Abstract
Semantic role labeling (SRL), which extracts shallow semantic relation representation from different surface textual forms of free text sentences, is important for understanding clinical narratives. Since semantic roles are formed by syntactic constituents in the sentence, an effective parser, as well as an effective syntactic feature set are essential to build a practical SRL system. Our study initiates a formal evaluation and comparison of SRL performance on a clinical text corpus MiPACQ, using three state-of-the-art parsers, the Stanford parser, the Berkeley parser, and the Charniak parser. First, the original parsers trained on the open domain syntactic corpus Penn Treebank were employed. Next, those parsers were retrained on the clinical Treebank of MiPACQ for further comparison. Additionally, state-of-the-art syntactic features from open domain SRL were also examined for clinical text. Experimental results showed that retraining the parsers on clinical Treebank improved the performance significantly, with an optimal F1 measure of 71.41% achieved by the Berkeley parser.
Introduction
Natural language processing (NLP) technologies are important for unlocking information embedded in narrative reports in electronic health record (EHR) systems. Although various NLP systems have been developed to support a wide range of computerized medical applications, such as bio-surveillance and clinical decision support, extracting semantically meaningful information from clinical text is still a challenge.
In the biomedical domain, semantic relation extraction systems, such as LSP1, MedLEE2, MedEx3 for clinical text and SemRep4, 5 for biomedical literature, have shown good performance and been widely used in different applications. These early-stage systems were often based on manually extracted patterns, following the sub-language theory6. According to the sub-language theory, the language of a closed domain (e.g., medicine and biomedicine) has special syntactic patterns as well as a limited number of main semantic types. Therefore, possible semantic relations could be identified by restricted constraints of syntactic and/or semantic patterns7. However, a careful examination of syntactic alterations that express the same semantic relations in biomedical text reveals that even in a semantically restricted domain, syntactic variations are common and diverse8. Thus, the coverage and scalability of manually extracted patterns may not be sufficient for those syntactic variations. In recent years, promoted by increasing challenges held by different portals (e.g., BioCreative, BioNLP, i2b2 and SemEval)9, more and more automatic information extraction systems have been built for different biomedical subdomains using data-driven statistical methods, such as machine learning algorithms. However, diverse syntactic variations still remain as an essential problem to extract semantic information from biomedical text, especially for clinical text, which contains more fragments and ill-formed grammars.
One potential solution to this problem is semantic role labeling10 (SRL) (also known as shallow semantic parsing), which focuses on unifying variations in the surface syntactic forms of semantic relations11. Specifically, the task of SRL is to label shallow semantic relations in a sentence as predicate argument structures (PAS)11. A predicate usually refers to a word indicating an event or a relation, and arguments (ARGs) refer to syntactic constituents representing different semantic roles in the event or relation. For each predicate, arguments representing the most important semantic roles are labeled with numbers, usually from ARG0 to ARG5. In addition, arguments representing modifiers of events (i.e., location, time, manner, etc.) are labeled as ARGMs. Taking the sentence “She should decrease the prednisone by 1-mg weekly” in Figure 1 as an example, the verb phrase “decrease” is the predicate indicating the event; the noun phrase “She” represents the role of ARG0, indicating the initiator/executor of the action “decrease”; the noun phrase “the prednisone” represents the role of ARG1, indicating the receptor of the action “decrease” (i.e. the entity decreased); while the prepositional phrase “by 1-mg weekly” represents the manner of how to decrease the prednisone (ARGM-Manner).
Figure 1.
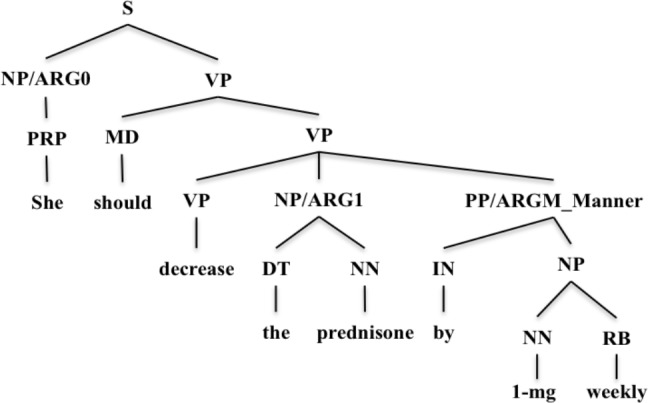
A syntactic parse tree with semantic roles added (ARGs).
Shallow semantic relations, or PASs are usually applied as features for machine learning algorithms, sentence structural representations in kernel-based models or inference rules in different applications, including question answering, text summarization and information extraction12–15, etc. Especially, PASs have been investigated in various biomedical sub-domains16–19 and made positive contributions in semantic information extractions, such as extracting drug-drug interactions from biomedical literature20 and temporal relations from clinical text 21.
Generally, a typical SRL system is built by using machine-learning methods based on annotated corpora. Since semantic roles are formed by syntactic constituents, two corpora are needed to build SRL systems, namely a corpus of syntactic parse trees and a corresponding corpus of semantic roles annotated on it. The most widely used large-scale corpora in open domain are the Penn Treebank22 and the SRL corpus PropBank11 developed on it. Many state-of-the-art syntactic parsers23–25 have been developed and applied to SRL in open domain 26–28. Some previous studies attempted to adapt these parsers (e.g., the Stanford Parser) to clinical text using medical lexicons29, 30 Recent years have also seen emerging efforts for syntactic annotation guidelines and corpora of clinical text31, 32 For example, the MiPACQ corpus (a multi-source integrated platform for answering clinical questions) annotated syntactic trees for 13,091 sentences following the Penn Treebank Style32. Furthermore, several SRL corpora were developed for clinical text following the PropBank Style. The available corpora were of different genres and note styles, including operative notes33, radiology notes32 from the SHARP Area 4 project (Strategic Health IT Advanced Research Projects), colon cancer pathology and clinical notes from the MiPACQ corpus32 and the THYME corpus34 (Temporal Histories of Your Medical Events). Based on those corpora, studies have been conducted to investigate SRL techniques for clinical text from EHRs. Albright et al. (2013)32 and Zhang et al. (2014)35 developed SRL systems on the MiPACQ corpus using dependency parse trees and constituent parse trees, respectively. Wang et al. (2014) built a SRL system on operative notes using an adapted parser36.
Given that semantic roles are formed by syntactic constituents in the sentence, an effective parser to first recognize those syntactic constituents is critical for developing a practical SRL system37. Furthermore, an effective feature set to describe the syntactic patterns between the predicate and the argument is also essential to SRL26. Although previous works have compared different syntactic parsers and representations for biomedical event extraction from
literature38, there are no formal evaluations and comparisons of state-of-the-art parsers23, 32, 36 and features32, 35, 36 for SRL in the medical domain.
In this study, we evaluated the SRL performance of three state-of-the-art constituent syntactic parsers: the Stanford parser23, the Charniak parser24 and the Berkeley parser25, using the MiPACQ corpus. We focused on constituent parse trees here because they could be directly converted to dependency parse trees39. The purposes of this study were two-fold: (1) to evaluate the SRL performance of existing state-of-the-art English parsers on clinical text, both the original parsers developed on Penn Treebank and parsers retrained on the clinical Treebank were examined; and (2) to validate the effectiveness of state-of-the-art syntactic features for SRL in the open domain40 and the biomedical domain19, 41 on clinical text. To the best of our knowledge, this is the first comprehensive study that investigates the influence of syntactic parsing and features for SRL on clinical text using multiple state-of-the-art parsers.
Methods
Dataset
This study used the MiPACQ dataset for SRL experiment. MiPACQ is built from randomly selected clinical notes and pathology notes of Mayo Clinic related to colon cancer26. Layered linguistic information is annotated in MiPACQ, including part of speech (POS) tags, syntactic Treebank, PASs for SRL, named entities, and semantic information from Unified Medical Language System. The syntactic Treebank annotations in MiPACQ follow the Penn Treebank guidelines, and the predicate-argument structure annotations for SRL follow PropBank guidelines. 13,091 sentences are annotated with syntactic trees. Among them, 6,145 sentences in MiPACQ are annotated for SRL, including 722 verb predicates with 9,780 PASs and 415 nominal predicates with 2,795 PASs.
The basic SRL system
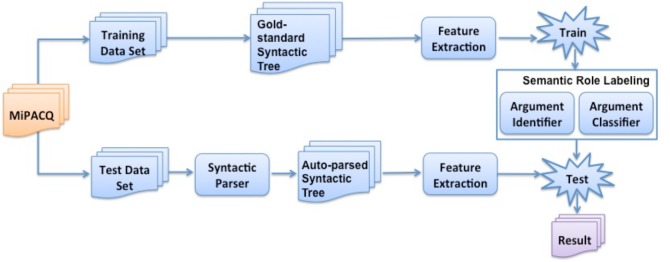
Figure 2 shows the study design for SRL of clinical text. Basically, the SRL system can be partitioned into the training stage and the testing stage. In the training stage, gold-standard syntactic trees of the training data set annotated in MiPACQ are used for feature extraction. A SRL task consists of two sub-tasks, the argument identification sub-task and the argument classification sub-task. First, a binary non-Argument vs. Argument classifier is built as the argument identifier on the entire dataset for all the predicates, instead of building one model per predicate. For argument classification, a multi-class classifier is built to assign semantic roles to arguments of all the predicates. In the testing stage, syntactic trees automatically generated by the syntactic parser are used for feature extraction. For each predicate, the argument candidates first go through the argument identifier. If one candidate is identified as an argument, it will go through the argument classifier that assigns the semantic role.
Figure 2.
Study design for semantic role labeling of clinical text
Comparing syntactic parsers and features
Three widely used state-of-the-art syntactic parsers, the Stanford parser23, the Charniak parser24, and the Berkeley parser25 are investigated for their influence on the SRL performance in our study. Moreover, state-of-the-art features, most of which are syntactic features, commonly used in open domain40 and biomedical domain19, 41 are extracted and compared for use with clinical text.
Syntactic Parsers
Stanford parser23: Stanford parser was initially developed based on un-lexicalized probabilistic context-free grammar (PCFG). The lexicalized PCFG parser and the dependency parser shared a factored product model, where the preference of PCFG phrase structures and lexical dependencies are combined by efficient exact inference, using an A* algorithm.
Charniak parser24: Chamiak parser is a high-performance lexicalized parser. It achieved an F1measure of 91.0% on the Penn Treebank. This parser is constructed by re-ranking the 50-best parse trees generated from a coarse-to-fine generative parser, using a discriminative Maximum Entropy model.
Berkeley parser25: Berkeley parser is also based on PCFG. In addition to local correlations between syntactic tree nodes, this parser learns to capture the non-local correlations between tree nodes using an EM algorithm based model.
Features
Similar to previous work of SRL for biomedical literature 41, 19 and clinical text35, we adopted the common features used in current state-of-the-art SRL systems. The features include baseline features from the original work of Gildea and Jurafsky (2002)42, advanced features taken from Pradhan et al. (2005)10 and feature combinations from Xue and Palmer (2004)26.
The features can be categorized into three major groups: (1) basic features include the lexical and syntactic features of the predicate and the argument; (2) context features include features of the syntactic nodes surrounding and the syntactic paths between the predicate and the argument; (3) feature combinations are feature tuples formed of two unitary features from the previous two groups. The complete feature set is described in Table 1. Except for the lemma of the predicate word and the relative position between the argument and the predicate, all the rest of the features are at the syntactic level and need to be extracted from the parse tree. For a more clear illustration, Table 2 lists the specific features extracted for the argument candidate “1-mg weekly” of the predicate “decrease “ in the example sentence shown in Figure 1.
Table 1.
Feature list of semantic role labeling
| Feature Group | Description |
|---|---|
| basic features | |
| Predicate | Lemmatization of the predicate word Voice of the verb predicate, i. e., active or passive |
| Argument | Syntactic head, first word, last word of the argument phrase and their POS tags Syntactic category of the argument node Whether the argument is a preposition phrase Enriched POS of prepositional argument nodes (e. g., PP-for, PP-in) |
| Relative position | Relative position of the argument with respect to the predicate (before or after) |
| Context features | |
| Production rule of predicate | Production rule expanding the predicate parent node |
| Syntactic category of argument neighbors | Syntactic categories of the parent, left sister and right sister of the argument node |
| Path | Syntactic path linking the predicate and an argument |
| No-direction path | Like Path, but without traversal directions |
| Partial path | Path from the argument to the lowest common ancestor of the predicate and the argument |
| Syntactic frame | Position of the NPs surrounding the predicate |
| Feature combinations | Predicate and head word of the argument Predicate and Syntactic category of the argument Predicate and relative position Predicate and path |
Table 2.
An example of features extracted for semantic role labeling.
| Sentence: She should decrease the prednisone by 1-mg weekly Predicate: decrease Argument candidate: 1-mg weekly | |
|---|---|
| Feature Group | Feature value |
| Basic features | |
| Predicate | decrease active |
| Argument | hw_1-mg, hw_pos_NN, fw_by, fw_pos_IN, lw_weekly, lw_pos_RB PP Yes PP-by |
| Relative position | after |
| Context features | |
| Subcategory of predicate | VP→VB–NP→PP |
| Syntactic category of argument neighbors | scp_VP, scl_NP, scr_null |
| Path | PP↑VP↓VB |
| No-direction path | PP_VP_VB |
| Partial path | PP↑VP |
| Syntactic frame | Position of the NPs surrounding the predicate |
| Feature combinations | decrease_1-mg decrease_PP decrease_after decrease_VB↑VP↓PP |
In the syntactic path feature, ↑ indicates a link from a child node to its parent, and ↓ indicates a link from a parent node to its child.
Experiments
PASs with at least one argument were used for the experiment. We used the open source toolkit, Liblinear,43 as implementations of the support vector machine algorithm. For each implemented method, all parameters were tuned for optimal performance.
Experiments and systematic analysis were conducted as follows:
Evaluate SRL performance of parsers with their default settings: In this experiment, we directly applied the three parsers to process all sentences of the test dataset. All the parsers were invoked with their default settings and models, which had been trained on the Penn Treebank.
Evaluate SRL performance of parsers re-trained on the clinical Treebank: To assess if the annotation of clinical Treebank could improve the performance of SRL, we applied three parsers retrained on the MiPACQ Treebank. We conducted ten-fold cross validation evaluation for each parser. The cross-validation involved dividing the clinical corpus equally into 10 parts, and training the parser on 9 parts with testing on the remaining part each time. We repeated the same procedure 10 times, one for each part, and then combined the results from the 10 parts to report the performance.
Evaluate SRL performance of each syntactic feature: To validate if syntactic features commonly used in the open domain were effective for clinical text, we conducted multiple runs of experiments, adding one new syntactic feature into the feature set for each run. The experimental results were compared to check the effectiveness of each feature.
Evaluation
Precision (P), recall (R) and F1-measure (F1) were used as evaluation metrics for argument identification (AI) and combined SRL task. Precision measures the percentage of correct predictions of positive labels made by a classifier. Recall measures the percentage of positive labels in the gold standard that were correctly predicted by the classifier. F1-measure is the harmonic mean of precision and recall. During the process of argument classification (AC), the boundaries of candidate arguments are already identified by the argument identification step. Therefore, the accuracy (Acc) of the classifier was used for evaluation, which is defined as the percentage of correct predictions with reference to the total number of candidate arguments correctly recognized in the argument identification step. Ten-fold cross validation was employed for performance evaluation.
Results
Table 3 illustrates the performance of semantic role labeling systems, which were trained on the gold standard syntactic trees and tested on the parsing results of Stanford, Charniak and Berkeley, as well as the gold standard syntactic trees, respectively. For these experiments, the whole feature set described in Table 2 was used. The original parsers trained on the Penn Treebank produced relatively lower performance. Charniak got the lowest F1-measure of 61.40%, whereas Berkeley outperformed the other two parsers with a F1-measure of 68.15%. After retraining on the clinical Treebank, the performance of all three parsers increased significantly, with the optimal F1-measure of 71.38% achieved by Berkeley. Testing on the gold standard parse trees yielded a F1-measure of 82.13%.
Table 3.
Performance of semantic role labeling systems trained on the gold standard syntactic trees and tested on the parsing results of Stanford, Charniak and Berkeley, and the gold standard syntactic trees, respectively (%)
| Parser | Model | AI | AC | AI+AC | ||||
|---|---|---|---|---|---|---|---|---|
| P | R | F1 | Acc | P | R | F1 | ||
| Stanford | Original | 70.42 | 82.17 | 75.84 | 88.16 | 62.09 | 72.44 | 66.86 |
| Retrained | 75.74 | 85.27 | 80.22 | 88.02 | 66.67 | 75.05 | 70.61 | |
| Charniak | Original | 67.75 | 74.54 | 70.98 | 86.51 | 58.61 | 64.49 | 61.40 |
| Retrained | 74.20 | 87.09 | 80.12 | 87.97 | 65.27 | 76.61 | 70.48 | |
| Berkeley | Original | 72.88 | 83.09 | 77.64 | 87.78 | 63.97 | 72.93 | 68.15 |
| Retrained | 76.72 | 85.37 | 80.81 | 88.33 | 67.77 | 75.40 | 71.38 | |
| Gold Standard | 91.41 | 91.60 | 91.51 | 89.75 | 82.04 | 82.21 | 82.13 | |
AI: argument identification AC: argument classification
To investigate whether syntactic features commonly used in the open domain are also effective for clinical text, multiple experiments were conducted by adding one new syntactic feature incrementally for each run. Table 4 lists the SRL performance of both the gold standard corpus and the parse results of the retrained Berkeley. As the baseline, the first run adopted all the basic features of predicate, argument and their relative position. Numbers in parenthesis show the changes to F1-measure of argument identification and accuracy of argument classification by adding each new feature. As illustrated in Table 4, all the syntactic features effective in the open domain were also helpful for argument identification of clinical text. The F1-measure was improved consistently from 20.07% and 17.47% to 91.51% and 80.81% for the gold standard corpus and the retrained Berkeley parser, respectively. In addition to the basic features, phrase types of argument neighbors, and the three path features made the most contribution to argument identification. In contrast, for argument classification, the basic features already yielded an accuracy of 86.74% for the gold standard corpus and an accuracy of 83.78% for the retrained Berkeley. Since the path features between the predicate and an argument dropped the accuracy slightly, we conducted additional experiments by removing those features for argument classification, which improved the overall F1-measure of our SRL systems to 82.14% (vs. 82.13%) for the gold standard corpus and to 71.41% (vs. 71.38%) for the retrained Berkeley parser.
Table 4.
Semantic role labeling performance of testing on the gold standard syntactic trees and parsing results of retrained Berkeley by adding one new feature each time (%)
| Feature Group | Test Corpus | AI | AC | AI+AC | ||||
|---|---|---|---|---|---|---|---|---|
| P | R | F1 | Acc | P | R | F1 | ||
| Baseline - Predicate+Argument+ Relative position | Gold | 60.57 | 12.04 | 20.07 | 86.74 | 54.83 | 10.94 | 18.23 |
| Auto-parsed | 57.56 | 10.30 | 17.47 | 83.78 | 52.97 | 9.48 | 16.07 | |
| Production rule of predicate | Gold | 59.73 | 13.99 | 22.67 (+2.60) | 87.20 (+0.46) | 54.54 | 12.77 | 20.69 (+2.46) |
| Auto-parsed | 58.34 | 10.91 | 18.38(+0.91) | 84.54 (+0.76) | 53.49 | 10.00 | 16.85 (+0.78) | |
| Phrase type of argument neighbors | Gold | 58.62 | 23.48 | 33.52 (+10.85) | 87.70 (+0.50) | 52.87 | 21.17 | 30.22 (+9.53) |
| Auto-parsed | 51.20 | 20.73 | 29.49 (+11.11) | 85.58 (+1.04) | 46.11 | 18.67 | 26.56 (+9.71) | |
| Path | Gold | 88.93 | 55.97 | 68.70 (+35.18) | 87.66 (-0.04) | 80.45 | 50.63 | 62.14 (+31.92) |
| Auto-parsed | 75.24 | 57.10 | 64.91 (+35.42) | 85.69 (+0.11) | 66.20 | 50.24 | 57.11 (+30.55) | |
| No-direction path | Gold | 88.58 | 60.62 | 71.97 (+3.27) | 87.58 (-0.08) | 79.66 | 54.51 | 64.72 (+2.58) |
| Auto-parsed | 74.38 | 61.09 | 67.07 (+2.16) | 85.44 (-0.25) | 64.97 | 53.35 | 58.58 (+1.47) | |
| Partial path | Gold | 91.13 | 91.19 | 91.16 (+19.19) | 87.60 (+0.02) | 79.85 | 79.90 | 79.87 (+15.15) |
| Auto-parsed | 76.87 | 84.92 | 80.69 (+13.62) | 85.41 (-0.03) | 65.60 | 72.47 | 68.86 (+10.28) | |
| Syntactic frame | Gold | 91.14 | 91.31 | 91.22 (+0.06) | 87.61 (+0.01) | 79.81 | 79.95 | 79.88 (+0.01) |
| Auto-parsed | 76.58 | 85.52 | 80.80 (+0.11) | 85.43 (+0.02) | 65.42 | 73.07 | 69.03 (+0.17) | |
| Feature combinations | Gold | 91.41 | 91.60 | 91.51 (+0.29) | 89.75 (+2.14) | 82.04 | 82.21 | 82.13 (+2.26) |
| Auto-parsed | 76.72 | 85.37 | 80.81 (+0.01) | 88.33 (+2.92) | 67.77 | 75.40 | 71.38 (+2.35) | |
AI: argument identification AC: argument classification
Discussion
Effective syntactic parsers and features are critical to establish a practical SRL system26,37. This study takes the initiative to make a formal evaluation and comparison of SRL performance on a clinical text corpus MiPACQ, using three state-of-the-art syntactic parsers and common syntactic features used in open domain. Experimental results demonstrate that retraining parsers on clinical corpora could improve the SRL performance significantly, with an optimal F1-measure of 71.41% achieved by the Berkeley parser. Despite the telegraphic type of clinical text, state- of-the-art syntactic features in open domain also proved to be effective for clinical text.
In terms of SRL errors caused by syntactic parsers, a major category was that the parsers did not recognize a large number of syntactic constituents acting as arguments (Original Stanford: 1175, Charniak: 1377, Berkeley: 1262). Nevertheless, retraining parsers on the clinical Treebank reduced such errors greatly (Retrained Stanford: 887, Charniak: 973, Berkeley: 816). Another major type of syntactic problems that caused SRL errors was the essential syntactic structure ambiguities44. For example, the sentence “He continues to note the sensation of bilateral leg numbness and pins and needle sensation with walking” contains conjunctive structures linking two phrases “the sensation of bilateral leg numbness”, and “pins and needle sensation”. It’s hard to determine if the prepositional phrase “with walking” only modifies the “pins and needle sensation” or both phrases.
Despite the unique characteristics of clinical text, such as fragments and ill-formed grammars, all the state-of-the-art syntactic features in the open domain contributed positively to clinical text, except for path features that dropped the accuracy of argument classification slightly. One possible reason for the decreased performance is that the specific semantic role of an argument in clinical text is dependent not only on syntactic paths but also on the clinical lexicon and relations. As an example, in the phrase “an advanced breast cancer treated with radiation therapy”, “an advanced breast cancer” is annotated as ARG2 (illness or injury) in the gold standard. However, it was mistakenly labeled as ARG1, because the extracted syntactic path features were similar to those of ARG1 in the corpus.
Our study has the following limitations. In this study, only the SRL performance of pathology notes and clinical notes are investigated. In the future, we plan to extend this study to other types of clinical notes such as operative notes, to assess the generalizability of our findings. In addition to parsers retrained on clinical text, parsers adapted to clinical text using domain lexicons and grammars could also be explored for SRL30. Although this study mainly focused on syntactic features, existing clinical knowledge45, such as semantic types36 and relations will be further examined as features for SRL in the next stage. Besides, there are other widely used parsers in open domain such as OpenNLP46, LingPipe47 and Gate48 in addition to the three parsers explored in this study. An extension of our study to other parsers will be conducted in the further, so that to reach the optimal SRL performance on clinical text.
Conclusion
This study made a formal evaluation and comparison of SRL performance on a clinical text corpus, MiPACQ, using three state-of-the-art parsers, the Stanford parser, the Berkeley parser, and the Charniak parser and state-of-the-art syntactic features from the open domain. Experimental results validated the effectiveness of retraining parsers with a clinical Treebank, with an optimal F1-measure of 71.41% achieved by the Berkeley parser. The results also demonstrated that common syntactic features in open domain could contribute positively to the clinical text.
Acknowledgement
This study was supported by grants from the NLM 2R01LM010681-05, NIGMS 1R01GM103859 and 1R01GM102282. We would like to thank the MiPACQ team for the development of the corpora used in this study.
References
- 1.Sager N. Natural language information processing: Addison-Wesley Publishing Company. Advanced Book Program. 1981 [Google Scholar]
- 2.Chen ES, Hripcsak G, Xu H, Markatou M, Friedman C. Automated acquisition of disease–drug knowledge from biomedical and clinical documents: an initial study. Journal of the American Medical Informatics Association. 2008;15(1):87–98. doi: 10.1197/jamia.M2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. Journal of the American Medical Informatics Association. 2010;17(1):19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. Journal of biomedical informatics. 2003;36(6):462–77. doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Kilicoglu H, Shin D, Fiszman M, Rosemblat G, Rindflesch TC. SemMedDB: a PubMed-scale repository of biomedical semantic predications. Bioinformatics. 2012;28(23):3158–60. doi: 10.1093/bioinformatics/bts591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris ZS, Harris Z. A theory of language and information: a mathematical approach. Oxford: Clarendon Press; 1991. [Google Scholar]
- 7.Friedman C, Kra P, Rzhetsky A. Two biomedical sublanguages: a description based on the theories of Zellig Harris. Journal of biomedical informatics. 2002;35(4):222–35. doi: 10.1016/s1532-0464(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 8.Cohen KB, Palmer M, Hunter L. Nominalization and alternations in biomedical language. PLoS One. 2008;3(9) doi: 10.1371/journal.pone.0003158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C-C, Lu Z. Community challenges in biomedical text mining over 10 years: success, failure and the future. Briefings in bioinformatics. 2016;17(1):132–44. doi: 10.1093/bib/bbv024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pradhan SS, Ward WH, Hacioglu K, Martin JH, Jurafsky D. Shallow Semantic Parsing using Support Vector Machines. In: Susan Dumais DM, Roukos S, editors. HLT-NAACL 2004: Main Proceedings; Boston, Massachusetts, USA: Association for Computational Linguistics; 2004. pp. 233–40. [Google Scholar]
- 11.Palmer M, Gildea D, Kingsbury P. The Proposition Bank: An Annotated Corpus of Semantic Roles. Comput Linguist. 2005;31(1):71–106. [Google Scholar]
- 12.Shen D, Lapata M. Using Semantic Roles to Improve Question Answering. Proceedings of the 2007 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning (EMNLP-CoNLL). Association for Computational Linguistics; 2007; Prague, Czech Republic. pp. 12–21. [Google Scholar]
- 13.Zhang R, Li W, Liu N, Gao D. Coherent narrative summarization with a cognitive model. Computer Speech & Language. 2016;35:134–60. [Google Scholar]
- 14.Moschitti A, Quarteroni S, Basili R, Manandhar S. Exploiting syntactic and shallow semantic kernels for question answer classification. Annual meeting-association for computational linguistics 2007; 2007; p. 776. [Google Scholar]
- 15.Luo Y, Uzuner Ö, Szolovits P. Bridging semantics and syntax with graph algorithms—state-of-the-art of extracting biomedical relations. Briefings in bioinformatics. 2016:bbw001x. doi: 10.1093/bib/bbw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Surdeanu M, Johansson R, Meyers A, Màrquez L, Nivre J. The CoNLL-2008 Shared Task on Joint Parsing of Syntactic and Semantic Dependencies; Proceedings of the Twelfth Conference on Computational Natural Language Learning; 2008; Stroudsburg,. PA, USA. Association for Computational Linguistics; pp. 159–77. [Google Scholar]
- 17.Wattarujeekrit T, Shah PK, Collier N. PASBio: predicate-argument structures for event extraction in molecular biology. BMC Bioinformatics. 2004;5(1) doi: 10.1186/1471-2105-5-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bethard S, Lu Z, Martin JH, Hunter L. Semantic role labeling for protein transport predicates. BMC bioinformatics. 2008;(9) doi: 10.1186/1471-2105-9-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahlmeier D, Ng HT. Domain adaptation for semantic role labeling in the biomedical domain. Bioinformatics (Oxford, England) 2010;26(8):1098–104. doi: 10.1093/bioinformatics/btq075. [DOI] [PubMed] [Google Scholar]
- 20.Yaoyun Z, Heng-Yi W, Jun X, et al. Leveraging Syntactic and Semantic Graph Kernels to Extract Pharmacokinetic Drug Drug Interactions from Biomedical Literature. The 2015 International Conference on Intelligent Biology and Medicine; 2015; Indianapolis, IN, USA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'souza J, Ng V. Classifying temporal relations in clinical data: a hybrid, knowledge-rich approach. Journal of biomedical informatics. 2013;46:S29–S39. doi: 10.1016/j.jbi.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kingsbury P, Palmer M. From TreeBank to PropBank. LREC’02. 2002 [Google Scholar]
- 23.Klein D, Manning CD. Accurate unlexicalized parsing. Proceedings of the 41st Annual Meeting on Association for Computational Linguistics - Volume 1. Association for Computational Linguistics; 2003; Sapporo, Japan. [Google Scholar]
- 24.Bikel DM. On the parameter space of generative lexicalized statistical parsing models. 2004 [Google Scholar]
- 25.Charniak E, Johnson M. Coarse-to-fine n-best parsing and MaxEnt discriminative reranking. Proceedings of the 43rd Annual Meeting on Association for Computational Linguistics. Association for Computational Linguistics; 2005; Ann Arbor, Michigan. [Google Scholar]
- 26.Xue N, Palmer M. Calibrating Features for Semantic Role Labeling Proceedings of EMNLP 2004. In: Lin D, Wu D, editors. Association for Computational Linguistics; 2004; Barcelona, Spain. pp. 88–94. [Google Scholar]
- 27.Merlo P, Musillo G. Semantic Parsing for High-precision Semantic Role Labelling. Proceedings of the Twelfth Conference on Computational Natural Language Learning; 2008; Stroudsburg, PA, USA. Association for Computational Linguistics; pp. 1–8. [Google Scholar]
- 28.Meza-Ruiz I, Riedel S. Jointly Identifying Predicates, Arguments and Senses Using Markov Logic. Proceedings of Human Language Technologies: The 2009 Annual Conference of the North American Chapter of the Association for Computational Linguistics; Association for Computational Linguistics; 2009; Stroudsburg, PA, USA. pp. 155–63. [Google Scholar]
- 29.Huang Y, Lowe HJ, Klein D, Cucina RJ. Improved identification of noun phrases in clinical radiology reports using a high-performance statistical natural language parser augmented with the UMLS specialist lexicon. J Am Med Inform Assoc. 2005 May-Jun;12(3):275–85. doi: 10.1197/jamia.M1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Pakhomov S, Ryan JO, Melton GB. Domain adaption of parsing for operative notes. Journal of biomedical informatics. 2015 doi: 10.1016/j.jbi.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan J-w, Yang EW, Jiang M, et al. Syntactic parsing of clinical text: guideline and corpus development with handling ill-formed sentences. Journal of the American Medical Informatics Association. 2013;20(6):1168–77. doi: 10.1136/amiajnl-2013-001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Albright D, Lanfranchi A, Fredriksen A, et al. Towards comprehensive syntactic and semantic annotations of the clinical narrative. Journal of the American Medical Informatics Association. 2013 Jan 25;:2013. doi: 10.1136/amiajnl-2012-001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Pakhomov S, Melton GB. Predicate argument structure frames for modeling information in operative notes. Studies in health technology and informatics. 2013;(192):783–7. [PMC free article] [PubMed] [Google Scholar]
- 34.Styler Iv WF, Bethard S, Finan S, et al. Temporal annotation in the clinical domain. Transactions of the Association for Computational Linguistics. 2014;2:143–54. [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang YT, Buzhou Jiang, Min Wang, Jingqi Wu, Yonghui Xu. Hua. domain adaptation for semantic role labeling of clinical text. Vol. 2014. Washington DC USA: AMIA; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serguei Wang YP, Ryan James O, Melton Genevieve B. Semantic Role Labeling for Modeling Surgical Procedures in Operative Notes. Vol. 2014. Washington DC USA: AMIA; 2014. [Google Scholar]
- 37.Punyakanok V, Roth D, Yih W-t. The Importance of Syntactic Parsing and Inference in Semantic Role Labeling. Comput Linguist. 2008;34(2):257–87. [Google Scholar]
- 38.Miwa M, Pyysalo S, Hara T, Tsujii Ji. Evaluating dependency representation for event extraction. Proceedings of the 23rd International Conference on Computational Linguistics; 2010. Association for Computational Linguistics; 2010; pp. 779–87. [Google Scholar]
- 39.De Marneffe M-C, Manning CD. The Stanford typed dependencies representation. 2008. Association for Computational Linguistics. 2008:1–8. [Google Scholar]
- 40.Pradhan S, Hacioglu K, Krugler V, Ward W, Martin JH, Jurafsky D. Support Vector Learning for Semantic Argument Classification. Mach Learn. 2005;60(1-3):11–39. [Google Scholar]
- 41.Tsai RT-H, Chou W-C, Su Y-S, et al. BIOSMILE: a semantic role labeling system for biomedical verbs using a maximum-entropy model with automatically generated template features. BMC bioinformatics. 2007;(8) doi: 10.1186/1471-2105-8-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gildea D, Jurafsky D. Automatic Labeling of Semantic Roles. Comput Linguist. 2002;28(3):245–88. [Google Scholar]
- 43.Fan R-E, Chang K-W, Hsieh C-J, Wang X-R, Lin C-J. LIBLINEAR: A library for large linear classification. The Journal of Machine Learning Research. 2008;9:1871–4. [Google Scholar]
- 44.Yang Jiang MH, Fan Jung-wei, Tang Buzhou, Denny Josh, Xu Hua. Parsing clinical text: how good are the state-of-the-art parsers? BMC Medical Informatics and Decision Making. 2015 doi: 10.1186/1472-6947-15-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aronson AR, Mork JG, Neveol A, Shooshan SE, Demner-Fushman D. Methodology for creating UMLS content views appropriate for biomedical natural language processing. AMIA Annu Symp Proc; 2008; pp. 21–5. [PMC free article] [PubMed] [Google Scholar]
- 46.Baldridge J. The opennlp project. 2005. URL: http://opennlpapacheorg/indexhtml (accessed 2 February 2012)
- 47.Carpenter B, Baldwin B. Text analysis with LingPipe 4. LingPipe Inc. 2011 [Google Scholar]
- 48.Cunningham H, Maynard D, Bontcheva K, Tablan V. GATE: an architecture for development of robust HLT applications. Proceedings of the 40th annual meeting on association for computational linguistics; 2002. Association for Computational Linguistics; 2002; pp. 168–75. [Google Scholar]