Abstract
Background
Identification of the types of strains of Mycobacterium tuberculosis (M. tuberculosis) complex causing tuberculosis (TB) could contribute to TB control program of specific geographic region as well as it could add knowledge onto the existing literature on TB worldwide. The objective of the present study was to identify the species and strains of M. tuberculosis complex causing pulmonary tuberculosis in central Ethiopia.
Methods
A health institution- based cross-sectional study was conducted on 338 smear positive TB cases visiting three hospitals between October 2012 and September 2013. Morning and spot sputum samples were collected before the starting of treatment regimens. Thus, a total of 338 pooled sputum samples collected from these cases. Samples were cultured on Löwenstein Jensen media and the isolates were identified by the region of difference (RD) 9 based polymerase chain reaction (PCR) and spoligotyping.
Result
Of the total isolates 98.6% of the isolates were identified to be M. tuberculosis while the remaining 1.4% were identified as M. africanum. Further, typing of M. tuberculosis using spoligotyping lead to the identification of 90 different strains of M. tuberculosis. Of these strains, 32 were clustered consisting of more than one isolate while the remaining 58 strains were unique consisting of single isolate. Thus, 79.3% (223/281) of the isolates were found in the clustered while only 20.6% (58/281) of the strains were unique. Forty-five of the spolgotyping patterns were registeredin the SITVIT2 or SpolDB4 database in while the remaining 45 were notfound in the database and hence were orphan strains. The dominant strains were SIT53, SIT149, and SIT54, consisting of 43, 37 and 34 isolates, respectively. Classification of the spoligotype patterns using TB-insight RUN TB-Lineage showed that 86.8, 6.4, 5, 1.4% ofthe isolatesbelonged to the Euro-American lineage, East-African-Indian, Indo-oceanic and M. africanum, respectively.
Conclusion
The identification of clustered and new strains using spolygotyping in present study does not give conclusive finding as spoligotyping has low discriminatory power. Thus, further identification of these isolates using mycobacterial interspersed repetitive unit-variable number tandem repeat (MIRU-VENTR) and or whole genome sequencing (WGS) recommended.
Keywords: Diversity of strain, M. tuberculosis, Central Ethiopia
Background
Tuberculosis (TB) is one of an infectious disease, affecting millions of people worldwide. According to the 13th Annual TB Report in 2014, there were 9.6 million new TB cases and 1.5 million TB deaths annually [1]. The spread of human immunodeficiency virus (HIV) and drug-resistant TB have exacerbated the situation.
Ethiopia has high rate of TB infection and the disease is one of major public health problems in the country [2]. According to a WHO report, the country is one among the world’s 22 countries with high TB burden [1]. The annual incidence of new TB cases was estimated to be 207/100,000 and the prevalence of TB in the country was 200/100,000 [1]. The country is one of the 27 high MDR-TB countries; ranked 15th with more than 5000 estimated MDR-TB patients each year [1, 2]. MDR TB was 1.6% of new cases and 12% of re-treatment cases [1].
Effective TB control program requires understanding of its epidemiology including the strains of M.tuberculosiscomplex (MTBC) circulating in the population [3]. Molecular epidemiological studies have been used to provide valuable information onthe spread of tubercle bacilli in outbreaks [4] and also contributed to study the transmission dynamics of TB [5]. Moreover, such studies can help in distinguishing exogenous reinfection from endogenous reactivation [6]. Additionally, molecular epidemiological studiescan be used to identify laboratory cross contamination [7] and to track the geographic distribution and spread of clones, including Multi drug resistant strains [8]. Furthermore, molecular typing has shown the large differences in pathobiological properties of MTBC species [9].
In Ethiopia, TB is a major public health problem and few molecular epidemiological studies have been done so far in some part of the country. While the genetic diversity of M. tuberculosis lineages in the country has been investigated [10–13], there is little or no data in some part of the country including this study area. The availability of such information will help study the phylogenetic characteristics of an organism, which in turn will provide new insight into the natural history of M. tuberculosis and Knowledge on the Mycobacterium tuberculosis strains circulating in the country communities is useful for epidemiology, transmission, and is essential in the control of the disease.
Thus, identificationthe types of strains of MTBC causing TB in a specific geographic region could contribute to the strengthening the TB control program of that specific geographic region, as it can alert personnel in the TB Control Program so that they can monitor the transmission of special strains such as drug resistant and virulent strains. In addition, identificationthe types of strains circulating in a specific geographic region could help in adding additional knowledge on the existing literature on TB worldwide. To our knowledge there is scarcity of information on strains of M. tuberculosis of circulating in central Ethiopia. Therefore, the objective of the present study was to identify the species and strains of MTBC causing pulmonary TB in central Ethiopia.
Methods
Study area
This study was performed at three different sites in central Ethiopia. These sites were Woliso and Atat towns and their surroundings in the southwest of Addis Ababa at a distance of 114 km and 187 km, respectively. The third site was Fiche town and its surrounding in the northwest of Addis Ababa at 115km. Sample collection was performed at hospitals located at these three sites, namely, St. Lukas, Atat and Fitche hospitals located at Woliso, Atat and Fitche tows, respectively. Sputum samples were collected from smear positive TB cases visiting these three hospitals.
Study design and study subjects
A health institution-based cross-sectional study was conducted on 338 smear positive TB patients visiting three hospitals, between October 2012 and September 2013. Three consecutive sputum samples (spot, early morning and spot) were collected from each of these 338 smear positive TB patient and pooled together for culturing. Sample collection was performed prior to the beginning of TB treatment. TB patients less than 18 years old excluded from this study. Socio-demographic data of the patients were obtained from the medical records of all patients.
Those samples positive for acid fast bacilli (AFB) by Ziehl-Neelsen (ZN) staining technique were collected labeled and samples from the individual patient pooled together. These sputum samples for culture were stored at −20°C and then transported in a cold box (at +4°C) to Aklilu Lemma Institute of Pathobiology (ALIPB), Addis Ababa, within a week for culture.
Culture
Morning and spot sputum samples were collected and processed for culture following the WHO Guideline [14]. Briefly, equal volume of 4% NaOH was mixedwith sputum sample and the mixture was centrifuged at 3000 rpm for 15 min at room temperature. After decanting the supernatant the sediment was neutralized with 2 N HClusing phenol red as an indicator. Neutralization was achieved when the color of the solution was changed from purple to yellow. Thereafter, 100 μl of the suspension was inoculated ontotwo sterile LJ medium slopes (which were enriched with either pyruvate or glycerol). The inoculated media were then incubated at 37°C in slanted positionfor 1 week and upright position for 4–5 weeks. The growth of the bacteria was read every week until the 8thweek of culture.
Preparation of specimens for molecular typing
Colonies were removed from the surface of LJ medium and suspended in 200 μl of sterile double distilled water. Thereafter, the colonies and water were mixed thoroughly and then, the mixture was heated at 80°C for 1 h in water bath. This is followed by centrifugation after which the supernatant was collected and used for amplification [15].
Region of difference (RD) 9-based polymerase chain reaction (PCR)
Identification of M. tuberculosis from the other members of M. tuberculosis complexspecies was done using RD9-based PCR. RD9-PCRwas performed on heat-killed cells to confirm the presence or absence of RD9 using three primers namely, RD9flankF, RD9 IntR, and RD9flankR. Amplification was done by standard thermo cycler (VWR Thermo cycler, UK). The PCR amplification mixture used consisted of 10 μlHotStarTaqMaster Mix (Qiagen, United Kingdom), 7.1μl distilled water, 0.3 μl of each three primers and 2 μl of DNA template(heat killed cells), giving a total volume of 20 μl. The PCR reaction was heatedat 95°C for 15 min after which it was subjected to 35 cycles consisting of 95 °C for one min, 55 °C for one minutes, and 72 °C for one minute. Thereafter, the reaction mixture was maintained at 72 °C for 10 min following which the product was removed from the thermocycler and run on agarose gel electrophoresis. For gel electrophoresis, 8 μl PCR products was mixed with 2 μl loading dye, loaded onto 1.5% agarose gel and electophoresed at 100 V and 500 mA for 45 min. The gel was then visualized using a computerized Multi- Image Light Cabinet (VWR). M. tuberculosis H37Rv, M. bovisbacilleCalmette-Guérin, and water were included as positive and negative controls. Interpretation of the result was based on bands of different sizes, as previously described by Parsons et al. [16].
Spoligotyping
Isolates that were positive for M. tuberculosis by RD9 PCR were further characterized by spoligotyping following the procedure described by Kamerbeeket al [17] and by observing the instructions of the spoligotype kit supplier (Ocimum Biosolutions Company, Iisselstein, and the Netherlands). The direct repeat (DR) region of the isolate was amplified by PCR using oligonucleotide primers (DRa and DRb) derived from the DR sequence [17]. The amplified biotinylated products were hybridized to a set of 43 oligonucleotides covalently bound to a membrane (Animal and Plant Health Agency, Great Britain). Bound fragments were incubated with streptavidinperoxidase conjugate and hybridizing DNA was detected by the enhanced chemiluminescence method, by exposure to X-ray film (Hyperfilm ECL, Amersham) as specified by the manufacturer’s instruction. The presence and absence of spacers was visualized on the film as black and white squares, respectively. Characterized strains of M. bovisand M. tuberculosis H37Rv were used as positive controls, whereas Qiagen distilled water (Qiagen company, Germany) was used as a negative control.
Use of SpolDB4 and Run TB-Lineage for the identification of strains and lineages
The results of spoligotyping were converted into octal and binary formats. These binary and octal formats of the strains were entered into query box so that the name of the strains are retrieved from the database if the spoligotype pattern of the strain in question fits the pattern that has already been registered in the SPolDB4 database [18] and at http://www.pasteur-guadeloupe.fr:8081/SITVITDemo/ [10]. If the pattern of the strain in question has not been registered in SPolDB prior to this study, the strain was considered as an orphan. In this study, an isolate is referred to as a colony that was grown on LJ media and found to be AFB-positive after staining with Ziehl Neelsen staining, whereas a strain is an isolate (s) with specific spoligotype pattern. Thus, a strain can consist of a single isolate or several isolates. A strain with a single isolate is termed as a unique strain while a strain with more than one isolate is considered as clustered strain. An online tool Run TB-Lineage http://tbinsight.cs.rpi.edu/run_tb_lineage.html was also used to predict the major lineages using a conformal Bayesian network (CBN) analysis and sub lineage using knowledge based Bayesian network (KBBN).
Statistical analysis
The statistical analysis was performed using STATA software version 12. Descriptive statistics were used to depict the demographic variables. Pearson chi-square was used to evaluate the association between source site of the isolate and clustering status of the isolates. Similarly, Fisher’s exact test was used to test of sourcesite is significantly associated with type of major lineage identified as well as with the type of dominant strain. Results were considered statistically significant whenever p-value was less than 5%.
Result
Demographic characteristics of the study participants
Among 338 smear positive sputa samples297 (87.9%) samples were confirmed as culture positive. A total of 297M. tuberculosis isolates were utilized to carry out RD9-PCRand spoligotyping analysis of which 281 gave valid spolygotyping data while the remaining 16 isolates did not give any pattern up on spolygotyping. One hundred thirty (46.2%) of 281 isolates obtained from female while 151 (53.7%) were isolated from males. Classification of the study participated on the basis of age showed that 99 (35.3%) were between 18 and 28 years of age, majority (47.7%; 134/281) were originated from Woliso and its surroundings (Table 1).
Table 1.
Background characteristics tuberculosis patients from whom the M. tuberculosis isolates were obtained
| Background characteristics | Number (%) of patients |
|---|---|
| Sex | |
| Male | 151(53.7) |
| Female | 130(46.3) |
| Age, Years | |
| 18–28 | 99(35.2) |
| 29–39 | 72(25.8) |
| 40–60 | 74(26.3) |
| > 50 | 36(12.8) |
| History of anti-tuberculosis treatment | |
| Previously treated | 10(3.6) |
| Not previously treated | 271(96.4) |
| Region | |
| Woliso | 134(47.7) |
| Fiche | 97(34.5) |
| Atat | 50(17.8) |
Region of difference (RD) 9-based polymerase chain reaction (PCR). The 281 isolates were analyzed using RD9 PCR and the result indicated that all isolates had intact RD9 implying that all the isolates were M. tuberculosis (Fig. 1).
Fig. 1.
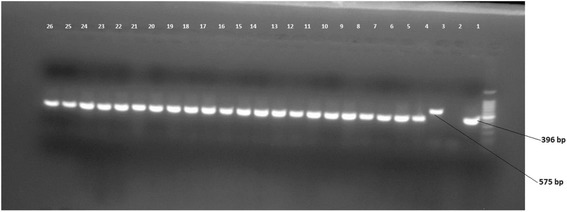
Picture of gel electrophoresis for RD9 deletion typing of M. tuberculosis isolates. Lane 1 is a ladder, lane 2 is M. Tuberculosis control, lane 3 is Negative control (molecular grade H2O), lane 4 is M. bovis control and lane 5–26 is culture isolate of M. Tuberculosis
Spolygotyping result
Spoligotyping of 281 isolates yielded 90 different spolygotype patterns. Of these, 32 strains were clustered containing of 223 (79.3%) isolates and 58 (20.6%) were unique strains (Tables 2 and 4). The overall diversity of the isolates was 32.4%. Out of the 90 spolygtype pattern, 45 were registered in the international data base (Table 2) and the remaining 45 were not found in the database (Table 3). The dominantly identified strains were SIT53, SIT149, and SIT 54 consisting of 43 isolates, 37 isolates, 34 isolates, respectively (Table 2). Classification of the spoligotype patterns using TB-insight RUN TB-Lineage showed that 86.8, 6.4, 5.3, 1.4% of the isolates belonged to the Euro-American lineage, East-African-Indian, Indo-oceanic and M. africanum, respectively (Table 2).
Table 2.
Description of 45 shared-types (SITs;n = 224 isolates) which have already been registered in the SITVIT2 or SpolDB4 database and corresponding spoligotyping defined lineages/sublineages starting from a total of 281 M. tuberculosis strains isolated in central Ethiopia
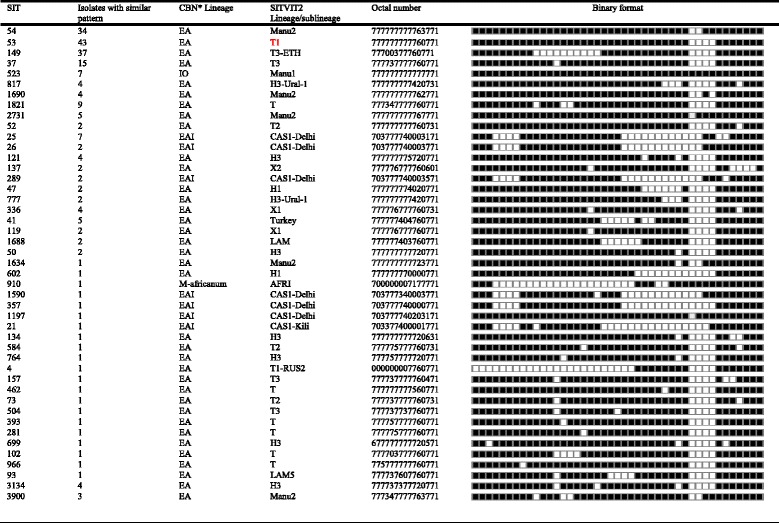
A total of 281 isolates grouped into 90 different spolygotype patterns. Of the 90 spolygtype pattern, 45 were registered in the international database while 45 of the strains were not found in the database. Of these 90 strains, 32 were clustered strains consisting of 223 (79.3%) isolates while 58 strains were found as single isolate. The dominantly found strains were SIT 53, SIT 149, and SIT 54 consisting of 43, 37 and 34 isolates, respectively. Classification of the spoligotype patterns using TB-insight RUN TB-Lineage showed that 86.8%, 6.4%, 5.3%, 1.4% of the isolates belonged to the Euro-American lineage, East-African-Indian, Indo-oceanic and M. africanum, respectively. EA Euro-American; EAI East-African Indian; IO Indo-Oceanic; MA M. africanum; MB M. bovis; CBN conformal Bayesian network; KBBN knowledge based Bayesian network
Table 4.
Distribution of strains and clustering rate in the Study Area (N = 281)
| Characteristics of the isolates | Number(%) of isolates in the study sites | P-value | ||
|---|---|---|---|---|
| Weliso | Atat | Fiche | ||
| Total spolygotyped isolates | 134 (47.7) | 50 (17.8) | 97 (34.5) | |
| Clustered isolates versus single | ||||
| Clustered | 112 | 41 | 70 | 0.163 |
| Single | 22 | 9 | 27 | |
| Clustering rate | 83.6% | 82% | 72.2% | |
| Major lineage by CBBN | ||||
| EA | 115 | 43 | 86 | 0.877 |
| EAI | 10 | 4 | 4 | |
| IO | 7 | 2 | 6 | |
| MA | 2 | 1 | 1 | |
| The three dominant Strains | ||||
| SIT 53 | 19 | 6 | 18 | 0.109 |
| SIT 149 | 17 | 9 | 11 | |
| SIT 54 | 21 | 8 | 5 | |
| Orphan strains | 21 | 10 | 26 | |
EA Euro-American, EAI East-African Indian, IO Indo-Oceanic, MA M. africanum, MB Mbovis, CBN conformal Bayesian network
Table 3.
Description of 45 orphan strains (N = 57) and corresponding spoligotyping defined lineages/sublineages recorded among M. tuberculosis strains starting from a total of 281 M. tuberculosis isolated in central Ethiopia
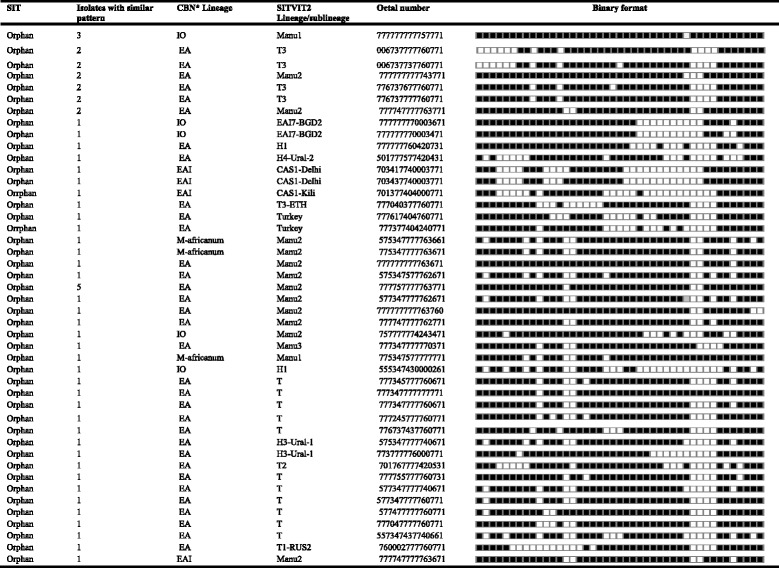
Of the 90 spolygtype pattern, 45 patters consisting of 57 isolates did not match with the patterns available in the database (orphans)
*CBN: conformal Bayesian network; EA EuroAmerican; IO Indo-Oceanic; EAI East African-Indian; MA M. africanum
Distribution of strains and lineages in the study area
Of the 281 isolates typed, 134(47.7) were originated from Woliso and its surroundings, with clustering rate of 83.6% (112/134), 97(34.5) were originated from Fiche with clustering rate of 72.2% (70/97). The remaining isolates were from Atat town with clustering rate of 14.6% (41/281). This finding did not show statistically significant difference in the proportion of clustering across the three source sites of the isolates (p-value = 0.163). Similarly, there was not significant association of the source site of the isolate with major lineage identified by CBBN (p-value = 0.877) as well as the type of dominant isolate (p-value = 0.109). The proportions of occurrence of the dominant lineage (Euro-American) at Woliso, Fitche and Atat towns were 85.5% (112/131), 89.1%(90/101) and 85.7% (42/49), respectively (Table 4).
Discussion
In the present study, 281 pulmonary TB cases were recruited from three towns and their surroundings in central Ethiopia for the isolation and identification of the species and strains of MTC causing pulmonary TB in the central Ethiopia. The isolation of was made from the sputum of patients on LJ medium while RD9-based PCR and spoligotyping used for identification of the isolates at the species and strain levels, respectively. An online tool Run TB-Lineage http://tbinsight.cs.rpi.edu/run_tb_lineage.html was also used for grouping lineages using a conformal Bayesian network (CBN) analysis and sub lineage using knowledge based Bayesian network (KBBN).
Out of the 90 different types of spolygotype patterns, 45 of the patterns which consisting majority of the isolates, matched with the patterns registered in the SITVIT2 database while the remaining 45 patterns did not match with the patterns registered in the SITVIT2 or SpolDB4 database. Similar studies that have been conducted in Ethiopia, and thus all the geographic regions of the country have not been covered and as a result all the circulating strains of M. tuberculosis have not yet been registered to the SITVIT2 database. Over 75% of the isolates were clustered strains with varies sizes of clustering while about a quarter of the isolates were found as unique strains. There are various assumptions with regard to such findings. Although spoligotyping has less discriminatory power in classifying strains, the finding of many isolates clustering in the same pattern could suggest the presence an on-going transmission of M. tuberculosis infection in the specific geographic region. On the other hand, the isolation of many unique strains could suggest the introduction of new strains into that specific region and these strains did not spread in that specific geographic region.
Similar to this study, SIT54 was dominantly isolated by earlier studies conducted in the Addis Ababa City (17), central Ethiopia (18) and in eastern central Ethiopia (19). This strain has been mainly reported to the SITVIT2 and SpolDB4 databasefrom South and East Asia, Middle East including Egypt and USA [10, 18]. Another interesting finding in the study was, similar to other studies conducted in Ethiopia earlier [11, 13] the ancestor strain SIT 523 was found consisting of good numbers of isolates. SIT 523 is characterized by the presence of all 43 spacers and is the ancestor strain of M. tuberculosis. It is less likely that strains keep all the 43 spacers intact for a long duration of time since they have to adapt to different pressures through changing their genetic makeup, and hence it is likely that the presence of 43 spacers intact could also be due to mixed infection, which needs further investigation using more powerful molecular techniques.
The M. tuberculosis isolated by the present study were belonged to four major lineages including the Euro-American, East-African-Indian, Indo-oceanic and the M. africanum. The dominant Lineage wasEuro- American Lineage consisting 86.8% of the isolates. This finding is in agreement with the earlier studies [11, 12, 19] conducted in different regions of Ethiopia. The 2nd and 3rd lineages under which the isolates grouped were East-African-Indian and Indo-oceanic lineage, respectively. Four isolates belonged to M. africanum Lineage. M. africanum has been reported to be an important cause of human TB in the west African countries including Guinea- Bissau [20], the Gambia [21], Sierra Leone [22], Senegal [23], Burkina Faso [24], Cameroon [25], Nigeria [26], and CôteD’Ivoire [27].
The limitation of this study is the use of only spoligotyping for typing of the isolates. Since the discriminatory power of spoligotyping is low, the finding of this study could lead to an overestimation of clustering because of the failure to differentiate between recent transmissions and mixed infections. In contrast to spoligotyping, MIRU-VNTR or WGS genotyping allow for a high-resolution and discrimination of the isolates for epidemiological studies and a valid phylogenetic strain classification [28–30].
Conclusion
The identification of clustered and new strains using spolygotyping in present study could not give conclusive report, as spoligotyping has low discriminatory power. Hence, further characterization of these isolates using MIRU-VNTR or WGS was recommended.
Acknowledgments
This study was jointly funded by the National Institute of Health (NIH, USA) through its H3Africa Consortium Program (Grant Ref. no. U01HG007472-01), Addis Ababa University through its Thematic Research Program, and Hawassa University. We would like to thank the staff member of TB laboratory at St. Lukas, Atat and Fiche Hospitals Hospital without whom this study could not have been completed.
Availability of data and materials
All of the data is contained within the manuscript
Authors’ contributions
ZB was involved in the design, data collection and laboratory work, and statistical analysis and interpretation of data and drafted the paper; AW was involved in the spoligotyping of the isolates ; YM, and GY was involved in critical revision of the paper;GM was involved in interpretation, and critical revision of the paper; GM was involved in statistical analysis, and critical revision of the paper; RP was involved in critical revision of the paper; GA was involved in the design, interpretation, and critical revision of the paper. All authors have read and approved the paper for submission.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Ethical clearance was obtained from Ethical Review Board of Natural Science, Addis Ababa University, Ethiopia (Ref. No. CNSDO/379/07/15). In addition, the purpose of the study was explained to all enrolled subjects in simple terms, and written informed consents were obtained from each of the study participants.
Abbreviations
- CBN
Conformal bayesian network
- CI
Confidence intervals
- DNA
Deoxo-nuclic acid
- DR
Direct repeats
- EA
Euro-American
- EAI
East-African -indian
- IO
Indo-occenic
- IS
Insertion sequence
- KBBN
Knowledge based bayesian network
- LJ
Löwenstein —jensen medium
- M. tb
Mycobacterium tuberculosis
- MA
M.-africanum
- MDR-TB
Multi drug resistance tuberculosis
- MIRU-VNTR
Mycobacterial interspersed repetitive unit-variable number tandem repeat
- MTBC
Mycobacterium tuberculosis complex
- PCR
Polymerase chain reaction
- RD
Regions of difference
- SIT
Shared international type
- SNPs
Single nucleotide polymorphisms
- TB
Tuberculosis
- WGS
Whole genome sequence
- WHO
World Health Organization
Contributor Information
Zufan Bedewi, Email: zufanw2006@yahoo.com.
Adane Worku, Email: adanewrk@yahoo.com.
Yalemtsehay Mekonnen, Email: yalemtshay.mekonnen@aau.edu.et.
Getnet Yimer, Email: getnet.yimer@aau.edu.et.
Girmay Medhin, Email: gtmedhin@yahoo.com.
Gezahegne Mamo, Email: gezahegnemamo@yahoo.com.
Rembert Pieper, Email: rpieper@jcvi.org.
Gobena Ameni, Email: gobena.ameni@aau.edu.et.
References
- 1.WHO . Global tuberculosis report. Geneva, Switzerland: WHO; 2015. [Google Scholar]
- 2.Federal Ministry of Health . Guideline for program and clinical management of drug resistant tuberculosis. Ethiopia. First. Addis Ababa: FMOH; 2009. [Google Scholar]
- 3.Van Soolingen D. Utility of molecular epidemiology of tuberculosis. Eur Respir J. 1998;11:795–797. doi: 10.1183/09031936.98.11040795. [DOI] [PubMed] [Google Scholar]
- 4.Paranjothy S, Eisenhut M, Lilley M, Bracebridge S, Abubakar I. Extensive transmission of Mycobacterium tuberculosis from 9 years old child with pulmonary tuberculosis and negative sputum smear. BMJ. 2008;337:1184. doi: 10.1136/bmj.a1184. [DOI] [PubMed] [Google Scholar]
- 5.Tostmann AS, Kik V, Kalisvaart NA, Sebek MM, Verver S. Tuberculosis transmission by patients with smear-negative pulmonary tuberculosis in a large cohort in the Netherlands. Clin Infect Dis. 2008;47:1135–1142. doi: 10.1086/591974. [DOI] [PubMed] [Google Scholar]
- 6.Dobler CC, Marks GB, Simpson SE, Crawford AB. Recurrence of tuberculosis at a Sydney chest clinic between 1994 and 2006: reactivation or reinfection? Med J Aust. 2008;188:153–155. doi: 10.5694/j.1326-5377.2008.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 7.Allix C, Supply P, Fauville-Dufaux M. Utility of fast mycobacterial interspersed repetitive unit-variable number tandem repeat genotyping in clinical mycobacteriological analysis. Clin Infect Dis. 2004;39:783–789. doi: 10.1086/423383. [DOI] [PubMed] [Google Scholar]
- 8.Mathema B, Kurepina NE, Bifani PJ, Kreiswirth BN. Molecular epidemiology of tuberculosis: current insights. Clin Microbiol Rev. 2006;19:658–685. doi: 10.1128/CMR.00061-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allix-Beguec C, Fauville-Dufaux M, Stoffels K, Ommeslag D, Walravens K. Importance of identifying Mycobacterium bovis as a causative agent of human tuberculosis. Eur Respir J. 2010;35:692–694. doi: 10.1183/09031936.00137309. [DOI] [PubMed] [Google Scholar]
- 10.SITVIT1Database. [http://www.pasteur-guadeloupe.fr:8081/SITVITDemo/]. Accessed 10 Sept 2012.
- 11.Diriba B, Berkessa T, Mamo G, Tedla Y, Ameni G. Spoligotyping of multidrug-resistant Mycobacterium tuberculosis isolates in Ethiopia. Int J Tuberc Lung Dis. 2013;17(2):246–250. doi: 10.5588/ijtld.12.0195. [DOI] [PubMed] [Google Scholar]
- 12.Garedew L, Mihret A, Mamo G, Abebe T, Firdessa R, Bekele Y, Ameni G. Strain diversity of mycobacteria isolated from pulmonary tuberculosis patients at DebreBirhan Hospital, Ethiopia. Int J Tuberc Lung Dis. 2013;17(8):1076–1081. doi: 10.5588/ijtld.12.0854. [DOI] [PubMed] [Google Scholar]
- 13.Mulugeta B, Ameni G, Bjune G, Couvin D, Rastogi N, and Abebe. Strain Diversity of Mycobacterium tuberculosis Isolates from Pulmonary Tuberculosis Patients in Afar Pastoral Region of Ethiopia. Biomed Res Int. 2014. [DOI] [PMC free article] [PubMed]
- 14.WHO . Laboratory services in tuberculosis control: culture. Part III. Geneva, Switzerland: WHO; 1998. [Google Scholar]
- 15.Yates M, Drobniewski F, Wilson S. Evaluation of a rapid PCR-based epidemiological typing method for routine studies of Mycobacterium tuberculosis. J Clin Microbiol. 2002;40:712–714. doi: 10.1128/JCM.40.2.712-714.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parsons ML, Brosch R, Stewart T, et al. Rapid and simple approach for identification of Mycobacterium tuberculosis complex isolates by PCR-based genomic deletion analysis. J Clin Microbiol. 2002;40:2339–2345. doi: 10.1128/JCM.40.7.2339-2345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35(4):907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, et al. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification population genetics and epidemiology. BMC Microbiol. 2006;6:23. doi: 10.1186/1471-2180-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Debebe T, Admassu A, Mamo G, Ameni G. Molecular characterization of Mycobacterium tuberculosis isolated from pulmonary tuberculosis patients in FelegeHiwot referral hospital, northwest Ethiopia. J Microbial. 2014;47(4):333–8. doi: 10.1016/j.jmii.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 20.Källenius G, Koivula T, Ghebremichael S, Hoffner S, Norberg R, et al. Evolution and clonal traits of Mycobacterium tuberculosis complex in Guinea-Bissau. J Clin Microbiol. 1999;37(12):3872–3878. doi: 10.1128/jcm.37.12.3872-3878.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jong B, Hill P, Brookes R, Gagneux S, Jeffries D, Otu J, et al. Mycobacterium africanum elicits an attenuated T cell response to early secreted antigenic target, 6 kDa, in patients with tuberculosis and their household contacts. J Infect Dis. 2006;193(9):1279–1286. doi: 10.1086/502977. [DOI] [PubMed] [Google Scholar]
- 22.Homolka S, Post E, Oberhauser B, George A, Westman L, Dafae F, Rüsch- Gerdes S, Niemann S. High genetic diversity among Mycobacterium tuberculosis complex strains from Sierra Leone. BMC Microbiol. 2008;8:103. doi: 10.1186/1471-2180-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viana-Niero C, Gutierrez C, Sola C, Filliol I, Boulahbal F, Vincent V, Rastogi N. Genetic diversity of Mycobacterium africanum clinical isolates based on IS6110-restriction fragment length polymorphism analysis, spoligotyping, and variable number of tandem DNA repeats. J Clin Microbiol. 2001;39(1):57–65. doi: 10.1128/JCM.39.1.57-65.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ledru S, Cauchoix B, Yaméogo M, Zoubga A, Lamandé-Chiron J, Portaels F, Chiron J. Impact of short-course therapy on tuberculosis drug resistancein South-West Burkina Faso. Tuber Lung Dis. 1996;77(5):429–436. doi: 10.1016/S0962-8479(96)90116-1. [DOI] [PubMed] [Google Scholar]
- 25.Niobe-Eyangoh S, Kuaban C, Sorlin P, Cunin P, Thonnon J, Sola C, Rastogi N, Vincent V, Gutierrez M. Genetic biodiversity of Mycobacterium tuberculosis complex strains from patients with pulmonary tuberculosis in Cameroon. J Clin Microbiol. 2003;41(6):2547–2553. doi: 10.1128/JCM.41.6.2547-2553.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cadmus S, Palmer S, Okker M, Dale J, Gover K, Smith N, Jahans K, Hewinson R, Gordon S. Molecular analysis of human and bovine tubercle bacilli from a local setting in Nigeria. J Clin Microbiol. 2006;44(1):29–34. doi: 10.1128/JCM.44.1.29-34.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niemann S, Kubica T, Bange F, Adjei O, Browne E, et al. The species Mycobacterium africanum in the light of new molecular markers. J Clin Microbiol. 2004;42(9):3958–3962. doi: 10.1128/JCM.42.9.3958-3962.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S. Proposal for standardization of optimized mycobacterial interspersed repetitive unitvariable- number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006;44:4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cowan L, Diem L, Monson T, Wand P, Temporado D, Oemig T, Crawford J. Evaluation of a two-step approach for large-scale, prospective genotyping of Mycobacterium tuberculosis isolates in the United States. J Clin Microbiol. 2005;43:688–695. doi: 10.1128/JCM.43.2.688-695.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Comas I, Coscolla M, Luo T, Borrell S, Holt K, Kato-Maeda M. Out-of-Africa migration and Neolithic co expansion of Mycobacterium tuberculosis with modern humans. Nat Genet. 2013;45:1176–1182. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All of the data is contained within the manuscript


