Abstract
We present an approach for manually and automatically classifying the resource type of medical questions. Three types of resources are considered: patient-specific, general knowledge, and research. Using this approach, an automatic question answering system could select the best type of resource from which to consider answers. We first describe our methodology for manually annotating resource type on four different question corpora totaling over 5,000 questions. We then describe our approach for automatically identifying the appropriate type of resource. A supervised machine learning approach is used with lexical, syntactic, semantic, and topic-based feature types. This approach is able to achieve accuracies in the range of 80.9% to 92.8% across four datasets. Finally, we discuss the difficulties encountered in both manual and automatic classification of this challenging task.
Introduction
The answers to medical questions can be found in a wide variety of resources. These resources include general medical knowledge (such as that found in textbooks or encyclopedias), highly specialized research knowledge (such as the biomedical literature), and even source specific to the given patient (such as the patient’s electronic health record (EHR) information). For automatic question answering (QA) systems that intend to answer a wide variety of medical questions, having the ability to properly target a specific resource (such as a corpus, sub-corpus, or database) would be very useful. Consider the following three questions from the datasets used in this study:
What kind of allergy does he have?
Is it necessary to wait until age 7 before doing an allergy evaluation?
Is there anything in the literature about allergy testing or desensitization to acyclovir?
While all these questions are topically concerned with allergies, they require different resources for answers. The first question is patient-specific, and the answer must come from the patient’s allergy list or notes in the health record. If the information is missing there, the patient himself must be questioned, as any attempt to look in a non-patient-specific source will yield useless results. The second question requires general medical knowledge, either in a textbook, practice guidelines, or manuals. The final question specifically asks for scientific literature, and would not be appropriately answered by a background knowledge data source. All three types of questions are commonly asked by clinicians at the point of care (indeed, all three of the above questions were asked by physicians and collected by Ely[1] and Patrick and Li[2]). A QA system designed to aid clinicians at the point of care, therefore, should be able to determine which resource contains the most appropriate answer to the question.
In this work we develop both manually annotated datasets and automatic methods for classifying questions by their intended resource. We focus on the three types of resources discussed above: patient-specific, general or background medical knowledge, and scientific research. To the best of our knowledge, this is the first work to address this particular facet of QA despite it being a critical task in developing general-purpose QA systems for clinicians. We have manually annotated over 5,000 questions from four existing question collections. A supervised machine learning method is then employed to classify questions using lexical, syntactic, semantic, and topic-based features. Finally, we discuss the difficulties in both manual and automatic resource classification for questions.
Background
The need for medical QA systems to handle a wide variety of questions has resulted in questions being classified along many dimensions: answer type, topic, relation, user, answerability, and now resource. These will be discussed in more detail below, but the type that impacts the most on resource choice (besides what we discuss in this paper) is the user. Specifically, the assessment of the user’s medical expertise. Users are often organized into one of two groups: consumers and professionals[3,4] (though it has been shown that this distinction is oversimplified[4]). Importantly, resource choice is based on the user profile[5]: while a physician might be directed to a resource such as UpToDate, a consumer might be better suited by MedlinePlus. This is largely orthogonal to the study of resource type presented here, however, we acknowledge that certain general knowledge sources are better suited to consumers, while others are better suited to professionals. The same might be said about patient-specific and research resources, where patient portals and consumer-focused research news sites such as HealthDay1 might be considered more appropriate for consumers. Thus, resource selection requires both understanding the user and type of knowledge source. For the purpose of this study, we focus entirely on the resource type.
As noted above, there are many other ways of classifying questions. It is important to understand the breadth of medical question classification schemes, as many have an impact on resource classification. The most common classification in the open domain is answer type, sometimes referred to as the expected answer type or even simply question type. The answer type is the semantic type of the answer, and is useful in candidate answer extraction. Answer types are more studied in non-medical questions[6,7,8], where factoid-style questions with entity answers are common. Answer type methods for medical questions have been proposed by Cruchet et al.[9], who studied a small number of English and French questions, and by McRoy et al.[10], who focus on cancer-related questions. In analyzing answer types, both Cruchet et al.[9] and Roberts and Demner-Fushman[4] have found that boolean (i.e., true/false, yes/no) questions are the most common surface type. However, as observed by those researchers, often a higher-level type is more appropriate. For example, the question “Can Group B streptococcus cause urinary tract infections in adults?” is a boolean question on the surface, but is really asking for cause/etiology. This higher level answer type has been called many names: medical type, topic, and commonly just question type. This is the most common type of question classification studied by researchers, and has a critical impact on resource as some types are more likely to be discussed in some resources than others (e.g., definition questions are appropriate for general knowledge resources, while comparative effectiveness of treatments is more likely to be discussed in research studies). Among the proposed methods for this general question type are Cruchet et al.’s bilingual system, Roberts et al.’s[11] consumer QA system, and several systems utilizing Ely’s[12] general topics[13,14,15], which includes types such as diagnosis, history, prognosis, and treatment/prevention. Many medical QA approaches have focused on identifying question templates, common question types with slots for specific items with the question. Cimino et al.[16] maps questions to templates such as “Pharmacologic_Substance treats Disease_or_Syndrome” using a rule-based approach. Patrick and Li[2] developed EHR question templates such as “Did the patient have X, T?” and utilized a multi-layered supervised machine learning approach to classify templates and slot components. A very common template approach for research questions is PICO (problem/population, intervention, comparison, and outcome). The PICO approach has been utilized by Demner-Fushman and Lin[17] and Schardt et al.[18], amongst others. Beyond these types of question classification, other approaches have studied (a) relation extraction[19] using SemRep[20], (b) disease classification[21], (c) anatomy classification[13], and (d) question answerability[22,23].
As mentioned, all these types of question classification have a relation to the best resource (patient-specific, general, research) in which to find the question’s answer. The previous work that is the most overtly overlapping with our own are the answer type approach by McRoy et al. and the answerability approaches by Yu and colleagues. McRoy et al.’s[10] answer types form a taxonomy that contains three top-level nodes: factual, patient-specific, and non-clinician. Patient-specific questions correspond to those considered here, while factual questions may include both general knowledge and research. Non-clinician questions deal with non-medical issues such as insurance and legal advice. We found such questions in our dataset as well, but exclude them from our study as we are focused on clinical questions. Similarly, the answerability classification performed by Yu and Sable[22] and Yu et al.[23] isolates patient-specific questions as “unanswerable” based on Ely’s taxonomy[12]. In all these cases, no distinction is made between research questions and general/background medical knowledge. Admittedly, this distinction is often unclear, so in the next section we attempt to develop an initial specification for when questions belong to different resource categories.
Data and Annotations
We utilize four different question sets:
Ely (n = 1,500): These questions come from the Clinical Questions collection2 which was collected by Ely et al.[1,24,25] and D'Alessandro et al.[26] and is maintained by the National Library of Medicine (NLM). The questions were collected from physicians, either during direct observation or during a phone interview. They are largely designed to represent the stream-of-conscious questions that physicians have on a day-to-day basis, the vast majority of which go unanswered. There are 4,654 questions in the collection, from which we randomly sampled 1,500.
MedPix (n = 1,666): These questions came from MedPix3, which is an image database with over 53,000 images from over 13,000 patients. Originally built by James Smirniotopoulos at the Uniformed Services University, it is now maintained by NLM. MedPix contains hundreds of cases that are associated with quiz-style multiple choice questions, allowing radiologists to receive CME credits. The question set contains patient-specific questions forcing users to review the patient’s image and note, as well as medical knowledge questions inspired by the patient but not requiring any patient-specific answer. These knowledge questions may involve general radiology knowledge, or require being familiar with recent research.
GARD (n = 1,476): These questions were submitted by consumers to the Genetic and Rare Diseases Information Center (GARD)4. The questions are then curated, answered by NIH experts, and maintained on the GARD website in an FAQ format. Despite being consumer questions, the nature of the subject matter makes them more similar to clinician questions than other types of consumer questions[4]. The questions were originally in a paragraph style, but manually decomposed into individual questions[27]. When isolated from the original request, the questions appear as either patient-specific (as they are about details provided in the full request) or for medical knowledge. Since they are topically concerned with genetic and rare diseases, the users mostly understand the scientific literature might be the only source of answers for the particular disease.
Li (n = 486): These questions were collected by Patrick and Li[2] and are included in Li’s dissertation[28]. They are organized by the taxonomy discussed in Patrick and Li. The focus of the questions are patients in the intensive care unit, and thus almost all are patient-specific. We use only the sub-set of questions, defined by the taxonomy, of clinical relevance (ignoring, e.g., questions about the hospital organization). As such, they need not be manually annotated in the manner of the above corpora. Instead, the existing taxonomy classification is utilized, where only a small number (7) of general information questions are included.
A selection of questions from these corpora can be seen in Table 2.
Table 2:
Most similar questions using question-level embeddings and LDA-based topics.
| Resource Type | Corpus | ||
|---|---|---|---|
| Question 1 | Does he have spleen lacerations? | PatientSpecific | Li |
| 1. Does she have ongoing haemolysis? | PatientSpecific | Li | |
| Embeddings | 2. Is he ventilated? | PatientSpecific | Li |
| 3. Did she have sepsis overnight? | PatientSpecific | Li | |
| Topics | 1. Does it matter whether you draw the prostate specific antigen before or after the rectal exam? | General | Ely |
| 2. What causes polyembryoma? | General | GARD | |
| 3. How are they treated? | General | GARD | |
| Question 2 | What causes scleromyxedema? | General | GARD |
| 1. What causes pilomatrixomas? | General | GARD | |
| Embeddings | 2. What causes nonpalpable purpura? | General | Ely |
| 3. What causes excessive sweating? | General | Ely | |
| 1. What is his neurological status? | PatientSpecific | Li | |
| Topics | 2. I am interested in finding out information related to dihydropyrimidine dehydrogenase (DPD) in children. | General | GARD |
| 3. What is the differential diagnosis of neonatal seizures? | General | Ely | |
| Question 3 | Is there anything in the literature about allergy testing or desensitization to acyclovir? | Research | Ely |
| 1. Is there anything in the literature about milk and molasses enemas? | Research | Ely | |
| Embeddings | 2. Which of these features of a “ring lesion” is least common in an abscess? | Research | MedPix |
| 3. Is there anything in the literature on elbow fat pad signs? | General | Ely | |
| 1. Is there a genetic test which can help to rule-out neurofibromatosis? | General | GARD | |
| Topics | 2. Is a sample for genetic testing are required? | General | GARD |
| 3. Any information you have about any current therapies could be helpful. | Research | GARD |
To annotate each question, we used the following guidelines to distinguish between patient-specific, general, research, and other questions.
- PatientSpecific: In these questions, the answer is either contained directly in the patient’s chart or information needs to be retrieved from the chart to answer the question.
- What is her latest A1c value?
- Is this rash shingles or staphylococal impetigo?
- General: In these questions, the answer should be contained in a general medical knowledge source, exemplified by a medical textbook. This could include textbooks for specialties and sub-specialties.
- What causes scleromyxedema?
- How do you do a paracentesis?
- What are the immunizations for an 18-year-old?
- What urological symptoms are associated with Beckwidth Weidemann Syndrome?
- Research: In these questions, the answer should be best found in a research-type source. This includes both original scientific articles (bench and clinical) and articles summarizing the state-of-the-art in some area, including reviews and practice guidelines. While guidelines could arguably be considered general knowledge, we include them as research because they are typically updated more frequently than textbooks and they are closely associated with review articles that are published in the scientific literature.
- What research is being done at present regarding CDPX1?
- What percent of patients with Congenital Adrenal Hyperplasia (CAH) will have upper genitourinary tract abnormalities (e.g., vesicoureteral reflux and hydronephrosis)?
- How are the newer medications for attention-deficit hyperactivity disorder being used?
- Is glimepiride better than other sulfonylureas for diabetes?
- Other: These are non-clinical questions. Sometimes they are financial in nature, other times they are marked as Other because it is too difficult to understand the question and they should not be considered part of the dataset.
- Mother paid $101 for 50-dose desmopressin spray. Can that be the correct cost?
- Can you help me?
- Which of the following is True?
All of the questions except the Li set were double-annotated by an MD/PhD (LR) and a medical librarian (SS). Table 1 contains the reconciled statistics for all four corpora: numbers (and percents) of PatientSpecific, General, Research, and Other questions. As can be seen, the three manually annotated corpora are heavily imbalanced toward general questions (one of the main reasons we chose to also include the almost entirely patient-specific Licorpus). Table 1 additionally contains the κ agreement for the three manually annotated corpora, ranging from 0.62 to 0.78. These inter-annotator agreement numbers are decent, but by no means great. This should serve to emphasize that resource choice is still difficult for humans. Half of the disagreements were General versus Research (Ely: 50%, MedPix: 51%, GARD: 41%). Many of the disagreements involved confusion of what to consider Other (Ely: 34%, MedPix: 26%, GARD: 50%), so we had to carefully re-evaluate what to consider part of the scope of this work. Most of these Other disagreements were due to peculiarities in the datasets themselves and so should not be a concern for a clinical QA system. For instance, the GARD questions were automatically extracted from longer questions, resulting in some elliptical questions such as “How?” and “Can you help me?”. In MedPix, some of the questions made little sense without knowing the multiple-choice answers. These are not the type of question to be asked without context to a clinical QA system, so these inter-annotator disagreements can safely be ignored.
Table 1.
Annotation data for the four corpora. The LI corpus was annotated using data from Li[28].
| Corpus | Questions | PatientSpecific | General | Research | Other | κ |
|---|---|---|---|---|---|---|
| Ely | 1500 | 183 (12%) | 1099 (73%) | 159 (11%) | 59 (4%) | 0.62 |
| MedPix | 1666 | 317 (19%) | 1246 (75%) | 54 (3%) | 49 (3%) | 0.78 |
| GARD | 1476 | 40 (3%) | 1175 (80%) | 149 (10%) | 112 (8%) | 0.63 |
| Li | 486 | 479 (99%) | 7 (1%) |
Methods
Here we present an initial approach for automatically classifying the resource type for medical questions. Our goal is to characterize the linguistic criteria necessary to distinguish between questions requiring different resource types. As such, we forgo exhaustive experimentation and instead experiment with a variety of features at different linguistic levels: lexical, syntactic, semantic, and topic.
Lexical: We consider two bag-of-words features and length-based lexical features. The bag-of-words features include a simple unigram feature and a subset of unigrams that do not belong to a medical concept. To recognize concepts, we utilize the latest version of MetaMap[29] and restricted concepts to a specific set of UMLS[30] semantic types: diap, cgab, acab, inpo, patf, dsyn, anab, neop, mobd, sosy, drdd, clnd, antb, phsu, nsba, strd, vita. Thus, this feature could be considered lexico-semantic as it incorporates a semantic resource. The purpose of the non-concept unigram feature is to ignore medical terms that may be used in any resource (such as diseases) and instead focus on words that help delineate resource type. Additionally, we experiment with word- and character-length features. Upon observation, Research questions tend to be longer (in words) and contain words that are longer (in characters) than General questions.
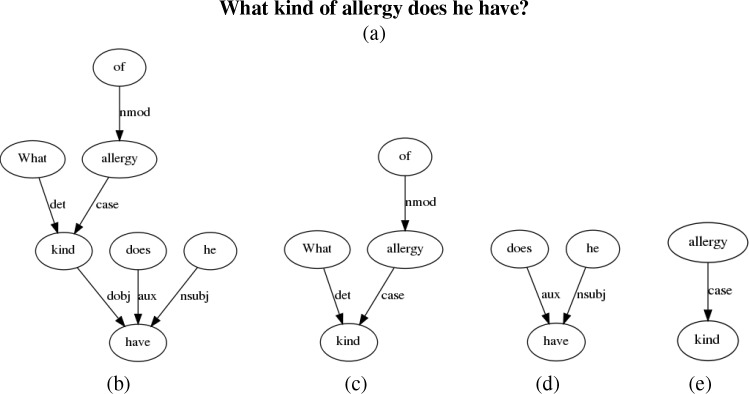
Syntactic: We consider a syntax feature based on the syntactic dependency parse. Syntax can help disambiguate different uses of the same word. For example, the word patient in “What is the diagnosis for the patient” (PatientSpecific) and “My patient has Hepatitis A and I'm wondering if there are any recent treatments?” (Research). We use the Stanford CoreNLP dependency parser[31]. As features, we consider subgraphs of the full dependency parse. Figure 1 shows a sample of dependency subgraphs for the question “What kind of allergy does he have?” Since these are even sparser than n-grams, we pre-calculate the most frequent subgraphs in each question corpus using the ParSeMiS implementation5 of the Gaston frequent subgraph mining algorithm[32] similar to the method employed in Luo et al.[33]. This still results in a large number of subgraphs, so we further filter based on the statistical association of a subgraph and an output class using Fisher’s exact test, taking all subgraphs above a threshold. All Fisher scores are calculated on the training set.
Semantic: We consider a concept feature based on MetaMap as well as distributional semantic features that represent each question in a semantic vector space. The concept feature indicates the semantic types present in the question, as different semantic types are more prevalent with different resource types. The distributional semantic features utilize paragraph2vec[34], a word sequence extension to the popular word2vec word embedding algorithm[35]. The gensim[36] implementation of paragraph2vec (known there as doc2vec) is used with default options. The vectors are pre-built using all questions as well as the 450,000-sentence health reference corpus used in Kilicoglu et al.[37]. We experiment with two versions of this feature: the first uses the embedding vector directly, while the second counts the output classes of the k-nearest neighbors (e.g., 5-NN) using cosine distance between the embedding vectors on the questions from the training set. Table 2 shows question similarity examples using embeddings based on PARAGRAPH2VEC.
Topics: We consider topic model features that represent questions as distributions over topics. We utilize the gensim version of Latent Dirichlet Allocation[38]. The unlabeled data and features are otherwise identical to the embedding features described above. Table 2 also shows question similarity examples using topics based on LDA.
Figure 1:
(a) Question, (b) Syntactic dependency tree, (c-e) Selection of subgraphs of dependency tree.
Using the above features, we utilize a single 3-class support vector machine (SVM) with a linear kernel[39]. Four datasets are used for evaluation: (1) Ely, (2) MedPix, (3) GARD, and (4) All = {Ely, MedPix, GARD, Li}. The LI dataset alone is too imbalanced for consideration. The feature set for each dataset is chosen from the features described above using a greedy search algorithm[40] on a development split of the data. The evaluation results reported in the next section are computed using a 5-fold cross validation of each corpus.
Results
The selected features for each dataset are as follows:
- Ely
-
–Non-concept unigrams
-
–Number of words < 8
-
–5-NN based on topic scores (200 topics)
-
–
- MedPix
-
–Unigrams
-
–Frequent subgraphs (less than log-prob -1.0)
-
–Semantic types in question
-
–Average character length of words
-
–
- Gard
-
–Non-concept unigrams
-
–5-NN based on topic scores (20 topics)
-
–
- All
-
–Unigrams
-
–Topic scores (200 topics)
-
–Number of words
-
–
As can be seen, different features were automatically selected for each dataset. A unigram feature (either the basic unigram or the non-concept unigram feature) was chosen as a base feature, while lexical, syntactic, semantic, and topic features were chosen to augment the bag-of-words feature. No dataset utilized the distributional semantic features, despite these being a good method for assessing similarity. It is thus likely that the distinguishing factor between resource type questions is quite localized (a single word or short phrase), while the similarity based on paragraph2vec is based on the entire question. In our experiments, the paragraph2vec-based vectors tended to better represent overall question similarity than the LDA-based vectors; however, the latter were chosen in three datasets (Ely, GARD, and All). Since LDA uses a bag-of-words assumption instead of the language model type assumption used by paragraph2vec, it tends to group words that are used in the same context but have very different semantic functions (this is why it is referred to as a topic model). This topical grouping effect might be more robust for this task than the full-question semantic similarity used by paragraph2vec.
Table 3 contains the results for each dataset. The baseline unigram feature alone greatly outperforms the weak baseline of choosing the most frequent resource type (Ely: +2.7%, MedPix: +14.28%, GARD: +4.55%, All: +6.95%). The selected features, however, only slightly outperform this stronger baseline feature (Ely: +1.88%, MedPix: +1.42%, GARD: +0.44%, All: +0.45%). This suggests that not only are simple lexical features the most important for classifying resource type, but the higher-level linguistic features are not easily representable with the features studied here (despite the fact that these are state-of-the-art features in many text classification tasks). In the Discussion below, we perform some error analysis to uncover the links between human disagreements and errors in the automatic system.
Table 3:
Automatic resource type classification results across four corpora.
| Feature Set | ELY | MedPix | GARD | All |
|---|---|---|---|---|
| Most Frequent Class | 76.27% | 77.06% | 86.14% | 71.88% |
| Unigram Bag-of-words | 78.97% | 91.34% | 90.69% | 78.83% |
| Best feature set | 80.85% | 92.76% | 91.13% | 79.28% |
Discussion
Providing relevant answers ultimately depends on finding the most appropriate source of information for a given question. As discussed in the Background, this involves not only the semantics of the question itself, but also the user’s background knowledge. In this work, however, we seek to isolate the resource choice from other considerations, grouping resources in a cross-sectional way (e.g., all general medical knowledge resources, regardless of whether the user is a high school student or highly-trained cardiologist). This reduces the need to jointly understand all the possible resource choice factors at once, which is especially useful if a system is designed to be used by a specific sub-group (e.g., clinicians).
At the highest level of granularity, the resources can be split into PatientSpecific (e.g., EHR or personal health record), General (e.g., textbooks and other background knowledge literature), and Research (e.g., journal articles, systematic reviews). Our results show that separating the literature (General and Research) into distinct buckets might not be feasible. Instead, one could consider a continuum ranging from the most basic background knowledge (e.g., Harrison’s Principles of Internal Medicine) to the most late-breaking research (e.g., the latest updates on the Zika virus in PubMed). While some questions are unambiguously background knowledge (e.g., “Is Williams syndrome inherited?”), and others unambiguously require research answers (e.g., Question (3)), many questions fall into a zone of ambiguity. In terms of manual annotation, the General vs. Research ambiguity resulted in many disagreements. If one only considers questions both annotators agree are either General or Research (i.e., ignore questions either annotator marked as PatientSpecific or Other), then inter-annotator agreement would be lower on two of the three datasets (Ely: 0.47, MedPix: 0.28, GARD: 0.67). In terms of automatic annotation, this was a large source of classifier error, such as the following mis-classifications:
-
(4)
Should a gestational diabetic have any antenatal fetal well-being testing done?
-
(5)
What are the best exercises for rotator cuff?
-
(6)
What is the success rate of various methods of endometrial biopsy? (that is, getting in)?
-
(7)
Is Keflex the drug of choice for a patient with positive streptococcal screen (allergic to penicillin, sulfa)?
Here, Questions (4) and (5) are General questions mis-classified as Research, while (6) and (7) are Research questions mis-classified as General. The choice of resource type is largely based on the granularity of information needed. The annotators felt that the first two questions concerned large populations and did not ask for detailed specifics, while the second two questions asks for details that often don’t make it into textbook-style knowledge resources (i.e., the exact success rate for (6) and the review of efficacy of various streptococcal drugs for (7)).
On the other hand, PatientSpecific questions are distinct and less ambiguous since they do not belong on such a continuum. However, there were difficulties when a question requires both patient-specific information and possibly external knowledge as well. For example:
-
(8)
What should I do about this bullous lesion found on sinus films?
Here, more knowledge from the patient chart is needed regarding the lesion, after which other knowledge sources might be required as well. We chose to consider such questions as PatientSpecific, but another option would involve a multi-label approach that might also label the question as General. In fact, this question was indeed mis- classified as General. Optimistically, however, one might claim that with an automatic QA system, users would not ask questions that required multiple information sources, so these questions should be less of a concern.
Perhaps, instead of using a 3-class resource type representation, the most ideal solution would be a large corpus of question-answer pairs. That is, instead of pre-deciding where a given answer is most likely to be found for a question, search a range of knowledge resources and identify the ones that actually contain an answer. The corpus created here was annotated in isolation from any possible answers (i.e., annotators did not look for an answer it multiple sources to see where one happened to be available). Rather, the annotators used their best judgement about where certain types of medical information would likely be found. With a large corpus of question-answer pairs, however, there is no need for such assumptions. The trade-off is obviously the additional cost of annotating answers over simply a 3-class determination in isolation. Our experience has indicated that annotating answers is incredibly time consuming[41], requiring approximately two orders of magnitude more time to find an answer than to simply classify a question in isolation. Further, rather than a 3-class classification problem, this would result in a far more granular representation which would either require far more annotations to achieve the same result, or an approach entirely different from the one presented here. Alternatively, a compromise solution could involve a more fine-grained hierarchical ontology of medical knowledge sources, encompassing many of the different types of publications (e.g., original research, systematic reviews, web articles). This could extend, for example, the MeSH article types to include other types of medical knowledge sources. Ambiguities could then be represented by selecting multiple nodes in the ontology.
Conclusion
We have presented our approach for classifying resource types of medical questions to aid QA systems in selecting the most appropriate data source. Three resource types were considered: patient-specific, general knowledge, and research knowledge. To develop the approach, four different corpora totaling over 5,000 questions were annotated with the three resource types, with moderate inter-annotator agreement. An initial automatic method was proposed utilizing state-of-the-art features, including dependency subgraph, distributional semantic, and topic modeling features. Nonetheless, basic lexical features provided the vast majority of the gain, suggesting that many existing state-of-the-art features cannot easily distinguish between these questions. Further advances, therefore, are needed not only in the automatic classification, but also in the representational methods used to identify the most appropriate medical resource.
Data Availability The annotated question corpora are available by request (kirk.roberts@uth.tmc.edu).
Acknowledgements
This work was supported by the National Library of Medicine (NLM) grant 1K99LM012104 (KR), as well as the intramural research program at NLM (LR, SS, DDF).
Footnotes
References
- 1.Ely JW, Osheroff JA, Ferguson KJ, Chambliss ML, Vinson DC, Moore JL. Lifelong self-directed learning using a computer database of clinical questions. J Fam Pract. 1997;45(5):382–388. [PubMed] [Google Scholar]
- 2.Patrick J, Li M. An ontology for clinical questions about the contents of patient notes. J Biomed Inform. 2012;45:292–306. doi: 10.1016/j.jbi.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Liu F, Antieau LD, Yu H. Toward automated consumer question answering: Automatically separating consumer questions from professional questions in the healthcare domain. J Biomed Inform. 2011;44(6) doi: 10.1016/j.jbi.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts K, Demner-Fushman D. Interactive use of online health resources: A comparison of consumer and professional questions. J Am Med Inform Assoc. 2016. [DOI] [PMC free article] [PubMed]
- 5.Proctor PM, Kan M, Lee SY, Zubaidah S, Yip WK, Jhao J, Arthur D, Li GM. eEvidence: Supplying Evidence to the Patient Interaction. In Connecting Health and Humans. 2009:488–492. [PubMed] [Google Scholar]
- 6.Hermjakob U. Parsing and Question Classification for Question Answering. Proceedings of the ACL Workshop on Open-Domain Question Answering; 2001; pp. 17–22. [Google Scholar]
- 7.Li X, Roth D. Learning question classifiers. Proceedings of the 19th International Conference on Computational Linguistics; 2002. [Google Scholar]
- 8.Roberts K, Hickl A. Scaling Answer Type Detection to Large Hierarchies. Proceedings of the Sixth International Conference on Language Resources and Evaluation; 2008. [Google Scholar]
- 9.Cruchet S, Gaudinat A, Boyer C. Supervised Approach to Recognize Question Tpe in a QA System for Health. In Stud Health Technol and Inform. 2008;136:407–412. [PubMed] [Google Scholar]
- 10.McRoy S, Jones S, Kurmally A. Toward automated classification of consumers’ cancer-related questions with anew taxonomy of expected answer types. Health Informatics Journal. 2015:1–13. doi: 10.1177/1460458215571643. [DOI] [PubMed] [Google Scholar]
- 11.Roberts K, Kilicoglu H, Fiszman M, Demner-Fushman D. Automatically Classifying Question Types for Consumer Health Questions. AMIA Annu Symp Proc; 2014; pp. 1018–1027. [PMC free article] [PubMed] [Google Scholar]
- 12.Ely JW, Osheroff JA, Ebell MH, Chambliss ML, Vinson DC, Stevermer JJ, Pifer EA. Obstacles to answering doctors’ questions about patient care with evidence: qualitative study. BMJ. 2002;324:1–7. doi: 10.1136/bmj.324.7339.710. PMC99056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi T, Shyu CR. Representing Clinical Questions by Semantic Type for Better Classification. AMIA Annu Symp Proc; 2006; [PMC free article] [PubMed] [Google Scholar]
- 14.Yu H, Cao Y. Automatically Extracting Information Needs from Ad Hoc Clinical Questions.. AMIA Annu Symp Proc; 2008; pp. 96–100. [PMC free article] [PubMed] [Google Scholar]
- 15.Cao Y, Cimino JJ, Ely J, Yu H. Automatically extracting information needs from complex clinical questions. J Biomed Inform. 2010;43:962–971. doi: 10.1016/j.jbi.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cimino JJ, Aguirre A, Johnson SB, Peng P. Generic queries for meeting clinical information needs. Bull Med Libr Assoc. 1993;81:195–206. [PMC free article] [PubMed] [Google Scholar]
- 17.Demner-Fushman D, Lin J. Answering Clinical Questions with Knowledge-Based and Statistical Techniques. Computational Linguistics. 2007;33(1):63–103. [Google Scholar]
- 18.Schardt C, Adams MB, Owens T, Keitz S, Fontelo P. Utilization of the PICO framework to improve searching PubMed for clinical questions. BMC Med Inform Decis Mak. 2007;7(16) doi: 10.1186/1472-6947-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hristovski D, Dinevski D, Kastrin A, Rindflesch TC. Biomedical question answering using semantic relations. BMC Bioinform. 2015;16(6) doi: 10.1186/s12859-014-0365-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. JBiomed Inform. 2003;36(6):462–477. doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 21.Roberts K, Kilicoglu H, Fiszman M, Demner-Fushman D. Decomposing Consumer Health Questions. Proceedings of the 2014 BioNLP Workshop; 2014; pp. 29–37. [Google Scholar]
- 22.Yu H, Sable C. Being Erlang Shen: Identifying Answerable Questions. IJCAI Workshop on Knowledge and Reasoning for Answering Questions; 2005. [Google Scholar]
- 23.Yu H, Sable C, Zhu HR. Classifying Medical Questions based on an Evidence Taxonomy. Proceedings of the AAAI 2005 Workshop on Question Answering in Restricted Domains; 2005. [Google Scholar]
- 24.Ely JW, Osheroff JA, Ebell MH, Bergus GR, Levy BT, Chambliss ML, Evans ER. Analysis of questions asked by family doctors regarding patient care. BMJ. 1999;319(7206):358–361. doi: 10.1136/bmj.319.7206.358. PMC28191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ely J, Osheroff J, Chambliss M, Ebell M, Rosenbaum M. Answering Physicians’ Clinical Questions: Obstacles and Potential Solutions. J Am Med Inform Assoc. 2005;12(2):217–224. doi: 10.1197/jamia.M1608. PMC551553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D'Alessandro DM, Kreiter CD, Peterson MW. An Evaluation of Information-Seeking Behaviors of General Pediatricians. Pediatrics. 2004;113:64–69. doi: 10.1542/peds.113.1.64. [DOI] [PubMed] [Google Scholar]
- 27.Roberts K, Masterton K, Fiszman M, Kilicoglu H, Demner-Fushman D. Annotating Question Types for Consumer Health Questions. Proceedings of the Fourth LREC Workshop on Building and Evaluating Resources for Health and Biomedical Text Processing; 2014. [Google Scholar]
- 28.Li M. Investigation, Design and Implementation of a Clinical Question Answering System. PhD thesis, University of Sydney; 2012. [Google Scholar]
- 29.Aronson A, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010;17:229–236. doi: 10.1136/jamia.2009.002733. PMC2995713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Methods of Information in Medicine. 1993;32(4):281–291. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manning CD, Surdeanu M, Bauer J, Finkel J, Bethard SJ, McClosky D. The Stanford CoreNLP Natural Language Processing Toolkit. Proceedings of 52nd Annual Meeting of the Association for Computational Linguistics: System Demonstrations; 2014; pp. 55–60. [Google Scholar]
- 32.Nijssen S, Kok JN. The Gaston Tool for Frequent Subgraph Mining. Proceedings of the International Workshop on Graph-Based Tools; 2004. [Google Scholar]
- 33.Luo Y, Sohani AR, Hochberg E, Szolovits P. Automatic Lymphoma Classification with Sentence Subgraph Mining from Pathology Reports. J Am Med Inform Assoc. 2014;21(5):824–832. doi: 10.1136/amiajnl-2013-002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Q, Mikolov T. Distributed Representations of Sentences and Documents. arXiv preprint arXiv 1405.4053. 2014.
- 35.Mikolov T, Sutskever I, Chen K, Corrado G, Dean J. Distributed Representations of Words and Phrases and their Compositionality. Advances in Neural Information Processing Systems. 2013:3111–3119. [Google Scholar]
- 36.Řehůřek R, Sojka P. Software Framework for Topic Modelling with Large Corpora. Proceedings of the LREC 2010 Workshop on New Challenges for NLP Frameworks; 2010; pp. 45–50. [Google Scholar]
- 37.Kilicoglu H, Fiszman M, Roberts K, Demner-Fushman D. An Ensemble Method for Spelling Correction in Consumer Health Questions. AMIA Annu Symp Proc; 2015; pp. 727–736. [PMC free article] [PubMed] [Google Scholar]
- 38.Blei DM, Ng AY, Jordan MI. Latent Dirichlet allocation. Journal of Machine Learning Research. 2003;3:993–1022. [Google Scholar]
- 39.Fan R, Chang K, Hsieh C, Wang X, Lin C. LIBLINEAR: A Library for Large Linear Classification. Journal of Machine Learning Research. 2008;9:1871–1874. [Google Scholar]
- 40.Pudil P, Novovičová J, Kittler J. Floating search methods in feature selection. Pattern Recognition Letters. 1994;15:1119–1125. [Google Scholar]
- 41.Deardorff A, Masterton K, Roberts K, Kilicoglu H, Demner-Fushman D. A protocol-driven approach to automatically finding authoritative answers to consumer health questions in online resources. Submission.