Abstract
The heterogeneity of clinical data is a key problem in the sharing and reuse of Electronic Health Record (EHR) data. We approach this problem through the combined use of EHR standards and semantic web technologies, concretely by means of clinical data transformation applications that convert EHR data in proprietary format, first into clinical information models based on archetypes, and then into RDF/OWL extracts which can be used for automated reasoning. In this paper we describe a proof-of-concept platform to facilitate the (re)configuration of such clinical data transformation applications. The platform is built upon a number of web services dealing with transformations at different levels (such as normalization or abstraction), and relies on a collection of reusable mappings designed to solve specific transformation steps in a particular clinical domain. The platform has been used in the development of two different data transformation applications in the area of colorectal cancer.
Introduction
Electronic Health Record (EHR) systems are called to play an increasingly important role in health information technology. The quantifiable benefits mentioned in a 2006 AHRQ report1 include: savings from data capture and access; information connectivity for stakeholders; and decision support, improving efficiency, safety, and quality of healthcare. In many cases (e.g. information exchange, and interoperability with decision support systems), a key issue is dealing with the heterogeneity of clinical data. As a matter of fact, clinical data sources may differ in the data models, schemas, naming conventions and level of detail they use2. Several works define some sort of virtual health record (VHR) to bridge these differences. In order to achieve standardization, a number of works base their VHR on a standard EHR architecture3. Some authors take a step further and use clinical information models based on EHR standards for this purpose4,5. These models could come in the form of openEHR archetypes, CEN/ISO EN13606 archetypes, Clinical Document Architecture (CDA) templates, or Detailed Clinical Models (DCMs). An important role of such models is providing structure and terminology-based semantics to data instances that conform to some EHR model.
Beyond this, the semantic web has been proposed as the natural technological space for the integration and exploitation of biomedical data6. The semantic web7 describes a new form of web content meaningful to computers, in which the meaning is provided by ontologies. In the context of EHR systems, ontologies enable the formal representation of the entities and relationships involved in an EHR extract, and of the associated background knowledge. Linked Open Data8 (LOD) is the most prominent semantic web initiative to develop the Web of Data, in which datasets would be semantically connected over the Internet. LOD principles propose semantic publishing and sharing of data using RDF and OWL languages. Based on these contents, automated reasoners can be used to infer new information or to check logical data consistency.
In line with the above ideas, in a previous work9 we solved an EHR-driven phenotyping problem by means of a data transformation pipeline to convert EHR data stored in a proprietary database, first into information models based on openEHR archetypes, and then into RDF/OWL extracts which could ultimately be combined with an OWL domain ontology for automated reasoning. In the case study, an abstraction phase including certain calculations (e.g. counting, negation) was done at the archetype (or data) level, while the classification tasks were performed at the ontological (or knowledge) one. The separation of concerns between the archetype and the ontology levels was identified as a key factor in achieving the interoperability goals for the project at issue. The decision will depend on the project itself as well as on the features of the clinical information models used and/or the ontology language chosen. On the other hand, we concluded that a wide range of interoperability scenarios could be envisaged. For example, in a particular clinical domain, different information models may be under consideration. As another example, the transformation to the ontological level may not be required at all, or it may be limited to a set of RDF triples representing the EHR extract.
In this context, this paper describes a proof-of-concept platform to explore alternative solutions to a range of clinical data transformation problems. The platform is built upon a number of web services dealing with the transformation at the different levels. For this purpose, it relies on a collection of reusable fine-grained mappings designed to solve specific transformation steps (e.g. openEHR archetype to RDF) in a particular clinical domain (e.g. colorectal cancer). The paper is structured as follows. First we elaborate on the services offered by the platform, which we have classified according to the kind of operations they perform on the data: normalization, abstraction, semantic publishing, and reasoning services. Next we describe the web-based platform that we have implemented based on these services to evaluate the feasibility and utility of the idea, and explain how it has been used in two different data transformation scenarios in the domain of colorectal cancer. Lastly, we present some concluding remarks and outline future work.
Web services for clinical data transformation
Normalization service
Dual model EHR architectures (EHRA) encompass a reference model and set of archetypes for modelling EHRs. The reference model represents the generic and stable properties of the EHR. Conversely, archetypes are used to define domain-specific information models such as blood pressure recording, discharge report or a lab result. Notice that any reference model, such as ISO 13606-1, openEHR reference model or HL7 CDA, may be used as basis for defining archetypes.
The normalization service deals with one of the main problems when adopting EHR standards: how to standardize existing data. This involves transforming EHR content into a data structure compliant with both reference models and archetypes. We face a problem known in the literature as the data exchange (translation or transformation) problem10. This problem is a difficult one, since it deals with differences and mismatches between heterogeneous data formats and models. In the case of the EHR this problem is even more complex. On the one hand, we have the legacy data that conform to a particular schema, and with local semantics. On the other hand, we have EHR reference models and archetypes that have been defined without any regard to the internal architecture or database design of EHR systems. The main requirements on the target instances are that they shall be compliant with the target archetype, be non-redundant and be an accurate representation of the source data (EHR data to be normalized).
The output of this kind of services is an XML instance of the target schema (archetype). Our normalization process requires the source schema expressed as an XML Schema, the target archetype and a declarative mapping relating both schemas11. The mapping language is based on tuple-generating dependencies10 (tgds). They are expressive enough to represent, in a declarative way, many of the schema mappings of interest. The tgds basically specify how to compute a value for an atomic attribute of the target schema (archetype) by using a set of atomic elements from the data source. In our setting these value correspondences are defined by a set of pairs, consisting of a transformation function and a filter. The simplest kind of transformation function is the identity function which copies an atomic source value into a target atomic attribute. This is the most common transformation function in normalization scenarios since often they only involve restructuring source data to make them compliant with the target schema. In those cases where it is necessary to transform source atomic values, a wide range of transformation functions are supported. As an example, Table 1 contains a simple mapping transforming gender codes. It transforms the local gender code in the source path /patient/gender into a normalized code (0 for male, 1 for female and 9 otherwise). Note that the order of the filter-transformation function pairs is relevant and only the first applicable function is executed, consequently the last filter acts as a ‘default’ condition.
Table 1.
Example of mapping expression normalizing local gender codes.
| Filter | Transformation Function |
|---|---|
| /patient/gender=’M’ OR /patient/gender=’m’ | 0 |
| /patient/gender=’W’ OR /patient/gender=’w’ | 1 |
| /patient/gender=0 OR /patient/gender=1 | /patient/gender |
| true | 9 |
Value correspondences allow us to hide much of the structural complexity of archetypes and reference model. Users do not need to thoroughly specify the logical relations between all the entities of the source and target schemas. It is only necessary to specify the navigation path of the attributes used in the mapping. The semantics of the data transformation is complemented by a default grouping semantics which can be customized in different ways to cope with complex mapping scenarios. Finally, taking into account the mapping specification, the archetype constraints, and the source schema, an XQuery script is generated. The script takes as input an instance of the source EHR data and generates an XML document that is compliant both with the archetype and the underlying reference model. As a main advantage, this approach makes possible to separate the mapping specification from its implementation.
For the specification of the mapping and the generation of the normalization XQuery script we rely on the LinkEHR platform12,13. The normalization service has been implemented as a web service that takes as input the XQuery script and the source data. The actual data transformation is performed by an XQuery engine.
Abstraction service
While the normalization service focuses on the transformation of EHR data according to a standardized information structure, without significant modifications, the abstraction service deals with the transformations required when recorded data must be interpreted before use on the basis of clinical considerations. These transformations are generally based on specialized domain knowledge, and are often substantial in nature (e.g. the notion of severe comorbidity can be inferred from the existence of a total of 19 specific problems, but may not be recorded as such). This is what has been called inferential abstraction by some authors14, and terminology abstractions or abstract definitions by others3. Interoperability of EHR systems and decision-support systems is a typical scenario with such abstraction requirements.
This service, as the normalization one, deals with data transformations. The main difference lies in the fact that target instances do not need to be an accurate representation of source data. For instance, the target data may not be expressed at same granularity level as in the source data. As a consequence a wide range of data transformation functions are required such as arithmetical or string functions, type conversion, and especially aggregation or terminology abstraction functions.
This service has three inputs: the source schema expressed as an XML Schema or as an archetype, the target archetype (not necessarily based on the same reference model as the source schema) and the mapping relating both schemas. The mapping formalism is the same as in the normalization service; the only differences are the support for a broader set of functions in value correspondences (such as aggregation), and that archetypes can be the source schema of the mapping.
For instance, to obtain the value for the attribute “severe comorbidity”, assuming that the presence/absence of a number of morbidities affecting the patient can be obtained from the source schema, we could perform an abstraction based on the Charlson comorbidity score of those morbidities. Note that this is a rather complex example because the attribute “Charlson score” may not be included either in the source schema. The mapping expression for “severe comorbidity” will produce the value true if the Charlson score is above a certain threshold, and false otherwise. The filter part of the mapping will contain an adequate comparison expression whereas the transformation function will be a constant (either true or false).
For the specification of conceptual abstractions over SNOMED CT we support the SNOMED CT Expression Constraint Language15. This language, recently developed by IHTSDO, can be used for the definition of computable rules that define sets of clinical meanings represented by either pre-coordinated or post-coordinated expressions. For instance the constraint:
< 19829001 |disorder of lung|: 116676008 |associated morphology| = << 79654002 |edema|
is satisfied only by disorders of lung which have an associated morphology of edema (or a subtype thereof). This constraint may be used to abstract from acute pulmonary edema or toxic pulmonary edema to pulmonary edema. Note that in contrast to mappings between raw EHR data and archetypes, mappings between archetypes have the potential of being reused as they are, since they often encode domain knowledge. We have extended the Archetype Definition Language (ADL) with support for this language for terminology binding.
The result of the mapping specification is again an XQuery script which takes as input an instance of the source schema and generates an XML document that is compliant with the target archetype. The LinkEHR platform is used both for the specification of the mapping and for the automatic generation of the XQuery transformation script. The abstraction service has been implemented as a web service and the data transformation is performed by an XQuery engine. We additionally use an execution engine16 for the SNOMED CT Expression Constraint Language (available at http://snquery.veratech.es) which is invoked by the XQuery scripts to perform the abstractions required.
Semantic publishing service
The objective of the semantic publishing service is to represent EHR data in RDF or OWL. The input to this service must be data coming from relational databases or XML EHR extracts. In the present work, we focus on the second case, so this service takes normalized XML EHR extracts as input, that is, the outputs of the normalization and/or abstraction services. By normalized we mean that the data must have been captured using archetypes and expressed in XML conforming to such archetypes. At this stage there are two possible output formats for the EHR data, RDF and OWL. The choice depends on the expected data exploitation use case. If the user desires to obtain a LOD-oriented representation, RDF will be the output format. RDF is oriented to semantic data representation, but with reasoning possibilities limited by the triple store used. OWL is the output format of choice for those scenarios in which automated reasoning is required. OWL is oriented to knowledge representation and its OWL DL level permits to use DL reasoning over the knowledge base.
The XML EHR extracts may not be sufficient to generate their semantic representation because they generally lack domain knowledge. Our transformation engine also uses (1) the OWL ontology that is used for creating the semantic content and acts as knowledge schema; and (2) the mappings between the archetypes used for representing the extracts and the OWL ontology, which define how the EHR data are transformed into OWL individuals. The mapping rules ensure that the EHR data are correctly transformed into the semantic format and prevent redundancy in the output dataset.
For instance, the mapping rules would describe how the information about an adenoma in an EHR extract should be represented in the OWL ontology. In this example, a rule could map the attribute size of the adenoma in the archetype to the corresponding datatype property in the OWL ontology that is associated with the class representing adenomas. Such rule would be systematically applied to all the EHR data instances to create OWL individuals of the adenoma class. This is an example of a basic mapping rule. Our approach includes three types of basic mapping rules, which permit to link archetype entities with OWL classes, datatype properties and object properties, respectively.
Our experience in semantic transformation reveals that the basic rules are not sufficient to get semantically rich datasets. Sometimes, we need to define rules that involve multiple input entities and one or more ontology classes. Besides, the data sources do not always include all the content that is needed to obtain a semantic representation of the data or, at least, such meaning is not made explicit either in the XML schema or in the corresponding table. Therefore, we need to provide additional information in the mapping rules to enrich the EHR data. This is solved in our approach by means of transformation patterns, which will generate additional OWL axioms. These patterns can be regarded as a set of OWL axioms that provides the semantics required and missing in the data source. The patterns are linked to archetype entities, allowing for a semantic transformation of data. Finally, the patterns are expressed in OPPL2 (http://oppl2.sourceforge.net). It should be noted that once the mapping rules are defined, they can be stored and reused in similar transformation processes. The function used in most of these mapping rules is identity, since the atomic values are not changed. The exception to this is the function to create URIs from atomic values. Besides, rules may have filters associated, which would only transform the data instances for which the conditions defined in the corresponding filter hold true.
The semantic transformation method is also able to prevent the duplicity of OWL individuals in case different XML extracts represent the same OWL individual. For this purpose, identity conditions are used, which define the properties that make an OWL individual unique. OWL semantics is applied in the transformation process, which permits to ensure that only data consistent with the ontology constraints are transformed. Besides, when RDF is the output format, the process ensures that an RDF dataset consistent with the OWL ontology is produced. In our approach, the semantic publishing service is provided by the semantic web integration and transformation engine SWIT17 (http://sele.inf.um.es/swit).
Reasoning service
The previous services provide means for data transformation and abstraction, and their outputs are datasets that correspond to EHR extracts in formats like XML, RDF and OWL. The secondary use of EHR data is becoming increasingly important in clinical scenarios, because it enables the exploitation of EHR content with different healthcare-related purposes. One example of secondary use is the classification of patients (e.g. according to levels of risk). This involves applying a series of classification rules which partitions the patients in the groups of interest defined by the rules.
The input to this service is an EHR extract. The reasoning service also uses a set of classification rules, which are applied to the EHR extract. The reasoning service is able to produce two different outputs, which depend on the way it is invoked: (1) an EHR data extract in OWL format, which contains all the information inferred by the reasoner; and (2) the OWL classes to which the patient described in EHR data extract belongs. Hence, the first option returns the whole OWL content whereas the second one targets very concrete RDF triples. As an example of the latter, in the case of a colorectal cancer screening protocol, only the level of risk of a given patient would be returned.
Currently, our reasoning service has been implemented for EHR extracts in RDF/OWL format, that is, the result of the semantic publishing service. However, the solution is generic in the sense that the reasoning service could be implemented for EHR data coming in other formats such as XML. The current service deals with classification rules expressed in OWL. This decision is motivated by the possibility of applying DL reasoning over OWL content, so that state-of-the-art tooling can be reused for this purpose.
This service imposes two major requirements on the OWL ontology used for data classification. The first requirement is that it must contain one class per group of interest. Each group is implemented as an OWL class with equivalentClass axioms, because they define the sufficient conditions for an OWL individual to be classified as a member of the class. This enables a DL reasoner to automatically partition the clinical data into the desired groups of interest. The second requirement is that the classification ontology must be aligned with the domain ontology used for representing the EHR data in OWL. The simplest way to achieve this is to define the groups of interest based on the domain ontology. Otherwise, the reasoner is still able to infer the corresponding classifications if equivalentClass or SubClassOf axioms are established between the classes of the two ontologies.
Finally, this service has been implemented as two web services: one returns the OWL ontology with the inferred information, and another one returns the classification of a given individual. The reasoning of the input ontologies and the classification is obtained by applying the Hermit reasoner18.
A proof-of-concept platform
In this work we have implemented a web-based platform that can be used to explore different alternatives for a clinical data transformation problem using the above web services. The platform works on a collection of reusable mappings designed for specific data transformation steps in a particular clinical domain. Based on the mappings, the platform offers a number of transformation alternatives or paths, made up of one or possibly more transformation steps, which can be then executed on sample data (see below for more details). The platform deals with this execution, by making the appropriate calls to the services involved (normalization, abstraction, etc.) and passing the output of each step to the input of the next one. Note that the possible transformation paths will vary depending on the available mappings. Also note that the mappings will determine the scenarios that can be explored (normalization only, normalization plus semantic publishing, etc.). The design of the platform is highly generic, so that new mappings can be easily added to explore different data transformation problems. At the time of this writing the platform includes a series of mappings related to the domain of colorectal cancer (see http://cliniklinks.upv.es/demoAMIA2016.html).
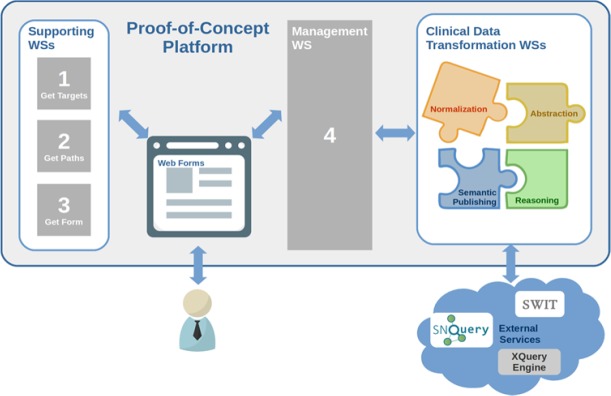
The following paragraphs detail the main steps when solving a data transformation problem with the platform, including the user interaction steps. They also describe the additional web services that have been implemented to support each step. The overall process and components are shown in Figure 1.
The platform displays automatically a list of all possible targets or destinations. These are retrieved from the currently available mappings through a specific-purpose web service. This action is performed in real time which facilitates the inclusion of new schemas and schema mappings. The user chooses the desired target.
Once the target has been established, another web service computes all possible transformation paths based on the mappings, in order to retrieve all the paths ending in the selected target. The mappings that define transformations between schemas and the schemas themselves may be distributed among servers in different organizations. The mappings and schemas can be interpreted as a directed acyclic graph, where the vertices are the schemas, and the edges are the mappings. A recursive in-depth search is performed which returns all the transformation paths from a source schema to the target one. The user selects which transformation paths to execute.
The user input for the execution of a transformation path is a clinical data instance based on the clinical data source schema. The instance may be provided directly from an EHR system or manually by clinicians by using digital data forms. Our proof-of-concept platform provides a web form based method to generate the input instance on the fly. The forms have been manually pre-designed according to the clinical data source schema. An additional web service automatically serves one form (each form is identified by the name of the first vertex in the selected transformation path) to the platform. The user fills out the displayed form to generate the source instance. In case multiple transformation paths are selected, the process would be much the same except for using multiple source instances (with different forms involved).
The platform includes a management web service that operates on the transformation paths and the data form contents. First, the management web service invokes another supporting web service that creates a valid source instance based on the form contents. With this instance, it then executes the corresponding web services for clinical data transformation (normalization, abstraction, semantic publishing, or reasoning transformations) in the required order. The management web service retrieves the output of each transformation step and hands it as input to the next one until the desired target is obtained, which is returned as result. It is also worth noticing that a single normalization or abstraction transformation may return a set of instances (e.g. in case the original source data contains information from more than one patient). This implies that all possible generated instances must be transformed so that further transformation steps are applied to each one of them.
Figure 1.
Overview of the platform architecture along with the main steps to execute a data transformation scenario
Application to the colorectal cancer domain
We have used our platform for the development of two different data transformation applications in the area of colorectal cancer. Colorectal cancer is an important cause of death in developed countries. Screening for colorectal cancer serves to identify people who may be at risk of developing the disease. Concretely, the framework of our work is the colorectal cancer screening program of the Region of Murcia (Spain), which uses screening protocols standardized in Europe and America to classify patients according to their level of risk. As a result of the application of these protocols, a database with data on more than 20,000 patients has been produced. Our goal is to demonstrate that the platform allows us to readily develop data transformation applications for different purposes, using EHR standards and semantic web technologies. In the first place, this requires a careful design of the different transformation steps, as well as of the models (archetypes, ontologies, etc.) to be used in each step, all this taking into account the needs of the project. Secondly, appropriate mappings must be defined for each transformation step, given the source and target of the transformation. Having defined the mappings, the platform will deal with the execution of the transformation pipeline, as described before. It should be noted that the mappings making up an application can be reused in other applications with similar transformation requirements.
In the rest of the section we describe the implementation of two applications using as starting point the database of Murcia’s colorectal cancer screening program. The first application corresponds to a basic normalization scenario, while the second is a more complex scenario implementing a colorectal cancer screening protocol. All the resources mentioned in the scenarios are available online (see http://cliniklinks.upv.es/demoAMIA2016.html).
Scenario 1: normalization to the CKM archetype for histopathology lab test results
This scenario corresponds to a single-step transformation performing a normalization. The input is the above database with the EHR extracts of the patients, and the output is a normalization of the previous database in terms of a standardized clinical information model. For this purpose we have chosen the openEHR-EHR-OBSERVATION.lab_test-histopathology archetype from the openEHR CKM repository19.
Normalization
As stated before, the normalization service requires at design time the specification of a high-level mapping between the source schema and the target archetype. The source schema is a nested schema describing anatomic pathology studies with their findings. There is a top-level set of studies, and each study record has a nested set of test records. Tests have an additional nesting where each test has a set of findings. Studies and test records include context information such as dates and performers. Finally, findings contain the details of the microscopic findings such as size or pathological staging. In the target schema (archetype), there is a top-level record that represents a test. Each test has a set of microscopic findings and a set of macroscopic findings along with other information items such as specimen details or test status. Since the source schema has an additional nesting level (study), a single source instance might produce several target instances.
The mapping specification was created using the LinkEHR mapping capabilities. Since our purpose was to make public the available data in the form of an XML document compliant with the openEHR reference model, we did not employ any transformation function neither normalized the local terminology. It was not necessary either to define a mapping for all the atomic data elements in the archetype since a great part of them are not present in source data. Note that archetypes are defined to be as generic as possible in order to accommodate any relevant piece of information.
Scenario 2: implementation of a colorectal cancer screening protocol using abstraction, semantic publishing and reasoning
In this scenario we have used the platform to implement the case study of a previous work9. The input to this clinical data transformation problem is the above-mentioned database with the EHR extracts of the patients, and the desired output is the level of risk for each patient. To take advantage of the platform’s capabilities for reasoning with OWL content, the EHR data will have to be transformed into OWL, to obtain the level of risk through classification reasoning. For this purpose, the semantic publishing service will be required.
Both the European and the American screening protocols define the level of risk by taking into account aspects like the amount of adenomas or the size of the largest adenoma, information which can be obtained from histopathologic findings. If this specific information is not included in the database, it will have to be calculated in terms of the services offered in the platform, that is, by means of either abstraction or reasoning services. For efficiency reasons, we consider more appropriate to perform operations like count or maximum as part of an abstraction step.
Hence, abstraction, semantic publishing and reasoning are the three services required in this scenario. Next, we describe the requirements and the inputs and outputs of each transformation step, including which inputs correspond to outputs of previous steps.
Abstraction
In the same vein as the normalization service, the abstraction one requires at design time the specification of a high-level mapping between the source and target schemas, the only difference is that the source schema can be also an archetype. In order to determine the patient’s risk level, we need to use concepts at two different levels of granularity: at finding and at test levels. For the representation of finding-level concepts, we defined a specialization of the archetype openEHR-EHR-OBSERVATION.lab_test-histopathology, named openEHR-EHR-OBSERVATION.lab_test-histopathology-colorectal_screening. This specialized archetype includes information about adenoma findings such as the type or maximum size of the recorded dimensions (width, breadth and height), the dysplasia grade and whether they are sessile and/or advanced. These information items are not stored directly in the database but can be calculated by performing different types of abstractions. In order to represent test-level concepts, concretely the maximum size of all adenomas and the number of adenomas, we developed a new archetype from scratch called openEHR-EHR-EVALUATION.colorectal_screening.v1.
Two mapping specifications were necessary, the first one between the database and the finding-level archetype and the second one between the finding-level archetype and the test-level archetype. The required abstractions were codified in the mapping specification and therefore are performed by the resulting XQuery script. As explained before, in our mapping formalism a value correspondence comprises a set of filter-transformation function pairs that allows complex conditional expressions (if-then-elseif …). Value correspondences proved to be powerful enough for defining the concept abstractions required in the first mapping (e.g. advanced adenoma). The second mapping required additionally the use of aggregation functions: max for the calculation of the maximum size of all adenomas, and count for the calculation of the number of adenomas. The archetypes and the mapping specifications were created using the LinkEHR archetype editing and mapping capabilities, respectively.
Semantic publishing
The semantic publishing service uses the mapping designed to transform the EHR data conformant with the archetypes of the previous step into the colorectal-domain OWL ontology, which provides the specific domain knowledge. The ontology itself is also used in this transformation step. The service takes as input the EHR extract produced by the abstraction service, and produces as output an OWL representation of that extract in terms of the colorectal-domain ontology.
Reusing an OWL ontology from the NCBO BioPortal (http://bioportal.bioontology.org) was our first option to promote the interoperability of the OWL content generated. We were not able to find any appropriate OWL ontology, so we had to develop one specifically for this use case. The development of the colorectal-domain OWL ontology was a manual process guided by the entities identified in the source database, in order to represent the domain knowledge that would be required to express the meaning of the EHR extracts in OWL format. This ontology includes classes for histopathology reports, types of findings, types of results of anatomical pathology tests, etc., and properties and axioms that define precisely the meaning of these entities. The ontology consists of 102 classes, 47 object properties, 42 datatype properties and 1693 logical axioms.
The mapping file specifying the correspondences between the archetypes and the colorectal-domain ontology was created using the SWIT tool. This mapping includes a transformation rule for each of the entities included in the archetypes resulting from the abstraction step. Note that we have only defined the transformation rules for those entities which we believe to be of interest for a semantic exploitation. Given that the mapping is defined between archetypes and the domain ontology, it is independent of the source database.
Reasoning
The reasoning service takes as input the EHR extracts in OWL format resulting from the application of the semantic publishing service. It operates by using the OWL classification ontology which contains the logical definition of the levels of risk according to the European and American protocols. The output of the reasoning service is the level of risk inferred based on both the input EHR extract and the classification ontology.
The OWL classification ontology was developed as an extension of the colorectal-domain one. Basically, the classification ontology defines five classes, which correspond to the risk groups specified in the protocols. The European protocol distinguishes high risk, intermediate risk, and low risk, whereas the American one only differentiates between high risk and low risk. The five classes are modeled in a similar way. First, each class is defined as a subclass of HistopathologyReport, which is a class of the domain ontology. Second, equivalentTo axioms are associated with each class, which define sufficient conditions for membership of the class. For example, the condition associated with both the LowRiskAmericanProtocol and LowRiskEuropeanProtocol classes is the same, and is described as follows: “A histopathology report that only contains at most 2 normal adenomas, but does not contain any advanced one”. This is implemented in the OWL classification ontology as follows: HistopathologyReport and (hasAdenoma only NormalAdenoma) and (max_size some integer[< 10]) and (number some integer[< 3]). The classes of interest were mainly defined by using the properties and classes of the domain ontology. The classification ontology introduces 16 new classes and 62 new logical axioms, but no additional object or datatype properties.
Conclusion
In this paper we introduce the proof-of-concept platform that we have implemented to explore different alternatives for a clinical data transformation problem. It is built upon specialized web services dealing with data transformations which work at different levels, namely: normalization, abstraction, semantic publishing and reasoning. More importantly, the platform uses a series of reusable mappings that might be distributed across servers in different organizations and supports multiple EHR information standards (such as HL7 CDA, ISO 13606 or openEHR). In principle the mappings restrict the range of transformation applications (and hence scenarios) that can be configured using the platform. However, the design is rather generic, so that new mappings can be easily added allowing for a wider range of scenarios.
We also describe two different data transformation applications that have been implemented using our platform and web services. It is important to stress that the different models (archetypes, etc.) to be used along the transformation process, and specially the mappings defined taking into account the source and target of each transformation step, are essential in this kind of implementations.
Mappings involve local (XML) schemas, archetypes and ontologies as source or target elements. Concretely, mappings can be defined between: (a) an XML schema and an archetype, (b) an XML schema and an OWL ontology, (c) two archetypes, and (d) an archetype and an OWL ontology. As described earlier, we follow a “specify and generate” approach for all types of mappings. In this approach, developers are responsible for defining high-level mappings using specific-purpose tools, namely LinkEHR for mappings resulting in archetypes ((a) and (c)) and an ontology alignment format for mappings to RDF/OWL ((b) and (d)). These definitions are then automatically compiled into an executable script that will be used to perform the actual transformation. With mapping reuse in mind, it becomes necessary to pay attention to quality assurance aspects.
In our platform, automated reasoning supports both the semantic publishing and reasoning services, that is, the generation of the semantic dataset and its exploitation. State-of-the-art approaches20–23do not use reasoning for guaranteeing the generation of consistent datasets, although reasoning has supported the transformation of clinical models into OWL24. Those approaches mainly exploit reasoning as part of SPARQL queries or use specific rule languages such as SWRL, which are options more limited in terms of reasoning than OWL DL. Since we were interested in classification, OWL DL was deemed the preferred alternative. SPARQL and SWRL seem interesting options for other exploitation services that could be included in the future in our platform.
As future work we intend to extend our platform with different mapping management functionalities. Examples include the development of mapping operators, such as the merging of mappings, as well as of criteria (e.g. quality criteria in terms of the amount of information transferred) that can be used to compare mappings25. Furthermore, we also foresee applications where the mappings (and/or models) listed in the platform are used in alternative ways. An example might be to use the mappings to determine the EHR data requirements for semantic publishing according to a given ontology. This will surely require including in the platform a precise characterization and/or a declarative description of the mappings. For instance, a formal description of the concept definition used in an abstraction mapping has proved very useful according to our previous experience4. Finally, we plan to carry out the necessary developments with a view to the utilization of the data transformation applications built with our platform in a clinical setting.
Acknowledgements
This work was supported by the Spanish Ministry of Economy and Competitiveness and the EU FEDER programme through grants TIN2014-53749-C2-1-R, TIN2014-53749-C2-2-R and PTQ-12-05620, and by Universitat Jaume I through project P1·1B2013-15.
References
- 1.Shekelle PG, Morton SC, Keeler EB. Costs and benefits of health information technology. Evid Rep Technol Assess (Full Rep) 2006;132:1–71. doi: 10.23970/ahrqepcerta132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sujansky W. Heterogeneous database integration in biomedicine. J Biomed Inform. 2001;34(2001):285–298. doi: 10.1006/jbin.2001.1024. [DOI] [PubMed] [Google Scholar]
- 3.Peleg M, Keren S, Denekamp Y. Mapping computerized clinical guidelines to electronic medical records: Knowledge-data ontological mapper (KDOM) J Biomed Inform. 2008;41(1):180–201. doi: 10.1016/j.jbi.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Marcos M, Maldonado JA, Martínez-Salvador B, Boscć D, Robles M. Interoperability of clinical decision-support systems and electronic health records using archetypes: A case study in clinical trial eligibility. J Biomed Inform. 2013;46(4):676–689. doi: 10.1016/j.jbi.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 5.González-ferrer A, Peleg M, Verhees B, Verlinden J. Data Integration for Clinical Decision Support Based on openEHR Archetypes and HL7 Virtual Medical Record. Process Support and KnowledgeRepresentation in Health Care. 2013:71–84. [Google Scholar]
- 6.Goble C, Stevens R. State of the nation in data integration for bioinformatics. J Biomed Inform. 2008;41(5):687–693. doi: 10.1016/j.jbi.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Berners-Lee T, Hendler J, Lassila O. The Semantic Web. Sci Am. 2001;284(5):34–43. [Google Scholar]
- 8.Berners-Lee T. Linked Data - Design Issues. 2009. http://www.w3.org/ Available at: http://www.w3.org/DesignIssues/LinkedData.html.
- 9.Fernandez-Breis JT, Maldonado JA, Marcos M, et al. Leveraging electronic healthcare record standards and semantic web technologies for the identification of patient cohorts. J Am Med Inform Assoc. 2013;20(e2):e288–96. doi: 10.1136/amiajnl-2013-001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fagin R, Kolaitis PG, Miller RJ, Popa L. Data exchange: Semantics and query answering. Theoretical Computer Science. 2005;336:89–124. [Google Scholar]
- 11.ten Cate B, Kolaitis PG. Structural Characterizations of Schema-mapping Languages. Commun ACM. 2010;53(1):101. [Google Scholar]
- 12.Maldonado JA, Moner D, Boscá D, Fernández-Breis JT, Angulo C, Robles M. LinkEHR-Ed: A multireference model archetype editor based on formal semantics. Int J Med Inform. 2009;78(8):559–570. doi: 10.1016/j.ijmedinf.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Maldonado JA, Costa CM, Moner D, et al. Using the ResearchEHR platform to facilitate the practical application of the EHR standards. J Biomed Inform. 2012;45(4):746–762. doi: 10.1016/j.jbi.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Rector AL, Johnson PD, Tu SW, Wroe C, Rogers J. Interface of Inference Models with Concept and Medical Record Models; Lecture Notes in Artificial Intelligence; 2001. pp. 314–323. [Google Scholar]
- 15.SNOMED CT Expression Constraint Language Specification and Guide, version 1.00. 2015.
- 16.Giménez-Solano V, Maldonado J, Salas-García S, Boscá D, Robles M. Implementation of an execution engine for SNOMED CT Expression Constraint Language; Medical Informatics Europe - MIE 2016, to appear; [PubMed] [Google Scholar]
- 17.Legaz-García M, del C, Miñarro-Giménez JA, Menarguez-Tortosa M, Fernández-Breis JT. Lessons learned in the generation of biomedical research datasets using Semantic Open Data technologies; Digit Healthc Empower Eur Proc MIE2015; 2015. p. 165. [PubMed] [Google Scholar]
- 18.Shearer R, Motik B, Horrocks I. HermiT: A highly-efficient OWL reasoner. In: Proceedings of the 5th International Workshop on OWL; Experiences and Directions (OWLED 2008); 2008. pp. 26–27. [Google Scholar]
- 19.openEHR foundation. Clinical Knowledge Manager - CKM. Available at: http://www.openehr.org/ckm/
- 20.Riazanov A, Klein A, Shaban-Nejad A, et al. Semantic querying of relational data for clinical intelligence: a semantic web services-based approach. J Biomed Semantics. 2013;4(1):1–19. doi: 10.1186/2041-1480-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lezcano L, Sicilia M-A, Rodriguez-Solano C. Integrating reasoning and clinical archetypes using OWL ontologies and SWRL rules. J Biomed Inform. 2011;44:343–353. doi: 10.1016/j.jbi.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Tao C, Jiang G, Oniki T a, et al. A semantic-web oriented representation of the clinical element model for secondary use of electronic health records data. J Am Med Informatics Assoc JAMIA. 2013;20(3):554–62. doi: 10.1136/amiajnl-2012-001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samwald M, Freimuth R, Luciano JS, et al. An RDF/OWL Knowledge Base for Query Answering and Decision Support in Clinical Pharmacogenetics. Stud Health Technol Inform. 2013;192:539–542. [PMC free article] [PubMed] [Google Scholar]
- 24.Legaz-García MC, Menárguez-Tortosa M, Fernández-Breis JT, Chute CG, Tao C. Transformation of standardized clinical models based on OWL technologies: from CEM to OpenEHR archetypes. J Am Med Informatics Assoc. 2015 doi: 10.1093/jamia/ocu027. ocu027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arenas M, Perez J, Reutter JL, Riveros C. Foundations of schema mapping management; Proc 29th ACM SIGACT-SIGMOD-SIGART Symp Princ Database Syst; 2010. pp. 227–238. [Google Scholar]