Abstract
Resolving word ambiguity in clinical text is critical for many natural language processing applications. Effective word sense disambiguation (WSD) systems rely on training a machine learning based classifier with abundant clinical text that is accurately annotated, the creation of which can be costly and time-consuming. We describe a double-loop interactive machine learning process, named ReQ-ReC (ReQuery-ReClassify), and demonstrate its effectiveness on multiple evaluation corpora. Using ReQ-ReC, a human expert first uses her domain knowledge to include sense-specific contextual words into the ReQuery loops and searches for instances relevant to the senses. Then, in the ReClassify loops, the expert only annotates the most ambiguous instances found by the current WSD model. Even with machine-generated queries only, the framework is comparable with or faster than current active learning methods in building WSD models. The process can be further accelerated when human experts use their domain knowledge to guide the search process.
Introduction
Clinical documents contain many ambiguous terms, the meanings of which can only be determined in the context. For example, the word “malaria” appearing in a clinician note may refer to the disease or the vaccine for the disease; the abbreviation “AB” may mean “abortion,” “blood group in ABO system,” “influenza type A, type B,” or “arterial blood,” depending on the context. Assigning the appropriate meaning (a.k.a., sense) to an ambiguous word, based on hints provided in the surrounding text, is referred to as the task of word sense disambiguation (WSD).1,2 WSD is a critical step towards building effective clinical natural language processing (NLP) applications, such as named entity extraction3,4 and computer-assisted coding.5,6
Among different approaches to inferring word senses in clinical text, supervised machine learning has shown very promising performance.7,8 Supervised machine learning methods typically build a classifier for each ambiguous word, which is trained on instances of these words in real context with their senses annotated, usually by human experts with required domain knowledge. To train an accurate WSD model, a large number of such annotated instances are needed,9 the curation of which can be costly as every instance has to be manually reviewed by domain experts. Many methods have been explored in the past to reduce this annotation cost.10–14 Among them, active learning, by inviting human experts to directly participate in the machine learning process, has proven to be an effective approach. The premise of active learning is its ability to reduce the number of judgment calls that human experts need to make while achieving the same results as having a fully annotated corpus, thus significantly reducing the amount of human labeling needed.14 As such, how to select the most informative instances to present to human experts to annotate is the key to success for the family of active learning based methods.
Existing active learning methods use different strategies to select the most informative instances for annotation.15 For example, some select the instance with the least confident prediction or the instance with competing label assignments. However, these strategies suffer from the “cold-start” problem: a number of precisely annotated examples for every sense are usually required to kick off the classifier. Further, a classical active learning procedure does not fully utilize the domain knowledge of human experts. For example, practicing physicians frequently write or read ambiguous words in their notes without any difficulties in conveying or understanding their meaning. They are able to do so largely because of the surrounding context of the ambiguous words; e.g., when AB is used as shorthand for “blood group in ABO system,” physicians know that it commonly appears as “blood type AB,” “AB positive,” or “AB negative.” These contextual words are strong indicators of the sense of an ambiguous word, which is invaluable to a WSD model but remains largely untapped by existing active learning methods.
In this paper, we demonstrate a method that capitalizes on human experts’ domain knowledge to improve the performance of interactive machine learning. We apply a framework that we recently developed, referred to as ReQ- ReC (ReQuery-ReClassify), to the problem of word sense disambiguation in clinical text. Originally designed for high-recall microblog and literature search,16’17 ReQ-ReC features a double-loop interactive search and classification procedure that effectively leverages the domain knowledge of human experts. In an outer loop (ReQuery) of the procedure, an expert searches and labels the instances of an ambiguous word along with sense-specific contextual words. Then, a ReQ-ReC system helps the expert compose additional search queries by suggesting other potentially useful contextual words. In an inner loop (ReClassify), the framework requests the expert to annotate the most informative instances selected from those retrieved by all previous queries and then use the results to update the classifier accordingly. An expert can flexibly switch between these two “teaching strategies:” (1) to generate initial examples of a particular sense by launching a keyword search, and (2) to provide fine-grained clarification by labeling the instances selected by the system. Empirical experiments on three different clinical corpora show that this framework is more effective in building accurate WSD models than current active learning methods, even if the expert solely relies on system suggested keywords.
Method
A. The ReQ-ReC Framework
A.I. Sample scenario
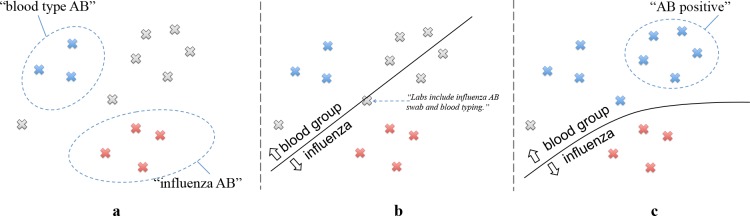
To illustrate how ReQ-ReC works, let us consider the following scenario. Suppose we have a set of clinical text snippets (e.g. sentences) all containing the word “AB,” which means either “blood group in ABO system” or “influenza type A, type B.” Our task is to assign the actual sense to each instance. Based on the domain knowledge, a human expert would know that if “AB” co-occurs with the phrase “blood type,” then it likely means “blood group in ABO system;” if it co-occurs with the word “influenza,” then it likely means “influenza type A, type B.” Naturally, the expert would use keywords “blood type AB” to retrieve a set of instances from the text corpus and label them as “blood group in ABO system;” she or he would then search for “influenza AB” and label the retrieved instances accordingly (Figure la). These context-sense pairs are used as an initial corpus to warm-start the first round of WSD model learning. The learned model will then be applied to predicting unlabeled instances and ask the expert to further clarify a few boundary cases, e.g. “Labs include influenza AB swab and blood typing.” (Figure lb). Determining the senses of these boundary cases would allow the model to capture the nuances in language use and quickly improve model accuracy. Later on, the expert may switch between searching for instances and labeling instances. After a few iterations, the expert may start to realize that in phrases such as “AB positive,” “AB” also means “blood group in ABO system.” Through a new search, she or he can quickly label another batch of instances of “AB positive,” which further improves the WSD model (Figure lc).
Figure 1.
An illustrative example of the searching and labeling process of the ambiguous abbreviation “AB.”
From this sample scenario several observations can be made. First, keyword search is a natural interface for domain experts to retrieve cases of ambiguous usage of words and to provide high-yielding, targeted annotation. This process can significantly reduce annotation cost, as human experts are only asked to label instances that are most informative to train the WSD model, while avoiding the need of labeling all instances in a corpus, most of which contribute little to improving the model performance. Additionally, search also benefits the learning algorithm: it provides a warm start in generating an initial model, and subsequent searches further refine the model by covering other potential senses of an ambiguous word or additional contextual words. Second, while classifying individual instances retrieved by keyword search is necessary for training the model, it is only able to produce a simplistic model, similar to rules. The ReQ-ReC framework therefore asks domain experts to also clarify boundary cases, which informs the model on how to weigh the nuances of language use in clinical text for better sense disambiguation. After being re-trained on these cases, the model becomes more robust and more accurate. In addition, answering these clarification questions might also inspire the human expert to come up with new search queries covering other potential senses of an ambiguous word or additional contextual words that might have not been thought about. Therefore, the two stages - keyword search and active classification - can be used iteratively to inform each other.
A.II. Anatomy of the ReQ-ReC framework
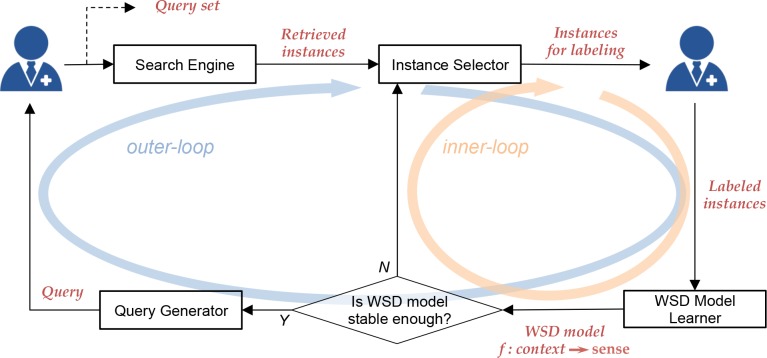
Generalizing from the above example, the double-loop procedure of the ReQ-ReC is illustrated in Figures 2. The procedure operates on an inverted index of the documents so that all keywords, including the ambiguous words and the contextual words, are searchable. The procedure maintains a set of search queries, a pool of retrieved instances, and a WSD model. To start, a human expert first uses her domain knowledge to compose a search query for each known sense, and then the system retrieves an initial set of contexts using the search function. The inner-loop kicks in there, in which the system iteratively presents a small number of instances selected from the current pool of retrieved instances to the expert and asks her/him to assign senses. The WSD model is consequently updated based on the accumulated annotations by the expert, which is then used to reclassify the pool of instances. After a few iterations of the inner-loop, the WSD model’s predictions stabilize on the currently unlabeled instances. At this point, the outer-loop of the system will kick in to recommend new search queries for each sense (the requeryprocess), aiming to retrieve more diverse instances with additional contextual words. These new search queries will be presented to the human expert for review and for further modification. Then, the system will retrieve a new set of instances using the new queries and add them to the existing pool of retrieved instances. After this requery process, the system will start a new inner-loop and continue to update the WSD model. The learning process ends when the expert is satisfied with the predictions made by the WSD model on those unlabeled instances in the newly retrieved pool.
Figure 2.
The ReQ-ReC framework.
The framework consists of the following key computational components.
Search. The framework uses a standard, Google-like search interface to retrieve instances containing ambiguous words. It can either leverage an existing clinical text search engine or build a just-in-time index over an unlabeled corpus while search is being performed. The search engine’s ranking function can use any retrieval models that take the input of a keyword query and outputs a certain number of instances from the index (using a vector space model, a boolean retrieval model, or a language modeling approach). After each search, the retrieved instances will be added to a pool, which becomes the basis of the next step, instance selection.
Instance selection. In every iteration of the inner-loop, the framework selects a small number (e.g. 5) of unlabeled instances from the current pool of retrieved instances and asks the expert to assign senses. At the beginning of the double-loop procedure, the framework can simply return the top instances ranked by the search engine’s retrieval model. With more and more instances being labeled by the human expert, the system will leverage this knowledge to update the WSD classifier and use active learning strategies to select the next instances for labeling.
WSD classification. Given an accumulated set of labeled instances, the WSD classification component learns or updates a multiclass classifier (such as a random forest or a support vector machine) and reclassifies the pool of retrieved instances.
Query expansion. When the classifier appears to be achieving a stable prediction on the pool of retrieved instances, the system proceeds to expand the pool in order to cover more contexts in which a sense may appear. This is done through constructing a new query for each sense and retrieving a new set of instances from the target corpus. Query expansion can be done using different methods, such as the Rocchio’s method or semantic term matching.18”20 The human expert may either approve this query, edit it, or compose one by her own.
ReQ-ReC is a general framework and each of the key components above can be instantiated in many different ways. In the following subsection, we describe specific implementations of each component.
A.III. Instantiating the ReQ-ReC framework
Search. In our current research implementation of the ReQ-ReC framework, we use the Lucene Package to build a search index for each ambiguous word.21 Instances are tokenized with Lucene’s StandardAnalyzer. To preserve the original form of ambiguous words (“nursing,” “exercises”) and negations (“no,” “without”), we do not perform stemming or stopword removal. We use the Dirichlet prior retrieval function with the parameter u set to 2000, a typical setup in information retrieval literature.22
WSD classifier. We use logistic regression with linear kernel for WSD classification, implemented by the LIBLINEAR package.23 If an ambiguous word has two senses, we build a binary classifier; otherwise we build a one-versus-rest multiclass classifier. Logistic regression classifiers output well-calibrated probability predictions for each sense y and each instance x, which will be used by active learning algorithms (θ is the classification model parameter). We use presence/absence of the all unigrams appeared in the instance as features. For the L2-regularization hyperparameter C, we set it to 1.0 across all ambiguous words. This setting is comparable to previous reported studies.14
-
Instance selection. In the inner-loop, there are multiple possible methods for selecting the next instance for labeling:
- Random Sampling. The algorithm simply selects an instance from the unlabeled pool uniformly at random.
- Least Confidence. The algorithm selects the instance x with the least predicted probability , where is the most probable sense. Intuitively, the model has little confidence in predicting the sense of instance x as y*, therefore it is most uncertain about the sense of x In this case, expert advice would be needed.
- Margin. The algorithm selects the instance x with the least predicted , where y1 and y2 are the most and second most probable senses. Intuitively, the model may not be able to determine if y1 or y2 is the appropriate sense, therefore it needs further clarification from the human expert.
- Entropy. The algorithm selects the instance x with the highest prediction entropy. High entropy means that the current WSD model considers any sense assignment as almost equally probable. Expert advice is thus needed to resolve the confusion.
In our implementation, we use the margin based active learning strategy to select instances. Note that all four methods can be launched without the search component, which in effect reduces the ReQ-ReC into a classical active learning system. In the evaluation experiments reported in this paper, these methods will be used as baselines for comparison.
Query expansion. In the outer-loop, a new query can either be automatically generated by the system and reviewed and improved by human experts, or be composed manually. In this study, we consider the following two extreme strategies: (a) the system automatically generates a new query based on the current status of the WSD model with no human input; and (b) the human expert composes new queries solely based on her or his domain knowledge. These two strategies represent the “worst” scenario and a “desirable” scenario of ReQ-ReC. We use the Rocchio’s method to automatically generate the next query qy for every sense y.18 The basic premise of Rocchio’s method is to learn a new query vector that is related to sense y and far away from other senses.
In fact, we hope that the new query qy will not be too close to the known contexts in which sense y may appear. This would allow the framework to suggest to human experts other contexts of the sense that might not have been thought of. To achieve this goal, we use the “diverse” method developed for high-recall retrieval,17 which generates a new query that balances its relevance to the sense and the amount of diverse information it introduces to the current pool of instances. In the rest of the paper, this strategy is referred to as “machine-generated” queries or the “worst case” of ReQ-ReC.
We also simulate the scenario where human experts use domain knowledge to include contextual words into search queries. To do this, we rank all the contextual words, words appearing in at least one instance of the ambiguous word, by the information gain, i.e. the reduction of uncertainty on the sense of the ambiguous word after seeing a contextual word.24 Top-ranked contextual words are considered as informative and used as search queries to warm- start the initial model learning. In our experiment, the simulated expert guides the first 6 queries using the top 30 contextual words.† As a simulation of domain knowledge, information gain is computed based on the entire set of labeled instances. Note that information gain is only a crude measure for selecting informative contextual words; human experts can do better with their domain knowledge. This simulation would result in an underestimate of the true performance of ReQ-ReC. We denote this scenario as ReQ-ReC with “simulated expert” queries.
Evaluation Methodology
B.I. Evaluation corpora
In this study, we used three biomedical corpora to evaluate the performance of the ReQ-ReC framework.
The MSH corpus contains MEDLINE abstracts automatically annotated using MeSH indexing terms.8 Originally, it has 203 ambiguous words, including 106 abbreviations, 88 words, and 9 terms that are a combination of abbreviations and words. Following previous work,14 we only included ambiguous words that have more than 100 instances so we have sufficient data for training and evaluation. This results in 198 ambiguous words.
The UMN corpus contains 75 ambiguous abbreviations in clinical notes collected by the Fairview Health Services affiliated with the University of Minnesota.25 500 instances for each abbreviation were randomly sampled from a total of 604,944 clinical notes. Each instance is a paragraph in which the abbreviation appeared. In this study, we excluded unsure and misused senses in training and evaluation.
The VUH corpus contains 25 ambiguous abbreviations that appeared in admission notes at the Vanderbilt University Hospital.26 Similar to the MSH corpus, we only retained 24 abbreviations that have more than 100 instances. Each instance is a sentence in which the abbreviation appeared.
The statistics of the three corpora are summarized in Tables 1. We can see that the MSH corpus has the richest context in an instance and the least skewed distribution of senses for an ambiguous word. Because our main goal in this study was to compare the effectiveness of different learning algorithms, we did not further tune the context window size for each corpus.
Table 1.
Summary statistics of three evaluation corpora.
| #Ambiguous words | Average #instances/word | Average #senses/word | Average #tokens/instance | Average percentage of majority sense | |
|---|---|---|---|---|---|
| MSH | 198 | 190 | 2.1 | 202.84 | 54.0% |
| UMN | 75 | 500 | 5.4 | 60.59 | 73.8% |
| VUH | 24 | 194 | 4.3 | 18.73 | 78.3% |
B.II. Metrics
In this study, we used learning curves to evaluate the cost-benefit performance of different learning algorithms. A learning curve plots the learning performance against the effort required in training the learning algorithm. In our case, learning performance is measured by classification accuracy on a test corpus and effort is measured by the number of instances labeled by human experts. For each ambiguous word, we divided its data into an unlabeled set and a test set. When a learning algorithm is executed over the unlabeled set, a label is revealed only if the learning algorithm asks for it. As more labels are accumulated, the WSD model is continuously updated and its accuracy continuously evaluated on the test set, producing a learning curve. To reduce variation of the curve due to differences between the unlabeled set and the test set, we ran a 10-fold cross validation: 9 folds of the data are used as the unlabeled set and 1 fold used as the test set. The learning curve of the algorithm on the particular ambiguous word is produced by averaging the 10 curves. The aggregated learning curve of the algorithm is obtained by averaging the curves on all ambiguous words in an evaluation corpus.
To cope with the cold start problem of active learning algorithms, we randomly sampled one instance from each sense as the initial training set. To facilitate comparison, we used the same initial training set for random sampling and ReQ-ReC. The batch size of instance labeling was set to 1 for all learning algorithms, so that we could monitor the performance improvement by every increment in the training sample.
To summarize the performance of different learning algorithms using a composite score, we also generated a global ALC (Area under Learning Curve) for each algorithm on each evaluation corpus. This measurement was adopted in the 2010 active learning challenge.27 The global ALC score was normalized by the area under the best achievable learning curve (constant 1.0 accuracy over all points).
Results
We evaluated six interactive WSD algorithms (one trained on randomly sampled instances, three trained using active learning methods, and two using the worst case and the simulated expert case of ReQ-ReQ) on three biomedical text corpora (MSH, UMN, and VUH).
Tables 2 shows the global ALC scores for each learning algorithm on different evaluation corpora. ReQ-ReC with simulated expert queries consistently outperforms all other methods on all three corpora. On the MSH and VUH corpora, even the worst case of ReQ-ReC achieves higher ALC scores than all existing active-learning algorithms. On the UMN corpus, the worst case of ReQ-ReC is slightly outperformed by the margin active learning algorithm. Compared to other active learning methods, the worst case of ReQ-ReC has the highest ALC scores for 164 out of 297 words across three corpora (55.22%) (129/198 in MSH, 20/75 in UMN, and 15/24 in VUH). With simulated expert queries, ReQ-ReC has the highest ALC scores for 206 out of 297 words across the three corpora (69.36%) (156/198 in MSH, 35/75 in UMN, and 15/24 in VUH).
Table 2.
Average ALC scores for six learning algorithms.
| Random | Least Confidence | Margin | Entropy | ReQ-ReC worst case | ReQ-ReC expert | |
|---|---|---|---|---|---|---|
| MSH | 0.862 | 0.899 | 0.900 | 0.899 | 0.904 | 0.913 |
| UMN | 0.854 | 0.885 | 0.893 | 0.878 | 0.889 | 0.894 |
| VUH | 0.863 | 0.871 | 0.872 | 0.870 | 0.878 | 0.885 |
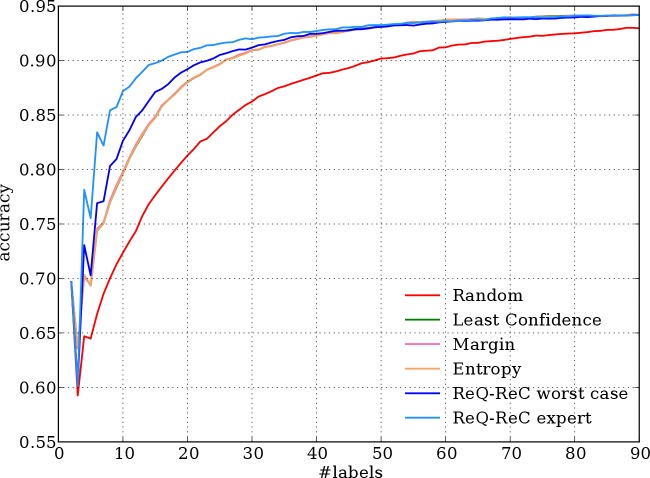
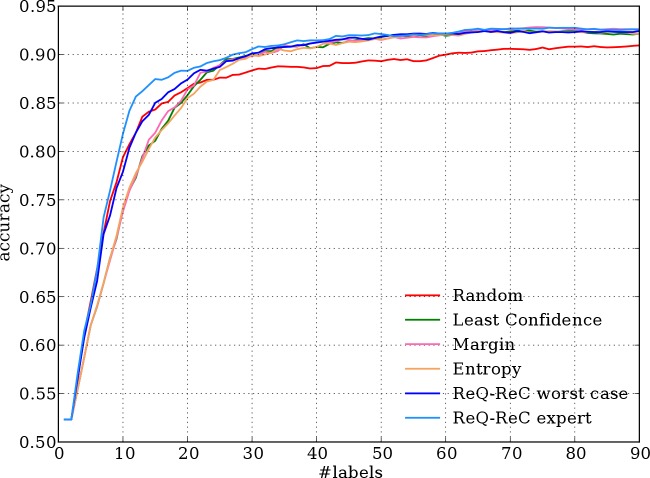
Figures 3, 4, and 5 shows the aggregated learning curves of all algorithms on three evaluation corpora, respectively. Results on the MSH corpus present the clearest patterns: the two ReQ-ReC methods learn faster than other algorithms, especially in the beginning stage (first 30 labels). The learning curves of three active learning algorithms are almost identical and much higher than that of random sampling, as previously reported.14 To achieve 90% accuracy, the best active learning algorithm requires 26 labels on average, while ReQ-ReC with simulated expert queries requires only 17 labels, saving 35% labeling effort.
Figure 3.
Aggregated learning curves of 198 ambiguous words in the MSH corpus.
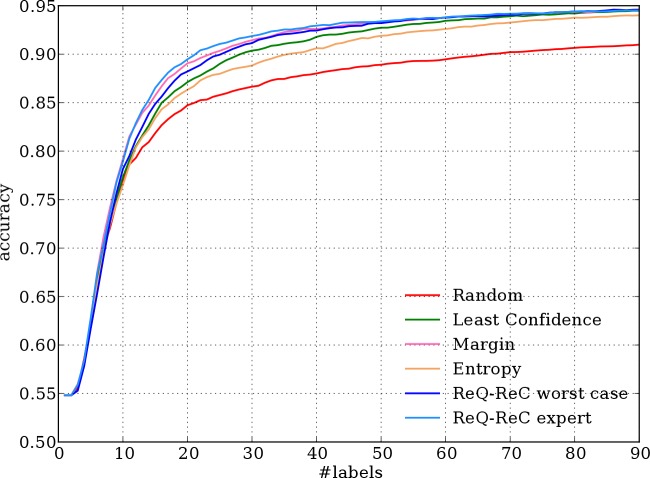
Figure 4.
Aggregated learning curves of 75 ambiguous words in the UMN corpus.
Figure 5.
Aggregated learning curves of 24 ambiguous words in the VUH corpus.
Patterns on the other two corpora are less significant, due to highly skewed sense distributions. In general, ReQ-ReC with simulated expert queries still achieves the best learning curve than other methods, but with a smaller margin, followed by an active learning algorithm on the UMN corpus and by the worst case of ReQ-ReC on the VUH corpus. Surprisingly, on the VUH corpus, random sampling learns faster than active learning methods at the very beginning. The benefit of active learning kicks in after 20 labels.
Discussion
The goal of inviting human experts into the machine learning process is to achieve large performance gains with relatively small labeling effort.15 An active learning process tries to select the next instance such that it brings in as large amount of fresh information as possible for the model to learn from, therefore giving rise to large gains. When asking for the next label, an active learner prefers to ask those instances that represent an unexplored subpopulation and/or instances whose labels the current model is still uncertain about. In contrast, a passive learner randomly picks the next instance from the unlabeled set, regardless of whether it overlaps with a previously labeled one, or whether the model can accurately guess its label, neither of which make the best use of the labeling effort.
WSD model learning benefits considerably from expert queries as a warm start. When the first few queries are informative contextual words, they construct a pool of representative instances. The initial WSD model learned on this representative pool inherits the domain knowledge from the search queries. Human experts can do even better than the simulated expert in composing these queries. Even when the queries are machine-generated, the query expansion procedure also picks up potentially informative contextual words. On the other hand, active learning methods select instances from the entire corpus rather than a representative pool. In the initial learning stage, models are usually poor and their predictions are unreliable.28 Thus the “uncertain” instances selected by such predictions may not benefit the learning as much as the representative ones. As the model becomes more robust in the later learning stage, the clarification questions raised by active learning will make more sense and labeling these instances can better improve the model.
Different characteristics of text documents affect learning process.29 In biomedical papers that are formally written (the MSH corpus), an ambiguous abbreviation often appears with its full form for clarification purposes, e.g. “high- risk (HR)” and “heart rate (HR).” The co-occurrence of the abbreviation with its full form greatly makes it easier for both the annotation process and the W SD model. In contrast, an ambiguous abbreviation in clinical notes (the UMN and VUH corpora) is almost never expanded to its full form as abbreviations are typically used to save the time of input. A clinical abbreviation can have many senses that are used in many different contexts. As a result, the annotation process for clinical abbreviations requires extensive search and labeling. Compared to active learning, the ReQ-ReC framework can better assist human experts in building clinical WSD models.
When an ambiguous word has many senses, the sense distribution is often highly skewed: one or two major senses cover more than 90% use cases, while many other senses are rarely used. As we can see in Tables 1, word senses of the two clinical corpora are highly skewed (for more than 4 senses, a majority guess has above 70% accuracy).
Skewed sense distribution presents challenge to machine learning.30 Without abundant labeled instances, it is difficult to learn a WSD model that accurately identifies a rare sense. The classification model will bias towards predicting the major senses and hurt the recall of the rare sense, which becomes an issue for high-stake events such as a rare disease. A straightforward way to cope with the rare sense learning problem is to harvest and label more data for the rare class, for which the first step is to search using contextual words. ReQ-ReC, originally designed for high-recall information retrieval, can be useful in searching for more rare senses.
This study has several limitations. First, in this study we assume the senses of an ambiguous word are known upfront and one instance is already available for each sense, which is a standard setup in the active learning literature. In reality human expert may have knowledge of some but not all of the senses; it is more natural to discover senses on the fly. Second, instead of using the simple bag-of-unigram features, we can use more elaborate features for WSD, e.g. part-of-speech tags, medical concepts (extracted by MetaMap), and word embedding. This could further improve the WSD performance. Third, the framework is only evaluated through simulated experiments and is not evaluated with real users.
Conclusion
In this paper, we describe a novel interactive machine learning framework that leverages interactive search and classification to rapidly build models for word sense disambiguation in clinical text. With this framework, human experts first use keyword search to retrieve relevant contexts in which an ambiguous word may appear to enable targeted, high-yielding annotation. This interactive active learning process, capitalizing on human experts’ domain knowledge, could therefore significantly reduce the annotation cost by avoiding the need to have a fully annotated corpus. Experiments using multiple biomedical text corpora show that the framework delivers comparable or even better performance than current active learning methods, even if human wisdom is not used to aid in the search process (i.e., all search queries are automatically generated by the algorithm). In future work, we will conduct more evaluation studies to assess the performance of the framework using real-world scenarios and real human experts.
Acknowledgements
We thank the anonymous reviewers for their constructive comments. This work is supported in part by the National Institutes of Health under grant NLM 2R01LM010681-05 and by the National Science Foundation under grant number IIS-1054199.
Footnotes
The first two queries use the top 10 words; the next two queries use the next top 10 words, and so forth.
References
- 1.Ide N, Veronis J. Introduction to the special issue on word sense disambiguation: the state of the art; Computational linguistics.; 1998. Mar 1, pp. 2–40. [Google Scholar]
- 2.Schuemie MJ, Kors JA, Mons B. Word sense disambiguation in the biomedical domain: an overview. Journal of Computational Biology. 2005;12(5):554–65. doi: 10.1089/cmb.2005.12.554. [DOI] [PubMed] [Google Scholar]
- 3.Uzuner O, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. Journal of the American Medical Informatics Association. 2011 Sep 1;18(5):552–6. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang B, Cao H, Wu Y, Jiang M, Xu H. Clinical entity recognition using structural support vector machines with rich features. Proceedings of the ACM sixth international workshop on Data and text mining in biomedical informatics; ACM; 2012. Oct 29, pp. 13–20. [Google Scholar]
- 5.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. Journal of the American Medical Informatics Association. 2004 Sep 1;11(5):392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanauer DA, Mei Q, Law J, Khanna R, Zheng K. Supporting information retrieval from electronic health records: A report of University of Michigan’s nine-year experience in developing and using the Electronic Medical Record Search Engine (EMERSE). Journal of biomedical informatics. 2015 Jun 30;55:290–300. doi: 10.1016/j.jbi.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu H, Teller V, Friedman C. A Multi-aspect Comparison Study of Supervised Word Sense Disambiguation. Journal of the American Medical Informatics Association. 2004 Apr 2;11(4):320–31. doi: 10.1197/jamia.M1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jimeno-Yepes AJ, McInnes BT, Aronson AR. Exploiting MeSH indexing in MEDLINE to generate a data set for word sense disambiguation.; BMC Bioinformatics; 2011. Jun 2, pp. 12–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu H, Markatou M, Dimova R, Liu H, Friedman C. Machine learning and word sense disambiguation in the biomedical domain: design and evaluation issues.; BMC bioinformatics; 2006. Jul 5, 1 pp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H, Johnson SB, Friedman C. Automatic resolution of ambiguous terms based on machine learning and conceptual relations in the UMLS. Journal of the American Medical Informatics Association. 2002 Nov 1;9(6):621–36. doi: 10.1197/jamia.M1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu H, Kim W, Hatzivassiloglou V, Wilbur WJ. Using MEDLINE as a knowledge source for disambiguating abbreviations and acronyms in full-text biomedical journal articles. Journal of Biomedical Informatics. 2007 Apr;40(2):150–9. doi: 10.1016/j.jbi.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Leroy G, Rindflesch TC. Using symbolic knowledge in the UMLS to disambiguate words in small datasets with a naive Bayes classifier.; Medinfo; 2004. pp. 381–5. [PubMed] [Google Scholar]
- 13.Pakhomov S, Pedersen T, Chute CG. Abbreviation and acronym disambiguation in clinical discourse.. InAMIA Annual Symposium Proceedings; American Medical Informatics Association; 2005. p. 589. [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Cao H, Mei Q, Zheng K, Xu H. Applying active learning to supervised word sense disambiguation in MEDLINE. J Am Med Inform Assoc. 2013 Sep;20(5):1001–6. doi: 10.1136/amiajnl-2012-001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Settles B. Active learning literature survey. University of Wisconsin-Madison. Computer Sciences Technical Report 1648. 2009.
- 16.Li C, Wang Y, Mei Q. A user-in-the-loop process for investigational search: Foreseer in TREC 2013 microblog track.; In Proceedings of the 22nd Text REtrieval Conference (TREC 2013); 2013. [Google Scholar]
- 17.Li C, Wang Y, Resnick P, Mei Q. ReQ-ReC: High Recall Retrieval with Query Pooling and Interactive Classification.; In Proceedings of the 37th International ACM SIGIR Conference on Research & Development in Information Retrieval; 2014. pp. 163–172. [Google Scholar]
- 18.Rocchio J. Relevance feedback in information retrieval. In. In: Salton G, editor. The SMART Retrieval System: Experiments in Automatic Document Processing. Englewood Cliffs NJ: Prentice-Hall; 1971. pp. 313–23. [Google Scholar]
- 19.Aronson AR, Rindflesch TC. Query expansion using the UMLS Metathesaurus.; Proceedings of the AMIA Annual Fall Symposium; 1997. pp. 485–489. [PMC free article] [PubMed] [Google Scholar]
- 20.Manning CD, Raghavan P, Schutze H. Introduction to information retrieval. Chapter 9: Relevance feedback and query expansion.; Cambridge university press; 2008. Jul 12, [Google Scholar]
- 21.Apache Lucene Project. Available from. http://lucene.apache.org/
- 22.Zhai C, Lafferty J. A study of smoothing methods for language models applied to Ad Hoc information retrieval.; Proceedings of the 24th annual international ACM SIGIR conference on Research and development in information retrieval; New York, NY, USA: ACM: 2001. pp. 334–42. [Google Scholar]
- 23.Fan R-E, Chang K-W, Hsieh C-J, Wang X-R, Lin C-J. LIBLINEAR: A library for large linear classification. The Journal of Machine Learning Research. 2008. pp. 1871–4.
- 24.Yang Y, Pedersen JO. A comparative study on feature selection in text categorization.; Proceedings of the Fourteenth International Conference on Machine Learning; San Francisco, CA, USA: Morgan Kaufmann Publishers Inc.; 1997. pp. 412–20. [Google Scholar]
- 25.Moon S, Pakhomov S, Liu N. Ryan JO, Melton GB. A sense inventory for clinical abbreviations and acronyms created using clinical notes and medical dictionary resources. J Am Med Inform Assoc. 2014 Mar;21(2):299–307. doi: 10.1136/amiajnl-2012-001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Y, Denny J, Rosenbloom ST, Miller RA, Giuse DA, Song M, et al. In: Proceedings of the 7th International Workshop on Data and Text Mining in Biomedical Informatics. New York, NY, USA: ACM; 2013. A Prototype Application for Real-time Recognition and Disambiguation of Clinical Abbreviations. pp. 7–8. [Google Scholar]
- 27.Guyon I, Cawley G, Dror G, et al. Results of the Active Learning Challenge.; JMLR: Workshop and Conference Proceedings; 2011. pp. 19–45. [Google Scholar]
- 28.Attenberg J, Provost F. Inactive learning?: difficulties employing active learning in practice. ACM SIGKDD Explorations Newsletter. 2011 Mar 31;12(2):36–41. [Google Scholar]
- 29.Savova GK, Coden AR, Sominsky IL, Johnson R, Ogren PV, de Groen PC, Chute CG. Word sense disambiguation across two domains: biomedical literature and clinical notes. Journal of biomedical informatics. 2008 Dec 31;41(6):1088–100. doi: 10.1016/j.jbi.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 30.He H, Garcia EA. Learning from imbalanced data. Knowledge and Data Engineering, IEEE Transactions on. 2009 Sep;21(9):1263–84. [Google Scholar]