive models could act as “in silico” screening tests for whether or not patients should receive testing. If the models are validated against a prospective dataset, they might find practical application via integration into the electronic medical record, running in the background as the clinician is entering data about a patient. At the time of test ordering, the physician would be presented with information regarding what tests are likely negative or potentially positive. As a clinical decision tool at the point of care, the results of this model might encourage providers to limit testing to only those cases where the results would change clinical management. The use of basic variables, as well as documented clinical symptoms, can reduce test volume for certain tests by up to nearly half with a low false negative rate. In addition, the use of customized panels based on the predictive models would further reduce test volume. While the actual cost of testing is not transparent at most institutions, the amount charged for performing a respiratory virus panel is estimated to be around $1500, although this varies by institution and the exact type of testing performed. Ordering clinicians and patients could therefore expect the average charge to drop to $1000 if customized panels were employed. Future work will focus on refining the text mining approach and concept identification, as well as exploring the concept of customized panels, based on the prediction results of the models.
Our study has several limitations. The study population included only patients who received respiratory virus testing and selection criteria were not based on diagnostic codes. As a result, the seasonal prevalence of test results may not be reflective the true prevalence of viral infections. Furthermore, the focus on pediatric patients only accounted for about 20% of total respiratory virus testing volume, whereas testing on adults comprises the majority of testing. In addition, the numerous exclusion criteria may limit the generalizability of the models. While the billing data are true independent variables, the variables collected by text mining with MetaMap may not be fully independent. Because the timing of when admission notes were written could not be controlled, there is the possibility that some notes may have been written after test results were communicated. We took several measures to prevent this, including selecting the oldest admission note on record and manually reviewing notes for mention of testing. The text mining approach recorded concepts as “mentioned” versus “unmentioned,” which may not fully capture the whole clinical picture for a patient, as pertinent positives or negatives may have been unknowingly omitted by the author of the HPI. Our study also did not differentiate between different strains of viruses, which some of the tests are able to do (i.e. influenza A and B, parainfluenza 1-3). Initially, we expected that concepts from clinical notes would provide more robust prediction performance than we encountered. The reasons are likely multifactorial. First, the sample size that we used was small, compared to other NLP and machine learning efforts, making it less likely that we would be able to find concepts that would be able to differentiate between viruses. Second, given the high variability between providers’ styles when writing notes, the values for many concepts were left blank and were not subsequently imputed. Finally, the clinical manifestations of upper respiratory viruses are very similar with few distinguishing symptoms (i.e. barky cough for parainfluenza, gastrointestinal symptoms with adenovirus, and myalgia for influenza), making it difficult for physicians or the program to accurately diagnose viruses based on clinical history and exam alone, thus giving value to respiratory virus testing as a means of making a final diagnosis.
Conclusion
The results presented here offer a new perspective on analyzing testing practices for respiratory viruses using data mining and natural language processing techniques. We find that in a tertiary academic children’s hospital, the majority of respiratory virus testing returns negative. Furthermore, our results suggest that additional clinical factors may be used in a clinical model to predict the likelihood of an infection and the need for further diagnostic testing. Text mining of clinical notes may augment the predictive power of future models, as demonstrated in our models of RSV. This work contributes to the growing body of evidence that diverse forms data in the electronic medical record, not just billing data, can be used productively to build models that aid physicians in decision making, and may help in reducing test volumes.
References
- 1.Krauss BS, Harakal T, Fleisher GR. The spectrum and frequency of illness presenting to a pediatric emergency department. Pediatr Emerg Care. 1991 Apr;7(2):67–71. doi: 10.1097/00006565-199104000-00001. journals.lww.com. [DOI] [PubMed] [Google Scholar]
- 2.Silka PA, Geiderman JM, Goldberg JB, Kim LP. Am J Emerg Med. 7. Vol. 21. Elsevier; 2003. Nov, Demand on ED resources during periods of widespread influenza activity; pp. 534–9. [DOI] [PubMed] [Google Scholar]
- 3.Campbell AJP, Wright PF. Parainfluenza Viruses. In: Nelson Textbook of Pediatrics. Elsevier. 2011:1125–6.e1.. [Google Scholar]
- 4.Hall CB. Respiratory syncytial virus and parainfluenza virus. N Engl J Med. 2001 Jun 21;344(25):1917–28. doi: 10.1056/NEJM200106213442507. [DOI] [PubMed] [Google Scholar]
- 5.Peltola V, Waris M, Osterback R, Susi P, Hyypiä T, Ruuskanen O. Clinical effects of rhinovirus infections. J Clin Virol. 2008 Dec 2;43(4):411–4. doi: 10.1016/j.jcv.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 6.Arnold JC, Singh KK, Spector SA, Sawyer MH. Undiagnosed respiratory viruses in children. Pediatrics. 2008 Mar 4;121(3):e631–7. doi: 10.1542/peds.2006-3073. [DOI] [PubMed] [Google Scholar]
- 7.Doan Q, Enarson P, Kissoon N, Klassen TP, Johnson DW. Rapid viral diagnosis for acute febrile respiratory illness in children in the Emergency Department. Cochrane Database Syst Rev. 2012 May;16(5) doi: 10.1002/14651858.CD006452.pub3. CD006452. [DOI] [PubMed] [Google Scholar]
- 8.Hall CB. Respiratory syncytial virus and parainfluenza virus. N Engl J Med. 2001 Jun 21;344(25):1917–28. doi: 10.1056/NEJM200106213442507. [DOI] [PubMed] [Google Scholar]
- 9.Ren L, Xiang Z, Guo L, Wang J. Viral infections of the lower respiratory tract. Curr Infect Dis Rep. 2012 Jun 30;14(3):284–91. doi: 10.1007/s11908-012-0258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landry ML, Ferguson D. SimulFluor respiratory screen for rapid detection of multiple respiratory viruses in clinical specimens by immunofluorescence staining. J Clin Microbiol. 2000 Feb 3;38(2):708–11. doi: 10.1128/jcm.38.2.708-711.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Syrmis MW, Whiley DM, Thomas M, Mackay IM, Williamson J, Siebert DJ, et al. A sensitive, specific, and cost-effective multiplex reverse transcriptase-PCR assay for the detection of seven common respiratory viruses in respiratory samples. J Mol Diagn. 2004 May 21;6(2):125–31. doi: 10.1016/S1525-1578(10)60500-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barenfanger J, Drake C, Leon N, Mueller T, Troutt T. Clinical and financial benefits of rapid detection of respiratory viruses: an outcomes study. J Clin Microbiol. 2000 Aug 2;38(8):2824–8. doi: 10.1128/jcm.38.8.2824-2828.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulert GS, Lu Z, Wingo T, Tang Y-W, Saville BR, Hain PD. Role of a respiratory viral panel in the clinical management of pediatric inpatients. Pediatr Infect Dis J. 2013 May;32(5):467–72. doi: 10.1097/INF.0b013e318284b146. journals.lww.com. [DOI] [PubMed] [Google Scholar]
- 14.Mills JM, Harper J, Broomfield D, Templeton KE. Rapid testing for respiratory syncytial virus in a paediatric emergency department: benefits for infection control and bed management. J Hosp Infect. 2011 Mar 31;77(3):248–51. doi: 10.1016/j.jhin.2010.11.019. [DOI] [PubMed] [Google Scholar]
- 15.Cismondi F, Celi LA, Fialho AS, Vieira SM, Reti SR, Sousa JMC, et al. Reducing unnecessary lab testing in the ICU with artificial intelligence. Int J Med Inform. 2013 May 28;82(5):345–58. doi: 10.1016/j.ijmedinf.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.López Pineda A, Ye Y, Visweswaran S, Cooper GF, Wagner MM, Tsui FR. Comparison of machine learning classifiers for influenza detection from emergency department free-text reports. J Biomed Inform. 2015 Dec;58:60–9. doi: 10.1016/j.jbi.2015.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doan S, Maehara CK, Chaparro JD, Lu S, Liu R, Graham A, et al. Building a Natural Language Processing Tool to Identify Patients with High Clinical Suspicion for Kawasaki Disease from Emergency Department Notes. Acad Emerg Med [Internet] 2016. Jan 30, Available from: http://dx.doi.org/10.1111/acem.12925. [DOI] [PMC free article] [PubMed]
- 18.Aronson AR, Lang F-M. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010 May 6;17(3):229–36. doi: 10.1136/jamia.2009.002733. jamia.bmjjournals.com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001 Feb 5;:17–21. ncbi.nlm.nih.gov. [PMC free article] [PubMed] [Google Scholar]
- 20.Pratt W, Yetisgen-Yildiz M. A study of biomedical concept identification: MetaMap vs. people; AMIA Annu Symp Proc; 2003. Jan 20, pp. 529–33. [PMC free article] [PubMed] [Google Scholar]
- 21.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA Data Mining Software: An Update.. SIGKDD Explor Newsl; ACM; New York, NY, USA. 2009. Nov, pp. 10–8. [Google Scholar]
- 22.Yang Y, Pedersen J. A comparative study on feature selection in text categorization; Fourteenth International Conference on Machine Learning; 1997. pp. 412–20. [Google Scholar]
- 23.Azhagusundari B, Thanamani A. Feature selection based on information gain. International Journal of Innovative Technology and Exploring Engineering. 2013;2(2) [Google Scholar]
- 24.Zhao H. Knowl Inf Syst. 3. Vol. 15. Springer-Verlag; 2008. Jun 1, Instance weighting versus threshold adjusting for cost-sensitive classification; pp. 321–34. [Google Scholar]
- 25.Quinlan JR. San Francisco. CA, USA: Morgan Kaufmann Publishers Inc; 1993. C4.5: Programs for Machine Learning. [Google Scholar]
- 26.Fawcett T. Pattern Recognit Lett. 8. Vol. 27. New York NY, USA: Elsevier Science Inc; 2006. Jun, ROC Graphs with Instance-varying Costs; pp. 882–91. [Google Scholar]
- 27.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982 Apr;143(1):29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 28.Bonner AB, Monroe KW, Talley LI, Klasner AE, Kimberlin DW. Impact of the rapid diagnosis of influenza on physician decision-making and patient management in the pediatric emergency department: results of a randomized, prospective, controlled trial. Pediatrics. 2003 Aug 5;112(2):363–7. doi: 10.1542/peds.112.2.363. [DOI] [PubMed] [Google Scholar]
- 29.Chapin K. Multiplex PCR for detection of respiratory viruses: can the laboratory performing a respiratory viral panel (RVP) assay trigger better patient care and clinical outcomes? Clin Biochem. 2011 May 1;44(7):496–7. doi: 10.1016/j.clinbiochem.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 30.Dundas NE, Ziadie MS, Revell PA, Brock E, Mitui M, Leos NK, et al. A Lean Laboratory: Operational Simplicity and Cost Effectiveness of the Luminex xTAG™ Respiratory Viral Panel. J Mol Diagn. 2011 Mar;13(2):175–9. doi: 10.1016/j.jmoldx.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fendrick AM, Monto AS, Nightengale B, Sarnes M. The economic burden of non-influenza-related viral respiratory tract infection in the United States. Arch Intern Med. 2003 Feb 24;163(4):487–94. doi: 10.1001/archinte.163.4.487. [DOI] [PubMed] [Google Scholar]
- 32.Papenburg J, Hamelin M-É, Ouhoummane N, Carbonneau J, Ouakki M, Raymond F, et al. Comparison of risk factors for human metapneumovirus and respiratory syncytial virus disease severity in young children. J Infect Dis. 2012 Jul 15;206(2):178–89. doi: 10.1093/infdis/jis333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Syrmis MW, Whiley DM, Thomas M, Mackay IM, Williamson J, Siebert DJ, et al. A sensitive, specific, and cost-effective multiplex reverse transcriptase-PCR assay for the detection of seven common respiratory viruses in respiratory samples. J Mol Diagn. 2004 May 21;6(2):125–31. doi: 10.1016/S1525-1578(10)60500-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pratt W, Yetisgen-Yildiz M. A study of biomedical concept identification: MetaMap vs. people; AMIA Annu Symp Proc; 2003. Jan 20, pp. 529–33. [PMC free article] [PubMed] [Google Scholar]
- 35.Aronson AR, Lang F-M. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010 May 6;17(3):229–36. doi: 10.1136/jamia.2009.002733. jamia.bmjjournals.com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meystre S, Haug PJ. Natural language processing to extract medical problems from electronic clinical documents: performance evaluation. J Biomed Inform. 2005 Dec 5;39(6):589–99. doi: 10.1016/j.jbi.2005.11.004. [DOI] [PubMed] [Google Scholar]