Abstract
Molecular genetics laboratory reports are multiplying and increasingly of clinical importance in diagnosis and treatment of cancer, infectious disease and managing of public health. Little of this data is structured or maintained in the EHR in format useful for decision support or research. Structured, computable reporting is limited by non-availability of a domain ontology for these data. The IHTSDO and Regenstrief Institute(RI) have been collaborating since 2008 to develop a unified concept model and ontology of observable entities – concepts which represent the results of laboratory and clinical observations. In this paper we report the progress we have made to apply that unified concept model to the structured recording of observations in clinical molecular genetic pathology including immunohistochemistry and sequence variant findings. The primary use case for deployment is the structured and coded reporting of Cancer checklist© and biomarker data as developed by the College of American Pathologists(CAP) with collaboration by the Royal College of Pathology(RCP).
Introduction
Molecular genetic pathology is a new scientific frontier exploding on the practice of clinical medicine. President Obama pushed the issue to the fore when he announced the national agenda for research into personalized medicine1. Unfortunately the extensive work in developing and managing information in genetic research has not translated into ontologies of use in support and documentation of clinical practice. Of the reference terminologies cited by the Office of the National Coordinator (ONC) as required by the US healthcare information architecture2, only LOINC3 has significant content addressing observables in molecular genetics. Unfortunately the LOINC reference terminology model applied to molecular genetics does not capture important details such as laboratory methods4 or support needs of the domain ontology that would support re-use of observational data in epidemiology, research and clinical decision making5. Diverse efforts within the informatics community to develop a clinically useful observables ontology6–11 have been informative but none have gained broad acceptance for integration into the ONC terminology architecture for the electronic health record (EHR).
Laboring behind the scenes in worldwide terminology management, harmonization efforts by the International Health terminology Standards Development Organization (IHTSDO) and Regenstrief Institute (RI)12 have been quietly working to expand the expressivity and utility of observable entities and clinical findings for use in molecular genetic structured clinical data. An observable entity is a concept with semantic overlap between LOINC and SNOMED CT (<<363787002|Observable entity|) and can best be described as a conceptual model for the results of an observation – administrative, clinical, laboratory or otherwise. Although RI has served the informatics community for years providing LOINC codes for laboratory medicine and molecular genetic pathology, the lack of a domain ontology for these concepts means that the there is no terminology support for queries of aggregation or for defining features such as methods4, 5. The harmonization work underway has developed a candidate unified concept model for genetic observables but the application of that work is not intuitive. This paper reports one small part of that effort focused upon the challenging issues of genetic biomarker observations in anatomic pathology supporting diagnosis and management of cancer.
The volume and types of molecular genetic data appearing in clinical medicine is growing exponentially. CAP first published cancer report protocols (checklists)13 in 1998 defining a minimum dataset for anatomic pathologists to report when they evaluate surgical specimens concluding a diagnosis of cancer. Currently CAP publishes 82 separate checklists for various tissue pathways. The checklists were expanded in 2013 to include tumor biomarkers which have become referent findings required for staging and planning treatment.
The number of these genetic observations – regarding either the tumor or the patient – important to outcomes in cancer treatment has grown with the science. An example we encountered while modeling the colorectal cancer check sheet was “detection of the BRAF V600E sequence variant in the resected tumor”. This genetic finding has been studied and clinically validated to predict response to certain treatments in both colorectal cancer and melanoma. Historically the clinical pathologist could only detect genetic sequence variations by analyzing extracted tumor DNA for specific mutations in single genes that code for the mutant proteins they formed. Those tests yielded limited analysis of genetic material and were expensive and time consuming. With the successful mapping of the human genome and improved technology, high throughput sequencing of human and neoplastic genomes became possible. Next generation sequencing (NGS) yields relatively rapid turn-around of much more genetic sequence data. Anatomic pathology today employs both protein and nucleotide sequence data in the diagnosis, prognostication and selection of targeted therapeutic regimens for cancer.
Primary use cases for a domain ontology of observables are query support of findings aggregated by genotypic variant, tissue of origin and histologic appearance. Query use cases implied by this expectation might include:
“Find all cases of malignancies that tested positive for the BRAF p.V600E (c.1799T>A) mutation”; “Find all patients who have tested positive for genetic predisposition to breast cancer”; “Find all cases of colon cancer in which no biomarker testing was performed”
The SNOMED CT concept model14 consists of a constrained set of relationships and accompanying target value sets allowed for incorporation within a computable concept definition. Allowed relationships and value sets are specified for domains of SNOMED CT, usually individual hierarchies. A SNOMED CT concept must also have at least one supertype (IS_A) relationship linking the concept within the hierarchy and a fully specified (context free) name which is the universal term denoting the concept. Each linguistic implementation of SNOMED CT can have one primary term and as many synonyms as required. When the concept model applied to the modelled SNOMED CT content is insufficient to Fully define a concept, the concept is declared as Primitive. The SNOMED CT ontology is subjected to description logic classification prior to publication as an editorial quality check and to compute the inferred relationships implied by the application of the concept model. The concept model is not complete for all segments of SNOMED CT. For years the Observable entities hierarchy (<<363787002|Observable entity|) has been published as a hierarchy of Primitives employing only stated supertype relationships.
The IHTSDO expects that extensions of SNOMED CT may be authored, compliant with the concept model, that represent material necessary for parochial or research needs not appropriate for the international release of SNOMED CT. In the US, the National Library of Medicine (NLM) develops and maintains the US extension to SNOMED that is required for Meaningful Use compliance by EHR vendors. The University of Nebraska maintains the Nebraska Lexicon© extension, dependent upon the US extension, which supports terminology needs of our Epic® implementation and terminology development we have been supporting for the community since 2004. We report in this paper our use of this terminology authoring environment to develop, test and deploy an observables ontology for molecular genetics requirements of the CAP cancer check sheets.
Concept model development
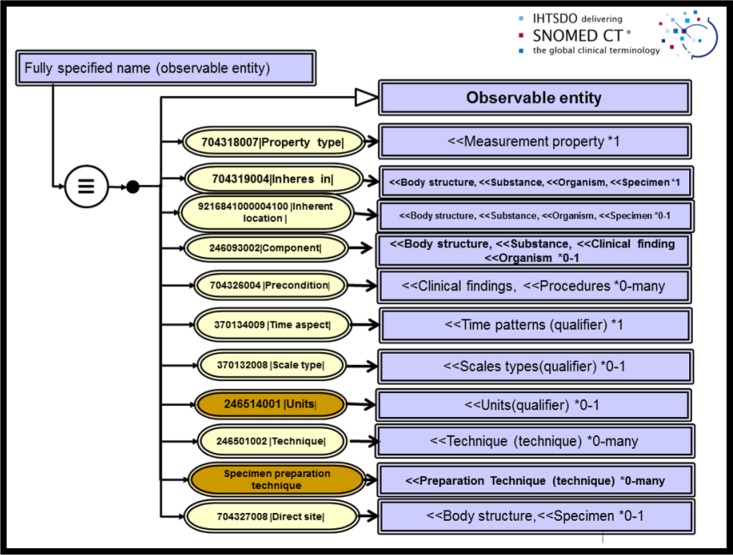
The Observables and Investigation Model Project was formed by the IHTSDO in 2008 to develop a computable concept model for the Observable entity domain and prepare a model for interoperation of content with LOINC data sets. In summer of 2015, the project convened a meeting of experts from the NLM, CAP and Health and Social Care Information Center (HSCIC) of UK to discuss details of application of the proposed concept model to the set of observables necessary to the structured reporting of cancer checklists and biomarkers. During the meeting, data elements contained in the CAP colorectal check sheet and the Royal College of Pathologists (RCP) counterpart were reviewed and analyzed for semantics. Pathologists provided expert direction regarding the clinical meaning of each element. Terminology experts then discussed the requirements and proposed templates for consistent application of a candidate concept model for observables. The deployment model for testing from that conference is pictured in Figure 1 for an Observable entity in anatomic and molecular pathology. Each attribute for refining the meaning of a concept is shown along with the valuesets of target concepts that are supported as well as the associated cardinality of the relationship. Non-defining relationships, called qualifiers, are pictured in brown in this figure.
Figure 1.
Harmonized observable concept model
Application of this model was reasonably straightforward for conventional observations in surgical pathology, requiring only model extensions of SNOMED to include 8 Properties and 4 Techniques. The Colon cancer checklist which we first modeled required 61 new observable entities for anatomic pathology.
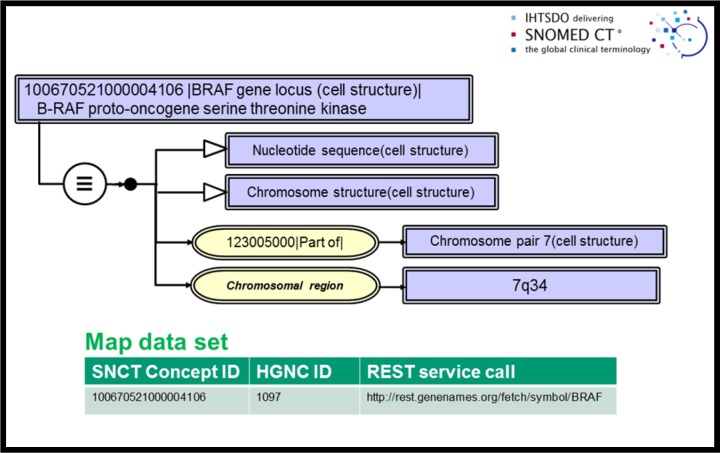
Consensus on application of this model to observables in molecular genetic pathology could not be developed in our first three meetings and a deployment strategy was only achieved at the Montevideo meeting of the IHTSDO in fall 2015. Criticisms emerging from initial discussions included: a) excessive numbers of primitives, b) insufficient semantic granularity in genetic structures and c) failure to support both protein-based and sequence-based observations. In order to meet these challenges we proposed employing Human Genome Nomenclature Committee15 resource data in our model by map reference to uniquely define nucleotide sequence entities (genes, microsatellites and other nucleotide segments) and to support the details of sequence data observations in Variant Call Format16. We also employed representation of numbers in our model for nucleotide references 17. We proposed that a gene locus and other sequence based data could be defined in an expanded concept model specifying the gene locus as cellular substructures, specifically nucleotide sequences. These data were further characterized using the reference naming of the Human Gene Nomenclature Committee (HGNC)15 and the genetic datasets which it cross references such as the annotated genome reference Ensembl19. An example concept model rendition for the B-RAF proto-oncogene as we defined it is included in figure 2. This particular concept definition employs sequence address data from the Genome Reference Consortium GRCh38 release in GenBank20, included in our publication by reference as a map data set. The map data which is shown in tabular form as an inset includes the HGNC reference number and a REST service call which will retrieve full HGNC reference data. Human protein products of gene transcription are currently implemented as primitive concepts in the 105590001|Substance| hierarchy.
Figure 2.
Cellular structure model for BRAF gene locus
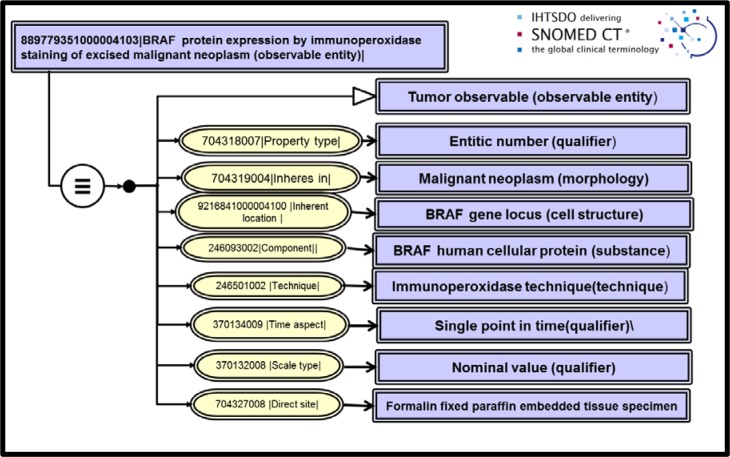
Once the reference genetic material was fully defined in SNOMED, we extended Techniques and Properties for molecular genetic pathology procedures. Immunohistochemistry observations could then be modelled. An example of an immunoperoxidase staining analysis for BRAF protein in a surgical tumor specimen is shown in figure 3. Immunoperoxidase and nuclear sequencing techniques are much more specific than the molecular procedures currently employed in LOINC 2.54 concept definitions. Therefore the observables concepts we have modeled for CAP checklists are generally semantic children of molecular genetic observables now in LOINC.
Figure 3.
Immunohistochemistry observable: BRAF protein expression by immunoperoxidase staining
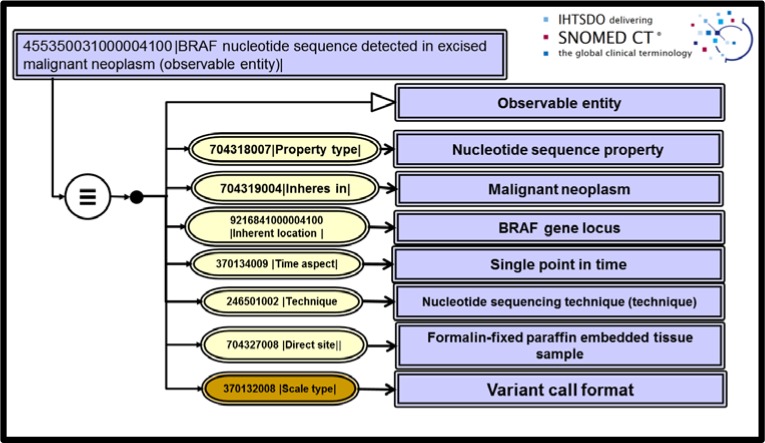
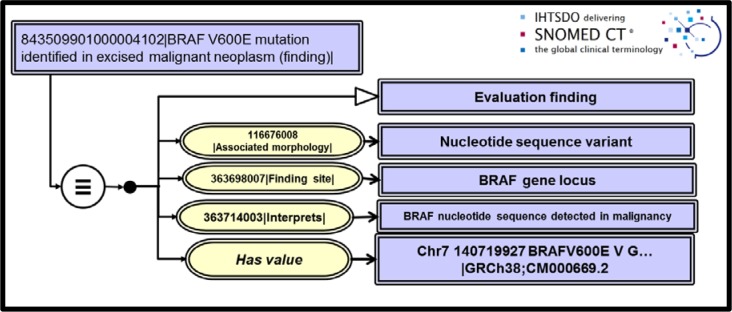
Nucleotide sequence observables were a particularly thorny issue we faced since input from our pathologist specialists required that we be able to store observations in our research databases with complete sequence data. An industry standard for identifying sequencing results has become the Variant Call Format16. These data files issue findings of sequence variants when compared to a reference standard genome and are typically ASCII files of a few kilobytes. The report data structures complies with the recommendations for description of sequence variants issued by the Human Genome Variant Society18. Once again, employing numbers as SNOMED CT values allowed us to extend findings data and to deploy Has value attributes in place of Has interpretation in an extended concept model for Clinical findings. Figures 4 and 5 show the Observable entity model for sequence data of the BRAF gene locus along with a positive clinical finding for the BRAF p. V600E(c. 1799T>A) mutation detected in the excised tumor.
Figure 4.
Nucleotide sequence observable for BRAF gene locus
Figure 5.
Observation (clinical finding) for detection of BRAF V600E mutation in an excised cancer
Application of the harmonized concept model and modelling of the SNOMED CT extension concepts to the recording needs of the check sheets was the first step in preparing a completely specified structured report for CAP cancer synoptics. For each observable entity representing a ‘question’ on the form, we further organized SNOMED CT valuesets of ‘answers’ that would populate the attribute-value pair of the HL7 OBX segment. For the example of “BRAF protein expression by immunoperoxidase staining in excised malignant neoplasm” presented in figure 3, we show the corresponding section of the CAP check sheet for colorectal cancer in figure 6. Included in the figure is an inset with the Observable entity concept identifier, fully specified name and the complete valueset of choices that should be selected by the pathologist in the anatomic pathology system.
Figure 6.
CAP check sheet for colorectal cancer; biomarkers segment for BRAF genetic observations
Results
Our deployment use cases are the 82 cancer and tumor biomarkers checklists published by CAP and supplemented by review with the RCP. There are many repeating observations across these checklists but our experience with deployment of our model for colon cancer required expansion of the Machine Readable Concept Model21 for the SNOMED CT hierarchies of Observables, Body structure and Clinical Findings. The magnitude of the new conceptual content in our Nebraska Lexicon© extension required for colon and breast cancer is summarized in table 1. This lists the number of new concepts by hierarchy which we developed for anatomic and molecular genetic pathology. The number of primitive concepts we required for each domain is also listed. In the fourth column we have listed an exemplar concept name from the content developed within that SNOMED CT hierarchy. This work has proceeded in collaboration with NLM, RI, IHTSDO, CAP and the RCP and was published on the NLM UMLS Knowledge Sources Server for public dissemination in July 2016. The work is available to all interested parties with a UMLS license.
Table 1:
Extension concept inventory for colon cancer checklist
| SNOMED CT hierarchy | Anatomic Pathology Concepts/Primitives | Molecular Genetic Concepts/Primitives | Exemplar molecular extension concepts |
|---|---|---|---|
| Observable entities | 61/1 | 32/3 | BRAF nucleotide sequence detected in excised malignancy |
| Body Structures | 10/9 | 29/3 | BRAF gene locus |
| Clinical findings | 6/2 | 7/3 | BRAF V600E variant identified in excised malignancy |
| Procedures | 2/1 | 0 | |
| Techniques | 4/4 | 7/7 | Pyrosequencing |
| Property types | 8/8 | 2/2 | Sequence property |
| Scale types | 0 | 9/9 | Variant call format |
| Situations | 1/0 | 0 | |
| Substances | 0/0 | 11/11 | BRAF human cellular protein |
| Attributes | 2/2 | 3/3 | |
| Qualifiers | 2/2 | 0 | |
| TOTALS | 88/29 | 100/41 |
Conclusion
As predicted, sequencing of the human genome has led to a proliferation of innovative scientific research with application to clinical medicine. We expect that new types of clinical knowledge will create challenges for recording, managing and using the clinical data that results from that research. While a number of efforts have been reported by the informatics community 3–11 to organize or develop terminology models to serve for recording of clinical genomic data, only LOINC offers substantial content within the suite of ONC US standards. Unfortunately, the LOINC model does not offer the features of a domain ontology that are desirable for best query and retrieval of clinical data.
We have presented in summary form the results of a two year collaboration with clinical and terminology standards developers which represents only a new snapshot of harmonization work between the IHTSDO and RI in process since 2008. This report focuses on Observable entities for molecular genetic observations because they represent new challenges to the application of the SNOMED CT/LOINC harmonized concept model. Such advances are a challenging but important component of a comprehensive terminology model for twenty-first century medicine.
CAP cancer check sheets offer one set of important use cases for clinical ontology development because they summarize anatomic and molecular genetic observations of established relevance for practice of precision medicine. We have deployed this model for testing and evaluation by other informatics centers and published the work in collaboration with the NLM. We expect that this will stimulate dialogue with the informatics terminology community on the ontology model we have deployed. We hope to scale the work across the rest of molecular genetic pathology observations for cancer and expand the work into microbiology and human germline genetic disease testing.
The model we present interacts and draws upon NCBI resources and ontologies supported by the Genome Reference Consortium22. We expect that the science in this field will continue to rapidly evolve and that new concepts and observations will emerge from that research requiring documentation in the EHR. The role of ONC terminologies and, more specifically clinical ontologies, should be to faithfully record those data and provide for query and re-use for purposes of clinical decision support, research and public health. We do not however, think it the role of SNOMED CT or LOINC to secondarily reproduce GRC reference data. For that reason, HGNC defining data sets are included in the model by map reference.
References
- 1.Interoperability. 2015. https://www.healthit.gov/policy-researchers-implementers/interoperabilityUpdated.
- 2.Huff SM, Rocha RA, McDonald CJ, et al. Development of the logical observation identifier names and codes (LOINC) vocabulary. J Am Med Inform Assoc. 1998;5(3):276–292. doi: 10.1136/jamia.1998.0050276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Obama B. The precision medicine initiative. 2015. https://www.whitehouse.gov/precision-medicine Updated.
- 4.Masys DR, Jarvik GP, Abernethy NF, et al. Technical desiderata for the integration of genomic data into electronic health records. J Biomed Inform. 2012;45(3):419–422. doi: 10.1016/j.jbi.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simpson RW, Berman MA, Foulis PR, et al. Cancer biomarkers: The role of structured data reporting. Arch Pathol Lab Med. 2015;139(5):587–593. doi: 10.5858/arpa.2014-0082-RA. [DOI] [PubMed] [Google Scholar]
- 6.Hoffman M, Arnoldi C, Chuang I. The clinical bioinformatics ontology: A curated semantic network utilizing RefSeq information; Pac Symp Biocomput; 2005. pp. 139–150. [DOI] [PubMed] [Google Scholar]
- 7.Sax U, Schmidt S. Integration of genomic data in electronic health records--opportunities and dilemmas. Methods Inf Med. 2005;44(4):546–550. [PubMed] [Google Scholar]
- 8.Hoffman MA. The genome-enabled electronic medical record. J Biomed Inform. 2007;40(1):44–46. doi: 10.1016/j.jbi.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 9.Murphy SN, Mendis ME, Berkowitz DA, Kohane I, Chueh HC. Integration of clinical and genetic data in the i2b2 architecture; AMIA Annu Symp Proc; 2006. p. 1040. [PMC free article] [PubMed] [Google Scholar]
- 10.Overby CL, Tarczy-Hornoch P, Hoath JI, Kalet IJ, Veenstra DL. Feasibility of incorporating genomic knowledge into electronic medical records for pharmacogenomic clinical decision support. BMC Bioinformatics. 2010;11(9) doi: 10.1186/1471-2105-11-S9-S10. S10-2105-11-S9–S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jing X, Kay S, Marley T, Hardiker NR, Cimino JJ. Incorporating personalized gene sequence variants, molecular genetics knowledge, and health knowledge into an EHR prototype based on the continuity of care record standard. J Biomed Inform. 2012;45(1):82–92. doi: 10.1016/j.jbi.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooperative agreement between the International Health Terminology Standards and Development Organisation and the Regenstreif Institute, Incorpororated. 2013. http://www.ihtsdo.org/resource/resource/104 Updated.
- 13.An overview of the College of American Pathologists cancer checklist. 2009. http://www.cap.org/apps/docs/committees/cancer/cancer_protocols/Overview_CAP_Cancer_Checlkists_090115.pdfUpdated.
- 14.International Health Terminology Standards Development Organization. SNOMED-CT Editorial Guide. January 2015 ed. Copenhagen: International Health Standards Terminology Development Organization; 2015. [Google Scholar]
- 15.HUGO gene nomenclature committee. 2015. www.genenames.orgUpdated.
- 16.VCF (variant call format) specifications. 2015. https://vcftools.github.io/specs.htmlUpdated.
- 17.Representation of numbers in SNOMED CT. 2016. https://confluence.ihtsdotools.org/download/attachments/15795743/Updated.
- 18.den Dunnen JT, Dalgleish R, Maglott DR, et al. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat. 2016;37(6):564–569. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]
- 19.Herrero J, Muffato M, Beal K, et al. Ensembl comparative genomics resources; Database (Oxford); 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.GRCh38.p7. 2016. http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/Updated.
- 21.International Health Terminology Standards Development Organization. SNOMED-CT Technical Implementation Guide. 2014 International Release (US English) ed. Copenhagen: International Health Terminology Standards Development Organization; 2014. [Google Scholar]
- 22.The genome reference consortium. 2016. http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/Updated.