Abstract
Classification of drug-drug interaction (DDI) from medical literatures is significant in preventing medication-related errors. Most of the existing machine learning approaches are based on supervised learning methods. However, the dynamic nature of drug knowledge, combined with the enormity and rapidly growing of the biomedical literatures make supervised DDI classification methods easily overfit the corpora and may not meet the needs of real-world applications. In this paper, we proposed a relation classification framework based on topic modeling (RelTM) augmented with distant supervision for the task of DDI from biomedical text. The uniqueness of RelTM lies in its two-level sampling from both DDI and drug entities. Through this design, RelTM take both relation features and drug mention features into considerations. An efficient inference algorithm for the model using Gibbs sampling is also proposed. Compared to the previous supervised models, our approach does not require human efforts such as annotation and labeling, which is its advantage in trending big data applications. Meanwhile, the distant supervision combination allows RelTM to incorporate rich existing knowledge resources provided by domain experts. The experimental results on the 2013 DDI challenge corpus reach 48% in F1 score, showing the effectiveness of RelTM.
Introduction
Potential drug-drug interactions, defined as the co-prescription of two or more drugs that are known to interact, are one of the primary causes of medical error1, 2 Some studies estimate that medical errors result in around 44,000 to 98,000 deaths per year in the United States3, and that 7,000 of those deaths are due to medication-related errors4. Therefore, it is significant to prevent or reduce such errors in order to improve the clinical diagnosis quality and thus save thousands of patients’ lives. The Institute of Medicine has noted that a lack of drug knowledge is one of the most frequent proximal causes of such errors5. A report in Australia6 mentioned that 75% of hospital admissions related to medication errors are preventable, which implies that drug-drug interactions are a preventable cause of morbidity and mortality. Yet, the consequences of drug-drug interactions in the community are not well characterized6. Indeed, health care providers often have inadequate knowledge of what drug interactions can occur, of patient specific factors that can increase the risk of harm from an interaction, and of how to properly manage an interaction when patient exposure cannot be avoided7,8.
One of the primary reasons for this knowledge gap is the lack of complete and authoritative source of DDI knowledge9. Rather, there are multiple sources, including DrugBank, DailyMed, National Drug File, Express Scripts DRUG DIGEST and Medscape for WebMD4, each tasked with extracting, evaluating, and staying up-to-date with pertinent DDIs reported in the literature, and drug product labeling10. However, the inconsistency of information found in these resources is a serious issue. A manual review by a clinical pharmacologist of 100 randomly selected potential interactions out of more than 300,000 automatically extracted interactions revealed that 40% were genuinely inconsistent11. The dynamic nature of drug knowledge, combined with the enormity of the biomedical literature, makes the task of collecting and maintaining up-to-date information on drug-drug interactions extremely challenging and time-consuming.
There is a strong need to approach this task with automated methods, supplemented with human efforts. Natural language processing (NLP) and information extraction methods for identifying and extracting DDIs have been increased attention in the last few years, and several attempts have already been made to develop methods for this task, showing good potentials for success. In 2011, the first shared task challenge for DDI extraction,
DDIExtraction-2011 invited participants to develop automatic methods to extract DDIs. Most of participants developed binary classification models to handle this task. In the 2013 challenge, the organizers further classified the DDI categories and correspondingly, participants expanded their binary classification work into multiple classifications. Some of them took two steps to do the classification, first, detecting whether there is a relation among a pair of drugs and second, assign specific category to it. In general, those approaches yielded results ranging from 40% to 68%. However, all those approaches deployed supervised machine learning frameworks and focused more on feature engineering. Nonetheless, at the time when the data is growing exponentially, the supervised approaches cannot meet the needs of real-world applications. In particular, the dependency on annotated corpus forms a bottleneck for those methods since human annotations are expensive and time-consuming. Besides, the model will easily become over-fitted due to limited annotated corpus. On the other hand, if we only use unsupervised methods, like K-means, the results will be hard to interpret and therefore arduous to cluster into consistent categories.
In light of this, we conjecture that using semi-supervised or distant-supervised approach to detect and classify drug- drug interactions from biomedical text can be a better alternative because it can address limitations of both supervised and unsupervised methods. Specifically, we design, implement and evaluate a Bayesian model complemented with knowledge-driven distant supervision. In alignment with the term topic modeling for text mining, we call it relation topic modeling (RelTM). This approach attempts to make assumptions on the generative process in discourses and uses Gibbs sampling12, 13 and Expectation-Maximization14 to infer the model parameters. Unlike supervised models such as support vector machines15, or logistic regression16, unsupervised Bayesian methods17–19 do not require human efforts such as annotation and labeling in data preparation stages, which is its advantage in today’s trending big data applications. Meanwhile, the distant supervision combination allows us to incorporate rich existing knowledge resources provided by domain experts.
Related Work
In this section, we discuss techniques related to both the DDI challenges and to our proposed framework. The state-of-art techniques for the DDI challenge participation in 2011 are from Segura et al20. They used a hybrid method that combines supervised machine learning, shallow parsing and syntactic simplification with pattern matching. The UMLS MetaMap tool (MMTx)20 was used to provide shallow syntactic parsing and a set of domain-specific lexical patterns were developed to extract DDIs. The experimental results showed that they achieved a precision of 0.51, a recall of 0.72 and an F1 score of 0.60 using SVM classifier. In the 2013 challenge, the system with the highest F1 score was proposed by the FBK-irst team21. Their system is a multi-phase relation extraction system. They used two separate phases for DDI extraction and classification. For DDI extraction, they removed less informative sentences and instances, and then trained a system on the remaining instances. A hybrid kernel classifier that contains a feature based kernel, a shallow linguistic kernel, and a Path-Enclosed Tree kernel was used in the first step. For classification of DDI, they trained four separate models for each class (one vs. all the other classes). The innovative part of this system is detecting less informative sentences, where a sentence is considered less informative if all drugs in a sentence fall under the scope of a negation cue (such as not). A negation detector system (focused on a limited set of negation cues, such as no, n’t and not) is used to identify and filter the less informative sentences. The remaining sentences are classified with the SVM Light-TK toolkit22, utilizing the Charniak-Johnson reranking parser23, 24, a self-trained biomedical parsing model, and the Stanford parser25. On the DDI-DrugBank test dataset, they obtained an F1 score of 0.68 and on the DDI-Medline test dataset, an F1 score of 0.40. Other high-performance systems usually took similar approaches with differences in the use of machine learning model and feature selections26–33. All of them employed gold-standard labels for supervised learning besides the utilization of rich features and knowledge resources. Their model developments and especially their efforts in feature engineering provide us food for thought and allows us to directly explore those resources for our usage.
As mentioned in the introduction, in order to reduce the dependency on human annotations, we plan to develop semi-supervised models. Our proposed method is inspired by research in areas other than DDI classification. The work which is most relevant to us is the study of relation discovery from news domain using generative models34. They proposed rel-LDA and type-LDA, the latter of which is quite similar to ours. Specifically, their type-LDA composed of a probability graphical model where each document comprises N pairs. Each pair consists of relation features, entity features for source argument and destination argument. This framework takes dependency path, trigger words, part-of-speech features and named entity tags as features. The pair here refers to a relation in the document. Their relations include authorOf, bornIn, founder, parent and so on. Similarly, we have a generative graphical model to generate DDI, relation features and drug features.
Nonetheless, there are a few important differences in our work. First, our approach is a combination of generative model and clustering method. A similarity function is integrated into the relation sampling. This is inspired by Blei et al’s correlated topic models35 and distance dependent Chinese restaurant process36, Ghosh et al’s spatial distance dependent Chinese restaurant process37 and Chang et al’s relation topic models for document networks38. All of these techniques share the common point that they attempt to accommodate random partitions of non-exchangeable data by taking distances or similarities between topics into consideration. We notice that DDI pairs of same categories share correlated features, many of which are implicit. Consequently, methods based only on cooccurrence may miss clustering them together while the introduction of similarity functions may make up for this weakness. This is an innovative extension of type-LDA. With this extension, we can add rich knowledge resources and further we can flexibly experiment diverse similarity measures to discriminate relations more accurately.
Second, our work borrows the concept of distant supervision39. Distant supervision for relation classification is based on the assumption that if two entities, which are relevant, are mentioned in one sentence, that sentence may imply a relation and thus can be used as evidence for that relation40. Based on this assumption, we employ two knowledge sources, Drugbank and DailyMed, which are the two main resources of drug-drug interactions all over the world to boost the performance of our framework. Accordingly, our work shares some similarities with the work of Riedel et al41, in which, they employ Freebase to train a semi-supervised graphical model to extract relations from New York Times corpus. Their semi-supervised graphical model is essentially an undirected factor-graph model, which makes use of Gibbs sampling to update confidence on a group of expressions involving two entities. With the confidence learning, a ranked list of expressions extracted from Freebased is used as evidence for relation extraction in New York Times corpus.
One more related work is our latest work on coreference resolution with non-parametric topic modeling. In that work42, the proposed infinite mixture model integrates definite sampling with maximum likelihood estimation of mention similarity to estimate the cluster of coreferring entities in clinical notes. That system achieves an F measure of 0.847 on i2b2 2011 coreference dataset. Our current work shares two common ideas. First, both works attempt to handle relation classification (one for identity relation, the other for drug-drug interaction) under the framework of Bayesian topic modeling; second, similarity functions are utilized to guide the sampling of the objective functions. The difference lies in that our current work is not non-parametrical and distant supervision is employed. In addition, the generative process of our previous work is mention-entity sampling and the observed variable is the mention while our current work is feature-entity-relation sampling. More complex sampling is involved in our current approach.
Methods
The task of relation extraction can be abstracted as mapping surface textual relations to underlying semantic relations. There are several components to a coherent relation type, including a tight small number of textual expressions (two drug mentions, or called relation pair) as well as constraints on the entities involved in the relation. Often, the textual expressions themselves may not include sufficient information for us to model the relation type and thus make reasonable predictions. Hence, the constraints will play quite a big role in constructing discriminant predictors. For supervised approaches, those constraints are utilized as features to learn classifiers. Differently, in our work, we deploy them as observed variables for our Bayesian model plus the textual expressions for entities. Through this way, we can assume that multinomial distributions exist among those observed variables and further we can set Dirichlet priors for relation pairs. In addition, we notice that drug mentions, constraints and relation features belong to different semantic space where relation pairs depend on both relation features, such as trigger words and relation constraints, like negation tagger and drug mentions while drug mentions depend on constraints related to them, such as syntactic roles. Therefore, they should be modeled at different levels.
Based on above observations, we present relation topic modeling (RelTM), a semi-supervised probabilistic generative model for inducing clusters of relation types and recognizing their textual expressions. The set of relation types is not pre-specified but induced from observed unlabeled data. The relation induction will be augmented with distant supervision by incorporating external resources-based similarity functions.
We know that relations can only hold between certain entity types, known as selectional preferences (Ritter et al., 2010; Seaghdha, 2010; Kozareva and Hovy, 2010). This model can capture the selectional preferences of relations to their arguments (drugs in our context). In the mean time, it clusters pair into relational clusters, and arguments into different entity clusters (drug types). Instead of using the relations types from the labeled data, we suppose the data consists of more clusters, which is called latent relations in the following sections. In other words, we expect a specific relation type may have more than one pattern of feature set. This allows each relational cluster to have several subclusters. These sub-clusters (latent relations) are merged in the categorizing step.
Our observed data consist of a corpus of sentences and each sentence is represented by a bag of relation pairs. Each pair represents an observed drug-drug interaction between two drugs and consists of three components: the relation features, the source drug and the destination drug. The relation pair is the primary observed random variable in our model and we construct our models so that clusters consist of textual expressions representing the same underlying relation type. Since unlike classical LDA where observed words are independent and shared by the whole collection of documents, it will be reasonable to assume that each word may be generated by some specific topic. In our framework, a set of drug mention features supports one drug and relation features and mention features jointly support one relation. Therefore, the generative process is somewhat different. Namely, all of its features are only sampled under one specific relation and all of mention features are only sampled under one specific drug entity as well.
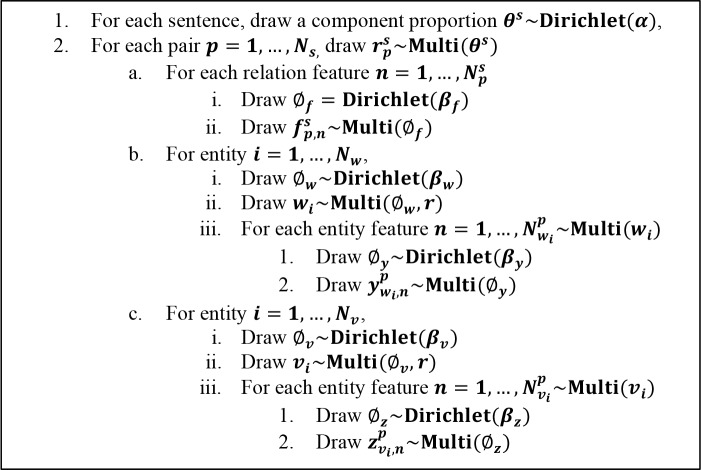
Generative Process
Before we start to introduce the generative process, some notations are explained. The symbol ~ stands for drawing the sample given a specific distribution. For example, the notation x~Multi(θ) represents sampling variable x from the multinomial distribution given parameter θ Correspondingly, x—Dirichlet(α) stands for sampling variable x from the multinomial distribution given parameter α Table 1 shows other notations used in this paper. Following the convention of NLP community, the real world objects mentioned in a discourse by noun phrases or pronouns are usually called entities, in our context, drugs, and the phrases or pronouns appearing in discourses are called mentions, namely drug instances. Mentions may consist of several words in free texts and each of such words is called a token. Each relation is composed of two drug mentions.
Table 1.
Notations of Variables
| Notation | Explanation |
|---|---|
| S | The total number of sentences |
| |R| | The total number of latent relations |
| N | A random variable to represent the total number of relation pairs in a sentence |
| |F| | The total number of features in each mention |
| «1:R | R-dimensional parameter of a Dirichlet distribution for relations |
| Parameters for R component distribution over relation features | |
| Parameters for R component distribution over drug entity 1 (w) or entity 2 (v) | |
| Parameters for F component distribution over features of drug entity 1 (y) or entity 2(z) | |
| θ1:R | R-dimensional parameters of relation distribution variables over a sentence |
| Parameters for R component distribution over the relation types | |
| Parameters for R component distribution over drug entity 1 (w) or entity 2(v) | |
| Parameters for F component distribution over features of drug entity 1 | |
| r1:N | Variables representing a sequence of relations in a specific sentence |
| f | Features of a given relation pair |
| {W, V}1:N | Variables representing hidden variable, the drug entity 1(w) or entity 2(v) in a specific sentence |
| {y,z}1:R | Finite set of observed variables that represents a specific features of drug entity 1(w) or entity 2(v) |
The plate diagram of graphical model of RelTM is illustrated in Figure 1. The generative process of relation features, drug entities and their entity features are described in Figure 2. Formally, there are N numbers of possible relations (N is a random variable) in each sentence. Each relation is represented by three distributions, namely, relation feature distribution, features of drug entity 1 and features of drug entity 2. When we detect the relations between two drugs, we first choose a relation type based on a relation type distribution, composed of |R| number of distribution with summation 1. Then we generate a bag of relation features one by one based on the chosen latent relation type as well as generate a pair of drug entities. Sequentially, drug entity 1 and the drug entity 2 are employed to generate mention features for specific drugs, namely, instances of entities. Essentially, we do all sampling with Dirichlet for priors and multinomial for their conjugate posteriors. Namely, each multinomial distribution is governed by symmetric Dirichlet distribution.
Figure 1.
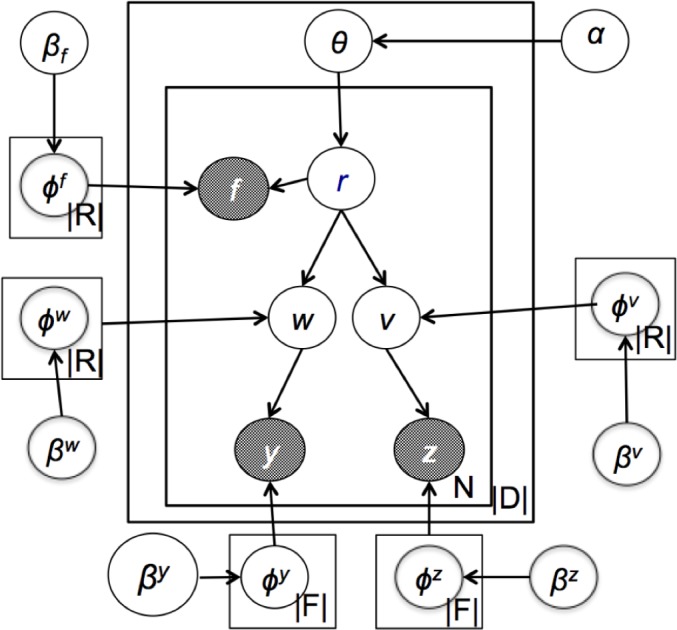
The Plate Diagram of DDI-LDA
Figure 2.
The generative process of DDI-LDA
The features of a relation pair consist of relation level features and mention level features. Relation level features include part-of-speech, dependency path, trigger, drug-bank indication (indications of drug-drug relations from the drug-bank database43), lexicon and POS features, mention level features include the drug mention itself, its named entity tag, i.e. the type of drug and its Unified Medical Language System (UMLS) concept unique identifier (CUI) and type unique identifier (TUI).
Strengthened Model with Similarity Measures
Although the current framework takes both relation features and mention features into considerations, it is found that co-occurrences hypothesis, the foundation of topic modeling, may lead to poor performance while evaluated by DDI evaluation metrics. Therefore, similarity measures are introduced to improve the system performance. We use Simple Matching Coefficient (SMC)2 as the similarity measure of two different relation pairs, which is widely used in data mining and statistics for quantifying the similarity of two binary vectors. The SMC of two different pairs can be simply represented by:
| Eq. 1 |
Specifically, after each sampling of relations for one pair of drugs is done, we selected high-discriminate features to compute similarity scores between current relation pair and previous relation pairs, instead of between two large feature sets. In this study, the trigger words in Drugbank and DailyMed are used. In other words, if two drug entities are mentioned with the same trigger verb in these two corpora, the similar feature count is incremented by one. After the count for each pair is obtained, it is divided by the size of the union of the entity feature sets. This step performs normalization to the co-occurrence feature count. Then the normalized feature counts of given relation assumptions are added to the probability vector in the inference stage.
Inferences
For simplicity and clarity, we omit the likelihood functions and their detailed derivations in this paper. We use collapsed Gibbs sampling to perform model inference. In collapsed Gibbs sampling, the distribution of a relation for its relation features and two drug mentions are based on values of mention features is computed. For the simplicity of the equation, we first define the following function:
| Eq. 2 |
where x ={y,z,w,v,f} and K ={|F|, |F|, |R|, |R|, |R|} respectively. Then we have:
| Eq. 3 |
In this is a count of how many features are assigned to relations in sentence s, excluding the count of current relation i. The first term is the distribution of current relation i sampled from the prior α. The second term is the product of distributions of a series of supporting feature vectors. They include the proportion of the mention features for drug entity 1 and for drug entity 2, the proportion of the drug entity 1 and drug entity 2 and the proportion of relation features in current relation i sampled from the prior α. And the third term is SMC term. For the simplicity of the derivations of Gibbs sampling, we separate the RelTM term and the SMC term in inference stage. That is to say, the RelTM is optimized during the inference stage, while the SMC term remains constant in Gibbs sampling.
| Eq. 4 |
| Eq. 5 |
The above two equations compute the distribution of the two drug entities. Similar to Eq. 2, the distribution of drug entity 1 is based on the counts of features assigned to all entities in sentence s, excluding the count of current entity i (term 1) and based on the distribution of all drug entities in sentence s (term 2). Similar is the distribution of entity 2.
Categorizing topics
For the DDI Drugbank dataset, because the ground truth relations have been manually labeled, intuitively, if an inferred latent relation is associated with many pairs in a particular category, the relation is likely to belong to that category. To capture this intuition, we match inferred latent relations to category q where assuming that all categories are equally important. We can estimate the probability of inferred latent relation type given category q as,
| Eq. 6 |
where denotes the learned probability of relation r given sentence s and sq denotes the subset of sentences in the DDI drugbank data collection that are labeled as category q.
Experimental Results
As a gold standard for all experiments in this task, we used DDIExtraction-2013 corpus provided by the challenge organizer, for development. It contains 142 Medline abstracts on the subject of drug-drug interactions, and 572 documents describing drug-drug interactions from the DrugBank database. The corpus includes 6976 sentences that were annotated with four types of pharmacological entities and four types of DDIs. The DDIs types are: advice, effect, mechanism, and int.
The four types of drug-drug-interactions are defined as follows.
Advice: the sentence notes a recommendation or advice related to the concomitant use of the two drugs. For example, “… UROXATRAL should NOT be used in combination with other alpha-blockers.”
Effect: the sentence states the effect of the drug interactions, including pharmacodynamic effect or mechanism of interaction. For example, “Quinoones may enhance the effects of the oral anticoagulant, warfarin, …”
Mechanism: the sentence describes a pharmacokinetic mechanism.
Int: the sentence mentions a drug interaction but doesn’t provide any additional information.
To demonstrate the effectiveness of our proposed model, we conducted a series of experiments on DDI Drugbank dataset. In our problem setting, we assume that all drugs are provided and sentences are the place all interaction among drugs take place. Any two drugs among sentences are potential relation pairs. Yet, we do not suppose relation labels between any two drugs in a sentence of the training data are known. Namely, we target to develop semi-supervised framework to assign labels to those unlabeled DDI pairs with the four categories introduced in Table 2.
Table 2:
Performance evaluation in DDI Drugbank dataset
| Category | Total Number of pairs | RelTM baseline | RelTM+SMC | ||||
|---|---|---|---|---|---|---|---|
| Precision | Recall | F1 | Precision | Recall | F1 | ||
| Mechanism | 827 | 0.325 | 0.302 | 0.313 | 0.427 | 0.317 | 0.364 |
| Effect | 1700 | 0.495 | 0.536 | 0.515 | 0.757 | 0.506 | 0.604 |
| Advise | 1322 | 0.386 | 0.210 | 0.272 | 0.604 | 0.376 | 0.464 |
| Int | 188 | 0.135 | 0.526 | 0.211 | 0.117 | 0.736 | 0.205 |
| Micro-average | 0.335 | 0.393 | 0.362 | 0.477 | 0.484 | 0.480 | |
Since we are using an unsupervised method in model development, we did not divide our dataset into training set and testing set. That is to say the experiments are carried out in the corpus described in Table 2. There are two models evaluated in this section: baseline RelTM model and the RelTM with similarity measure (SMC) model. The performance metrics is computed with the package provided by DDIExtractrion 2013 Challenge organizers. The performance of the two models mentioned above is shown in columns 3 to columns 5 and columns 6 to columns 8, respectively. Comparing these results, we can draw the conclusion that the combination of RelTM and distant supervision together improves the overall performance, which is indicated in the micro-average F1 score, from 0.3624 increasing to 0.480.
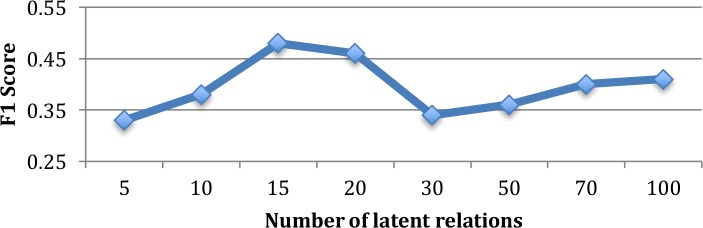
Since we are using a model with pre-specified number of latent relations, the same as other finite mixture model like LDA, the number of latent relations (topics) will also influence the system performance. Figure 3 demonstrates the relation between number of specified latent relations of our model and the micro-average F1 score. From the curve there are three trends worth mentioning. The first trend is when the number of latent relations is smaller than 15, the F1 score will increase with the increase of number of latent relations. This is because when the number of latent relations is less than the optimal number, the model may fail in splitting some similarity patterns of different DDI pairs. The optimal number of latent relations observed in the dataset is 15, based on experiments. The second trend is, after the optimal latent relation number, the F1 score will decrease with the number of relations increasing. This is due to the decreasing effectiveness of Gibbs sampling method we are using for the model inference. A similar pattern can also be found in conventional LDA model. However, as we continually obtained finer and finer clusters of DDI pairs, which applies after the relation number is larger than 50, the F1 score will slightly increase. This trend is because of the greedy manner of how we do the cluster labeling. As the clusters become smaller, the accuracy of cluster labeling will benefit the F1 score.
Figure 3.
Number of latent relations with respect to F1 score
Discussion
Due to the characteristic of Bayesian inferences, all the clusters are generated as random variables. It means they are generated according to the predicted probabilities of random variables in each category, instead of being directly assigned a label as in supervised methods such as support vector machine or random forest. The output results show our system can detect most of the strong category patterns, like the existence of the word “should” in advise and “effective” in effect in given sentences. However, while sampling the category label using category probabilities, most of the elements in the probability vectors are not strictly zero. Consequently, there is still possibility that DDI pairs with these strong patterns are not correctly classified, but the possibility of this kind of errors is within a low and acceptable range.
Another issue impacting the overall performance is the unbalanced category, in our study, int. In Table 2 we can find that there are only 188 instances of int pairs, which is much smaller than the second smallest category, which has 827 instances. Without any prior knowledge of category proportion, it is difficult for an unsupervised model to generate this small cluster, compared with other tunable supervised methods. As a result, our system generated more false positives in int categories than the other relatively large clusters. Therefore, the precision is lower that other categories, and it further impairs the micro-average F1 score. If we can utilize the prior knowledge of expected category proportion, this can be avoided by using unbalanced vector as Bayesian prior of RelTM instead of a scalar value we are currently using.
Although the performance of our system is not as good as the state-of-art supervised approaches, it does induce promising results, which shows that the utilization of distant supervised machine learning methods like LDA is promising in relation classification, even in challenging tasks as DDI classification. With the augmented similarity measures, which use domain knowledge in medical corpora, the results are improved. It is reasonable to believe that if more prior knowledge is used in the proposed model, then it has a potential to detect relations more accurately. This paves the way for us to deploy and extend our current system on large-scale unlabeled medical literatures and thus reduce the dependencies on the corpus annotations.
Conclusion and Future Work
In this paper, we proposed a topic modeling based distant supervised approach for the task of drug-drug interaction from biomedical text. Specifically, we proposed to design, implement and evaluate a Bayesian model complemented with knowledge-driven distant supervision. Under the assumption that the relations of drug-drug interaction are generated from proposed generative process, an inference algorithm using Gibbs sampling was also designed. A similarity measure was also introduced to strengthen the model and its effectiveness is proven by the experiments. The proposed approach does not require human efforts such as annotation and labeling in data preparation stage, which is its advantage in today’s trending big data applications. Meanwhile, the distant supervision combination allows us to incorporate rich existing knowledge resources provided by domain experts.
In future, we will address problems found in error analysis. In addition, we will extend the similarity function by incorporating updating rules and correlation between latent relations. One direction of exploration includes the release of symmetric Dirichlet prior to non-symmetric considering the unbalanced numbers of relation categories.
Acknowledgements
The authors gratefully acknowledge the support from the National Institute of Health (NIH) grant 1R01LM011934 and 1K99LM012021-01A1.
Reference
- 1.Rosholm J-U, Bjerrum L, Hallas J, Worm J, Gram LF. Polypharmacy and the risk of drug-drug interactions among Danish elderly. A prescription database study. Danish medical bulletin. 1998;45(2):210–3. [PubMed] [Google Scholar]
- 2.Gower JC. A general coefficient of similarity and some of its properties; Biometrics; 1971. pp. 857–71. [Google Scholar]
- 3.Kohn LT, Corrigan JM, Donaldson MS. To err is human:: building a Safer Health System; National Academies Press; 2000. [PubMed] [Google Scholar]
- 4.Reame NE, Wyman TL, Phillips DJ, de Kretser DM, Padmanabhan V. Net Increase in Stimulatory Input Resulting from a Decrease in Inhibin B and an Increase in Activin A May Contribute in Part to the Rise in Follicular Phase Follicle-Stimulating Hormone of Aging Cycling Women 1. The Journal of Clinical Endocrinology & Metabolism. 1998;83(9):3302–7. doi: 10.1210/jcem.83.9.5130. [DOI] [PubMed] [Google Scholar]
- 5.Kopp BJ, Erstad BL, Allen ME, Theodorou AA, Priestley G. Medication errors and adverse drug events in an intensive care unit: direct observation approach for detection. Critical care medicine. 2006;34(2):415–25. doi: 10.1097/01.ccm.0000198106.54306.d7. [DOI] [PubMed] [Google Scholar]
- 6.Runciman WB, Roughead EE, Semple SJ, Adams RJ. Adverse drug events and medication errors in Australia. International Journal for Quality in Health Care. 2003:i49–i59. doi: 10.1093/intqhc/mzg085. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y-F, Avery AJ, Neil KE, Johnson C, Dewey ME, Stockley IH. Incidence and possible causes of prescribing potentially hazardous/contraindicated drug combinations in general practice. Drug Safety. 2005;28(1):67–80. doi: 10.2165/00002018-200528010-00005. [DOI] [PubMed] [Google Scholar]
- 8.Hines LE, Murphy JE. Potentially harmful drug–drug interactions in the elderly: a review. The American journal of geriatric pharmacotherapy. 2011;9(6):364–77. doi: 10.1016/j.amjopharm.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Hines LE, Malone DC, Murphy JE. Recommendations for Generating, Evaluating, and Implementing Drug-Drug Interaction Evidence. Pharmacotherapy. The Journal of Human Pharmacology and Drug Therapy. 2012;32(4):304–13. doi: 10.1002/j.1875-9114.2012.01024.x. [DOI] [PubMed] [Google Scholar]
- 10.Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, et al. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic acids research. 2011;39:D1035–D41. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rastegar-Mojarad M, Harrington B, Belknap SM. Automatic detection of drug interaction mismatches in package inserts. Advances in Computing, Communications and Informatics (ICACCI); 2013 International Conference on; 2013. IEEE. [Google Scholar]
- 12.Heinrich G. Parameter estimation for text analysis. 2005.
- 13.Griffiths TL, Steyvers M, Blei DM, Tenenbaum JB. Integrating topics and syntax. Advances in neural information processing systems; 2004. [Google Scholar]
- 14.Blei DM, Ng AY, Jordan MI. Latent dirichlet allocation. Journal of machine Learning research. 2003;3:993–1022. [Google Scholar]
- 15.Cortes C, Vapnik V. Support vector machine. Machine learning. 1995;20(3):273–97. [Google Scholar]
- 16.Hosmer Jr DW, Lemeshow S. Applied logistic regression. John Wiley & Sons; 2004. [Google Scholar]
- 17.Ng V. Unsupervised models for coreference resolution; Proceedings of the Conference on Empirical Methods in Natural Language Processing; 2008. [Google Scholar]
- 18.Haghighi A, Klein D. Coreference resolution in a modular, entity-centered model; Human Language Technologies: The 2010 Annual Conference of the North American Chapter of the Association for Computational Linguistics; 2010. [Google Scholar]
- 19.Haghighi A, Klein D. Unsupervised coreference resolution in a nonparametric bayesian model; Annual meeting-Association for Computational Linguistics; 2007. [Google Scholar]
- 20.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program; Proceedings of the AMIA Symposium; 2001. American Medical Informatics Association. [PMC free article] [PubMed] [Google Scholar]
- 21.Chowdhury MFM, Lavelli A. USA: Atlanta, Georgia; 2013. FBK-irst: A multi-phase kernel based approach for drug-drug interaction detection and classification that exploits linguistic information; pp. 351–53. [Google Scholar]
- 22.Moschitti A, Quarteroni S, Basili R, Manandhar S. Exploiting syntactic and shallow semantic kernels for question answer classification. Annual meeting-association for computational linguistics. 2007.
- 23.Kahn JG, Lease M, Charniak E, Johnson M, Ostendorf M. Effective use of prosody in parsing conversational speech. Proceedings of the conference on human language technology and empirical methods in natural language processing. Association for Computational Linguistics. 2005.
- 24.McClosky D, Charniak E. Self-training for biomedical parsing. Proceedings of the 46th Annual Meeting of the Association for Computational Linguistics on Human Language Technologies: Short Papers; Association for Computational Linguistics; 2008. [Google Scholar]
- 25.Socher R, Bauer J, Manning CD, Ng AY. Parsing with Compositional Vector Grammars; ACL; 2013. [Google Scholar]
- 26.Thomas P, Neves M, Rocktäschel T, Leser U. WBI-DDI: drug-drug interaction extraction using majority voting; Second Joint Conference on Lexical and Computational Semantics (* SEM); 2013. [Google Scholar]
- 27.Tikk D, Thomas P, Palaga P, Hakenberg J, Leser U. A comprehensive benchmark of kernel methods to extract protein–protein interactions from literature; PLoS Comput Biol; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Airola A, Pyysalo S, Björne J, Pahikkala T, Ginter F, Salakoski T. All-paths graph kernel for protein-protein interaction extraction with evaluation of cross-corpus learning. BMC bioinformatics. 2008;9(11) doi: 10.1186/1471-2105-9-S11-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giuliano C, Lavelli A, Romano L. Exploiting shallow linguistic information for relation extraction from biomedical literature; EACL; 2006. [Google Scholar]
- 30.Vishwanathan S, Smola AJ. Fast kernels for string and tree matching; Kernel methods in computational biology; 2004. pp. 113–30. [Google Scholar]
- 31.Björne J, Kaewphan S, Salakoski T. UTurku: drug named entity recognition and drug-drug interaction extraction using SVM classification and domain knowledge; Second Joint Conference on Lexical and Computational Semantics (* SEM); 2013. [Google Scholar]
- 32.Rastegar-Mojarad M. Extraction and Classification of Drug-Drug Interaction from Biomedical Text Using a Two-Stage Classifier. 2013.
- 33.Rastegar-Mojarad M, Boyce RD, Prasad R. UWM-TRIADS: classifying drug-drug interactions with two-stage SVM and post-processing; Proceedings of the 7th International Workshop on Semantic Evaluation (SemEval); Atlanta, Georgia. June 2013. [Google Scholar]
- 34.Yao L, Haghighi A, Riedel S, McCallum A. Structured relation discovery using generative models. Proceedings of the Conference on Empirical Methods in Natural Language Processing; Association for Computational Linguistics; 2011. [Google Scholar]
- 35.Blei DM, Lafferty JD. A correlated topic model of science. Annals of Applied Statistics. 2007;1(1):17–35. [Google Scholar]
- 36.Blei DM, Frazier PI. Distance dependent Chinese restaurant processes. The Journal of Machine Learning Research. 2011;12:2461–88. [Google Scholar]
- 37.Ghosh S, Ungureanu AB, Sudderth EB, Blei DM. Spatial distance dependent Chinese restaurant processes for image segmentation; Advances in Neural Information Processing Systems; 2011. [Google Scholar]
- 38.Aggarwal CC, Zhai C. Mining text data: Springer Science & Business Media; 2012. [Google Scholar]
- 39.Go A, Bhayani R, Huang L. Twitter sentiment classification using distant supervision. CS224N Project Report; Stanford; 2009. 12 pp. [Google Scholar]
- 40.Mintz M, Bills S, Snow R, Jurafsky D. Distant supervision for relation extraction without labeled data. Proceedings of the Joint Conference of the 47th Annual Meeting of the ACL and the 4th International Joint Conference on Natural Language Processing of the AFNLP; Association for Computational Linguistics; 2009. [Google Scholar]
- 41.Riedel S, Yao L, McCallum A. Machine Learning and Knowledge Discovery in Databases. Springer; 2010. Modeling relations and their mentions without labeled text. pp. 148–63. [Google Scholar]
- 42.Liu S, Liu H, Chaudhary V, Li D. An Infinite Mixture Model for Coreference Resolution in Clinical Notes; Summit on Clinical Research Informatics; San Francisco, California. 2016. [PMC free article] [PubMed] [Google Scholar]
- 43.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic acids research. 2006;34:D668–D72. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]