Abstract
Background
After E. coli O157, E. coli O26 is the second most prevalent enterohaemorrhagic E. coli (EHEC) serotype identified in cases of foodborne illness in Australia and throughout the world. E. coli O26 associated foodborne outbreaks have drawn attention to the survival capabilities of this organism in a range of environments. The aim of the present study was to assess the ability of E. coli O26 to survive the effects of disinfectants, acids and antimicrobials and investigate the possible influence of virulence genes in survival and persistence of E. coli O26 from human and cattle sources from Australia.
Results
Initial characterization indicated that E. coli O26 are a genetically diverse group that were shown to belong to a number of pathotypes. Overall, 86.4% of isolates were susceptible to all antimicrobials tested with no significant differences in resistance observed between pathotypes. A representative subset of isolates (n = 40) were selected to determine their ability to survive disinfectants at proposed industry working concentrations and acid stress. Profoam, Kwiksan 22, and Topactive DES. were able to inhibit the growth of 100% of isolates. The remaining three disinfectants (Dairy Chlor 12.5%, Envirosan and Maxifoam) were not effective against the subset of 40 E. coli O26. Finally, elevated MICs (1,024 to 4,096 μg/ml) of acetic, propionic, lactic, and citric acids were determined for the majority of the isolates (85%).
Conclusions
Australian E. coli O26 isolates belong to a range of pathotypes that harbor differing virulence markers. Despite this, their response to antimicrobials, disinfectants and acids is similar confirming that stress response appears unrelated to the presence of EHEC virulence markers. Notwithstanding, the tolerance to disinfectants and the elevated acid MICs for EHEC and the other E. coli O26 pathotypes examined in this study may contribute to bacterial colonization on food contact surfaces and subsequent foodborne illness caused by this pathogen.
Keywords: E. coli O26, Virulence marker, Pathotype, Antimicrobial agent, Disinfectant, Organic acid
Background
Enterohaemorrhagic Escherichia coli (EHEC) strains have been associated with a number of food-borne outbreaks which have led to life threatening sequelae such as hemolytic uremic-syndrome (HUS) and hemolytic colitis (HC) [1–3]. Epidemiological surveillance indicates that E. coli of O157:H7 serotype is the most frequently encountered EHEC implicated in sporadic and outbreak cases of illness [4]. However, other non-O157 serotypes such as O26:H11, O45:H2, O103:H2, O111:H8, O121:H19, O145:H28 and their non-motile forms have emerged and are now considered an important cause of human infection resulting in HUS [5]. Among the non-O157 serotypes, E. coli O26:H11 is one of the major serotypes of concern [3, 6].
Although it is not completely understood which suite of bacterial virulence determinants are most necessary for E. coli O26 to cause disease in humans, isolates recovered from human clinical samples typically possess Shiga toxins 1 or 2 or both (stx 1, stx 2) encoded by lambdoid bacteriophages, the E. coli attaching and effacing gene (eae) which is located on the LEE island and is necessary for bacterial colonisation of the gut and formation of the A/E lesion, and enterohemolysin (ehx), a plasmid encoded virulence factor thought to work synergistically with stx and contribute to the pathogenicity of EHEC [7–10]. It has been recognised that cattle represent a major reservoir of E. coli O26 [11–13]. Exposure to this pathogen can occur by a variety of routes including contact with animals on farms, consumption of contaminated meat, milk and its derivatives, water, spinach, sliced watermelon, clover sprouts, blueberries and strawberries [11, 14–20]. The contamination of food products with either stx positive or stx negative E. coli O26 strains has been reported previously [21–25] and in some cases it has resulted in the recall of food products and a number of outbreaks [6, 18, 26]. In 2005, there was an outbreak in France due to consumption of unpasteurised cow cheese contaminated with EHEC O26 [27]. In 2007, EHEC O26 infections occurred amongst consumers of ice cream produced from pasteurised milk made and sold at a farm in Belgium [23]. Additionally, multistate outbreaks of EHEC O26 infections in the USA have been also reported by the CDC in 2010, 2011, 2013 and 2015 [16, 17, 28].
The detection of E. coli O26 isolates in various environments including food, processing equipment and food contact surfaces and identifying them as a causative agent for a number of food-borne outbreaks creates the need for implementing prevention strategies to control this pathogen. Food producers and processors can use a range of antimicrobial agents such as sanitizers and disinfectants to assist in controlling this organism. Resistance of E. coli strains recovered from various environments to a variety of antimicrobial agents has been reported [29–32]. Variations in the response of different strains of E. coli to a range of disinfectants was demonstrated by Skaloud et al., [31] and Beier et al., [32] and suggest that the stress response of a range of E. coli strains may be highly variable. To date, most studies have focused on the survival capability of E. coli O157:H7 and minimal information is known about the response of E. coli O26 strains to these types of stressors. It cannot be assumed that E. coli O26 will respond similarly to E. coli O157:H7 or other E. coli strains when exposed to antimicrobial agents. We hypothesised that EHEC O26 have enhanced ability to persist and survive antimicrobial intervention in their planktonic state and that this consequently contributes to EHEC incidence and infection. Therefore, the aim of the present study was to assess the ability of E. coli O26 recovered from human clinical cases and cattle sources to resist the effects of disinfectants, acids and antimicrobials in the planktonic state and investigate the possible association of virulence genes such as stx and eae with the survival of Australian E. coli O26 from human and cattle sources.
Methods
Bacterial strains
A total of 88 E. coli O26 isolates collected previously from human clinical (10) and cattle (78) sources between 1995 and 2013 were utilised in this study. E. coli O26 isolates were initially selected with broad criteria based on virulence profiles, time and source. Isolates stored at -80°C in protect bacterial preservers (Technical Service Consultants Ltd) were subcultured on tryptone soya agar (TSA; Oxoid, UK) and incubated at 37 °C overnight.
PCR
Whole cell suspensions were created by suspending a single colony in 200 μl sterile Milli-Q water and tested by PCR for the presence of stx (stx 1 and stx 2), eae, ehx, eae conserved fragment (ecf), and bundle forming pilus (bfpA), using the primers shown in Table 1. PCR master mix containing 10X Dream Taq™ Buffer (Thermo Fisher Scientific, Australia), 250 mM dNTPs (Thermo Fisher Scientific, Australia), 0.02 mg/ml bovine serum Albumin (Sigma-Aldrich, USA), 12.5 pmol forward and reverse primer (GeneWorks, Australia) and 1.25 U Taq DNA polymerase (GeneWorks, Australia) was used. PCR products were subjected to gel electrophoresis on 2% agarose gels for 45 min at 100 V with resulting bands then visualised using a UV transilluminator. The anticipated amplicon size for each PCR product is shown in Table 1.
Table 1.
PCR Primer sequences used in this study
| Primers | Sequence | Amplicon size | References |
|---|---|---|---|
| stx1-F | 5'-ATAAATCGCCATTCGTTGACTAC-3' | 180 | [4] |
| stx1-R | 5'-AGAACGCCCACTGAGATCATC-3' | ||
| stx2-F | 5'-GGCACTGTCTGAAACTGATCC-3' | 255 | [4] |
| stx2-R | 5'-TCGCCAGTTATCTGACATTCTG-3' | ||
| eae-F | 5'-GACCCGGCACAAGCATAAGC-3' | 284 | [4] |
| eae-R | 5'-CCACCTGCAGCAACAAGAGG-3' | ||
| hlyA-F | 5'-GCATCATCAAGC GTACGT TCC-3' | 534 | [4] |
| hlyA-R | 5'-AATGAGCCAAGCTGGTTAAGC T-3' | ||
| wzx O26-F | 5'-CGCGACGGCAGAGAAAATT-3' | 326 | [47] |
| wzx O26-R | 5'-ACAATCCAACCGAACCAAAC-3' | This study | |
| ecf-F | 5'-TATCAGCACCAAAGAGCGGGAACA-3' | 99 | [48] |
| ecf-R | 5'-CCCTTATGAAGAGCCAGTACTGAA-3' | ||
| rmlA 30snp-F | 5'-AAGTCGCAGGCTTGT-3' | 484 | This study |
| rmlA 30snp-R | 5'-CGAAGACCCGCTAAC-3' | ||
| BFPA300-F | 5'-GGAAGTCAAATTCATGGG-3' | 300 | [49] |
| BFPA300-R | 5'-GGAATCAGACGCAGACTGGT-3' |
Detection of rmlA SNP using RFLP
A single nucleotide difference (G→T) at position 30 within rmlA has been shown to be associated with the presence of stx in E. coli isolates [33]. Primers rmlA 30snp-F and rmlA 30snp-R were used to amplify a 484 bp portion of rmlA (Table 1). Amplified PCR products were digested for 4 h at 37 °C using the restriction enzyme AciI. PCR products were subjected to gel electrophoresis on 2% agarose gels for 45 min at 100 V with resulting bands then visualised using a UV transilluminator. Isolates harbouring the rmlA SNP yielded 10 bp (not visible), 161 bp and 313 bp fragments whereas those isolates without the SNP yielded 10 bp (not visible), 109, 161 and 204 bp fragments.
Antimicrobial Resistance (AMR)
To determine the AMR phenotype of E. coli O26 isolates a custom susceptibility panel AUSVN2 (TREK Diagnostics, UK) designed specifically for testing Australian Gram-negative isolates was used. All plates were inoculated and assessed using the Sensititre system (TREK Diagnostics). Antimicrobials that were tested are cefazolin, cefotaxime, ceftiofur, amoxicillin / clavulanic acid, nalidixic acid, cefoxitin, ciprofloxacin, meropenem, ceftriaxone, gentamicin, ampicillin, trimethoprim / sulfamethoxazole, chloramphenicol, kanamycin, tetracycline and streptomycin. The Clinical and Laboratory Standards Institute (CLSI) criteria were utilised to identify antimicrobial resistance breakpoints when available; otherwise European Committee on Antimicrobial Susceptibility Testing (EUCAST) and National Antimicrobial Resistance Monitoring System (NARMS) values were used. E. coli ATCC 25922 was used as a control strain.
Pulse-field gel electrophoresis (PFGE)
PFGE was performed using the standardised PulseNet protocol with chromosomal DNA of Salmonella ser. Braenderup H9812 digested with XbaI (Roche diagnostics, USA) used as a molecular size marker [34, 35]. PFGE gels were analysed using BioNumerics V7.5 (Applied Maths, Belgium).
Disinfectant and acid susceptibility
A total of six disinfectants and four acids were evaluated for their antimicrobial efficacy against E. coli O26 isolates. The disinfectants and acids tested were Topactive DES (Ecolab Pty Ltd, Australia), Dairy Chlor 12.5% (Campbell Cleantec, Australia), Maxifoam (Ecolab Pty Ltd, Australia), Envirosan (Ecolab Pty Ltd, Australia), Profoam (Jasol Australia, Australia), Kwiksan 22 (Ecolab Pty Ltd, Australia), acetic acid (Sigma-Aldrich, Mexico), citric acid (Sigma-Aldrich, Japan), lactic acid (Sigma-Aldrich, Japan), and propionic acid (Sigma-Aldrich, Japan). Disinfectants tested in this study are approved for use in Australian food industries and food processing areas. Each disinfectant was tested at concentrations spanning the recommended working concentrations. The active components of each of the disinfectants are as follows: Topactive Des.: hydrogen peroxide solution (<10%), acetic acid (<10%), amines, C12-16- alkyldimethyl (<10%), N-oxides (<10%), peracetic acid (<10%); Dairy Chlor: sodium hypochlorite (10- < 30%), sodium hydroxide (<10%); Maxifoam: potassium hydroxide (<10%), builder (<10%), alkaline salts (<10%), anionic surfactant (<10%), surfactants (<10%), hydrotrope (<10%), sodium hypochlorite (<10%) and scale inhibitors (<10%); Envirosan: dodecylbenzenesulfonic acid (<10%), propanoic acid, 2-hydroxy-,(s) (<10%); Profoam: quaternary ammonium compounds (0-5%), surfactants (10-30%); Kwiksan 22: quaternary ammonium compounds, benzyl-C8-18-alkyldimethyl, chlorides (10- < 30%). Evaluation of the effectiveness of the antimicrobial activities of disinfectants and acids and subsequent determination of the susceptibility profiles were performed on polystyrene microtiter plate using broth microdilution method as described previously [36, 37]. Briefly, a single colony from each isolate was streaked onto TSA agar plates and grown for 18 to 24 h at 37 °C. Working solutions for each disinfectant were prepared by diluting with Müller-Hinton broth (MHB; Oxoid, UK) then sterilising using a 0.45 μm syringe filtre (Sartorius Stedim Biotech GmbH, Germany). The working solutions were then two-fold serially diluted to achieve the test concentrations and pH for each concentration was determined. The disinfectant and acid ranges tested were: Topactive DES. (0.0156%-16%), Dairy Chlor 12.5% (0.0188%-12.5%), Maxifoam (0.025%-25.6%), Envirosan (0.025%-25.6%), Profoam: 0.078%-8%, Kwiksan 22 (0.0035%-3.6%), acetic acid (64-65,536 μg/ml), lactic acid (64-65,536 μg/ml), citric acid: (64-65,536 μg/ml) and propionic acid (64-65,536 μg/ml). E. coli O157:H7 Sakai strain was used as a control for the survival of isolates to disinfectant challenge and acid challenge assay.
Results
Characterization of E. coli O26
A total of 88 isolates were screened for the presence of PCR gene targets: stx 1, stx 2, eae, ehx, ecf, bfp and the rmlA SNP. A summary of the PCR screening and characterization of clinical and cattle isolates is shown in Table 2. All of the 88 isolates were found to be negative for stx 2 and bfp. Based on the results of the PCRs four distinct groupings were formed and subsequently referred to here as pathotypes: EHEC, potential EHEC (pEHEC), atypical Enteropathogenic E. coli (aEPEC) and Non-toxigenic E. coli (NTEC). The EHEC group includes all human clinical and 40 (51.2%) cattle isolates. Of the remaining cattle isolates, 33 (42.3%) were aEPEC, three (3.8%) were pEHEC, and two (2.6%) were NTEC.
Table 2.
Prevalence of genetic markers in E. coli O26 from clinical and cattle sources
| Pathotypes | Virulence makers | No of isolates | Source |
|---|---|---|---|
| EHEC | stx (stx 1), eae, ehx, ecf, SNP within rmlA | 50 | Clinical and cattle |
| pEHEC | eae, ehx, ecf, SNP within rmlA | 3 | Cattle |
| aEPEC | eae | 33 | Cattle |
| NTEC | Negative for all virulence markers tested | 2 | Cattle |
Antimicrobial Resistance (AMR)
A total of 88 isolates were assessed for their resistance to 17 antimicrobials. The distribution of minimum inhibitory concentrations (MICs) for each antimicrobial, concentrations tested and resistance breakpoints are presented in Table 3. Overall, there was a low level of resistance among the cattle isolates with 86.4% of all isolates susceptible to all antibiotics tested in this study. In total, 12 (13.6%) E. coli O26 isolates (10 EHEC and two aEPEC) exhibited resistance to at least one antimicrobial. Of the 12 isolates, four were resistant to only one antimicrobial with resistance to streptomycin or nalidixic acid observed in two and one EHEC isolates from cattle, respectively, and a single aEPEC isolate from cattle demonstrating resistance to tetracycline. Multidrug resistance was observed in three human clinical and five cattle isolates. The most common co-resistance phenotype observed was ampicillin-kanamycin-streptomycin-tetracycline (one cattle EHEC isolate and two human clinical EHEC isolates) and ampicillin-streptomycin (three cattle EHEC isolates) while resistance to both chloramphenicol-streptomycin and ampicillin-streptomycin-tetracycline were found in a human clinical EHEC isolate and a cattle aEPEC isolate, respectively. Although AMR appeared largely constrained to EHEC isolates with 20% (10/50) demonstrating resistance to at least one antimicrobial, the differences in AMR prevalence between pathotypes was found to not be significant (p = 0.05).
Table 3.
Distribution of antimicrobial MICs among E. coli O26 isolates from cattle and human sources
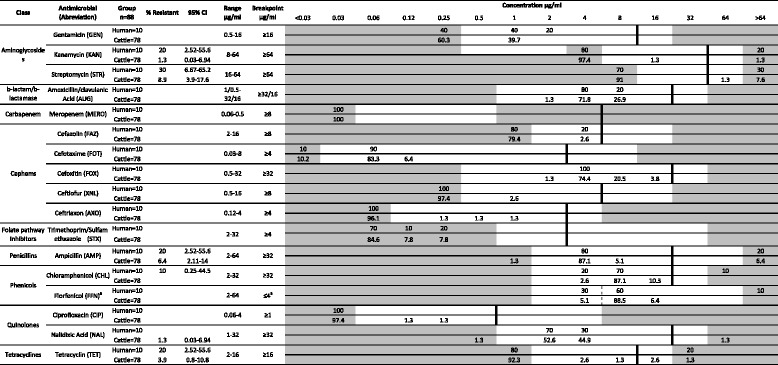
aOnly a susceptible breakpoint (≤4 μg/ml) has been established. Isolates with an MIC ≥8 μg/ml are reported as non-susceptible. Vertical lines indicate breakpoints for resistance. The white fields indicate the dilution range tested for each antimicrobial. Grey area indicates MIC values greater than or less than the tested concentration. Number of isolates is in percentage (%).*CI: Confidence intervals
PFGE analysis
Analysis of PFGE patterns revealed that the E. coli O26 strains in this study are highly diverse with similarity between isolates ranging from 71.4-100% (Fig. 1). Comparison of PFGE profiles of 88 isolates identified 75 distinct PFGE patterns at a similarity cut-off level of 100%. At a cut-off value of 90%, isolates could be grouped into 40 clusters, of which, 19 PFGE clusters were represented by a single isolate and the remaining 69 isolates grouped into 21 clusters containing between two and nine isolates. Of the 69 isolates, 44 had distinct PFGE patterns with the remaining 25 isolates splitting into 11 clusters of 2 indistinguishable isolates and 1 cluster of 3 indistinguishable isolates. Of note, two isolates that were unrelated temporally (one from cattle and one from human) produced indistinguishable PFGE patterns. PFGE patterns were classified into two main clusters designated A and B at a similarity level of 74%. Cluster A included 49 (98%) of EHEC, two pEHEC and one aEPEC isolate. Isolates grouped in cluster B were 97% (32/33) aEPEC as well as two NTEC, a single pEHEC and a single EHEC isolate. An association between clusters and AMR isolates were not found (P value > 0.05). To capture the diversity of E. coli O26 strains, a subset of O26 isolates (n = 40) representing various AMR profiles, a range of E. coli O26 pathotypes and a diverse PFGE pattern, were then chosen for subsequent tests.
Fig. 1.
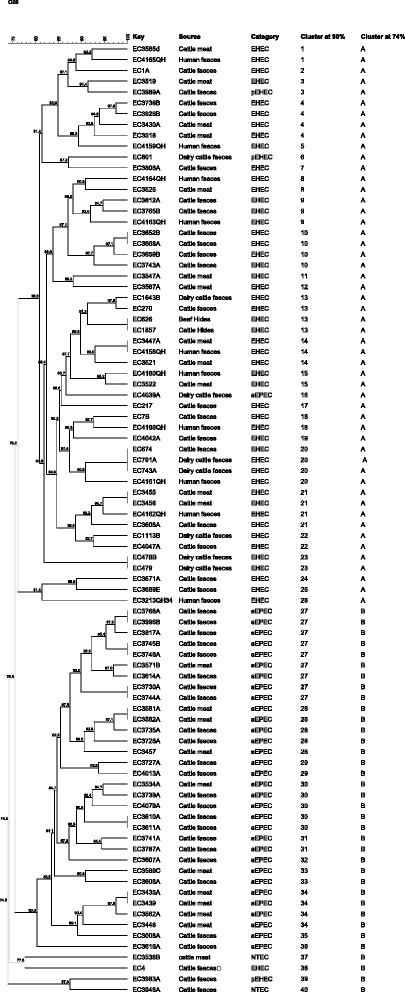
PFGE profiles and clusters of O26 isolates investigated in this study. All 88 isolates were analysed by PFGE with XbaI, and cluster analysis of the patterns was performed by BioNumerics V7.5 software using the Dice coefficient and unweighted pair group method (UPGMA). The degree of similarity (%) is shown on the scale at the top left of the figure. The cut-off level of 90% was chosen to assign isolates to the same cluster. At 74% similarity isolates were assigned to 2 clusters (a & b)
Susceptibility to disinfectants
The effectiveness of disinfectants currently used in the food industry against 40 E. coli O26 was examined in this study and results are shown in Table 4. The proposed industry recommended concentrations for Kwiksan, Profoam, Topactive DES, Dairy Chlor 12.5%, Maxifoam and Envirosan were 0.45, 1, 1, 0.3, 1.6, 0.4%, respectively. The most effective disinfectants with respect to their suggested working concentrations were Kwiksan 22, Topactive DES and Profoam with each able to inhibit the growth of the strains tested with MICs at or below the working concentrations. Dairy Chlor 12.5%, Maxifoam and Envirosan were less effective against the 40 E. coli O26 examined in this study with all strains able to grow at a concentration at or above the suggested working concentrations. Importantly, E. coli O26 isolates missing any or all EHEC virulence markers (i.e., aEPEC, pEHEC and NTEC) were able to survive the same concentrations of disinfectant tested in our study against EHEC isolates, showing the same MICs% as EHEC. When comparing the effective concentrations of disinfectants required for E. coli O26 isolates and the control strain E. coli O157:H7 Sakai, similar effectiveness was observed with elevated MICs demonstrated against Dairy Chlor 12.5%, (MIC = 2.4%), Maxifoam (MIC = 3.2%) and Envirosan (MIC = 1.6%). The remaining three disinfectants (Kwiksan 22, Topactive DES. and Profoam) that have shown to be effective against E. coli O26 were also effective against E. coli O157:H7 Sakai at the proposed industry working concentrations.
Table 4.
Distribution of disinfectant MICs among 40 E. coli O26 isolates from cattle and human sources
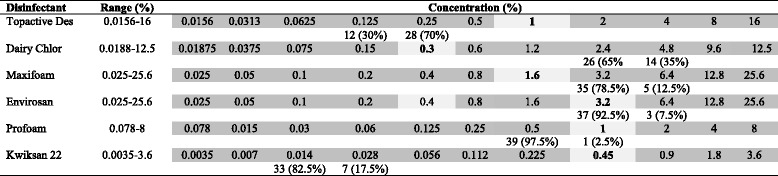
Light Grey fields indicate the recommended working concentrations for each disinfectant, Dark Grey fields indicate the dilution range tested for each disinfectant agents. Numbers in the white field indicates number and percentage of isolates susceptible to disinfectants at the tested concentration
Susceptibility to acids
The MIC distribution profiles of 40 E. coli O26 isolates against four acids (acetic, propionic, lactic, and citric acids) are shown in Table 5. For comparison of MICs of E. coli O26 to acids with different molecular weights, the values for molar MICs (MICsmolar) were used. Based on Weight/Volume (w/v) concentrations it appears that the order of acids with the most effect on the E. coli O26 strains is acetic acid and propionic then lactic followed by citric acid with MICs of 1,024, 1024, 2,048, and 4,096 μg/ml, respectively (Table 5). Recalculation of the MICs to molar values reveals that propionic, acetic, citric and lactic acids have MICsmolar of 13.82, 17.05, 21.3 and 22.7 mmole/ml, respectively, suggesting that propionic acid shows higher efficacy than acetic acid at retarding the growth of E. coli O26. When exposed to acids, the MICs for all 40 isolates occurred at an acetic acid pH and propionic acid pH that was much less acidic than that of the other two acids. The MICsmolar for 87.5% (35/40) and 12.5% (5/40) of isolates occurred at an acetic acid pH 4.08 and 4.42, respectively. For propionic acid the MICsmolar for 100% (40/40) of isolates occurred at pH of 4.55. When exposed to citric acid, the MICsmolar for 87.5% (35/40) and 12.5% (5/40) of isolates were observed at citric acid pH of 3.75 and 3.31, respectively, whereas the MICsmolar for 100% (40/40) of the isolates occurred at lactic acid pH of 3.67. No significant differences in susceptibility to acids was observed between pathotypes regardless of the type of acid assessed. Of note, the MICs of the tested E. coli O26 isolates in the acid challenge assay were comparable to the MIC values observed for the control strain of E. coli O157:H7 Sakai with MICs for acetic, citric, propionic and lactic acids of 512, 2048, 1024 and 2048 μg/mL observed.
Table 5.
MICsμg/ml distribution of acids for 40 E. coli O26 isolates from human and cattle
| Acids | Tested range (μg/ml) | MIC μg/ml | pH | No of isolates (%) |
|---|---|---|---|---|
| Acetic acid | 64-65563 | 1024 | 4.08 | 35 (87.5) |
| 512 | 4.42 | 5 (12.5) | ||
| Citric acid | 64-65563 | 4096 | 3.31 | 35 (87.5) |
| 2048 | 3.75 | 5 (12.5) | ||
| Lactic acid | 64-65563 | 2048 | 3.67 | 40 (100) |
| Propionic acid | 64-65563 | 1024 | 4.55 | 40 (100) |
Discussion
O26 is the second most prevalent serotype identified in cases of foodborne illness attributed to E. coli in Australia and throughout the world. A range of measures exist for food producers to limit the spread and transfer of these organisms, however little is known about the variability of response to these control measures by E. coli O26 isolates. Isolates included in this study could be categorised into four pathotypes (EHEC, pEHEC, aEPEC and NTEC) based on the presence or absence of EHEC associated markers (stx, eae, ehx). Whilst isolates belonging to the EHEC group are of most interest because of their link to human clinical disease, this study identified a number of pEHEC organisms that appear to differ from EHEC isolates through the absence of stx. The ability of EHEC to acquire and lose stx has been described previously [38] and consequently there is a need to consider the clinical impact of these isolates.
The development of antimicrobial resistance within E. coli and particularly EHEC O26 remains an ongoing concern. In this study, a low level of antimicrobial resistance was observed with 86.4% of isolates susceptible to all antimicrobials tested. These data are consistent with other studies that evaluated the AMR status of E. coli in Australian cattle populations at slaughter and in food purchased at retail [39, 40]. These studies determined that approximately >92% of isolates were susceptible to all antimicrobials tested. Furthermore, resistance to antimicrobials of critical or high importance in human medicine was not identified. As previously stated, comparison of the AMR results from different E. coli O26 pathotypes determined that although resistance was largely identified in EHEC isolates, these differences were not significant from any of the other pathotypes.
The use of PFGE enabled the identification of two distinct clusters at a similarity level of 75%. Cluster A was primarily composed of EHEC isolates and included a large proportion (98.7%) of isolates that were positive for the ecf and SNP within rmlA suggesting that these markers are notable features that could be used to define cluster A from cluster B. Interestingly, an individual aEPEC isolate grouped in PFGE cluster A and one EHEC isolate grouped into cluster B where the majority of isolates were aEPEC. This result may reinforce the hypothesis of emergence of EHEC and Non-EHEC by loss and gain of the stx gene. In fact, previous studies showed that conversion of EHEC O26 to stx-negative E. coli O26 is bidirectional where EHEC O26 lose stx genes converting to aEPEC and aEPEC O26 can be lysogenised with Stx-encoding phages to give rise to the emergence of EHEC [38]. Another explanation could be that aEPEC isolates located in the same cluster with EHEC may contain pathogenic O island (OI-122, OI-43. OI-48, OI-50 or OI-57) encoded genes which were found to be significantly associated with aEPEC that showed high similarity to EHEC irrespective of their virulence attributes [41, 42].
E. coli O26 strains that have been responsible for a number of foodborne outbreaks or isolated from a variety of food matrices and food producing animals draw attention to their tolerance to the environmental stresses applied in the food processing areas and food industry [19, 21, 25, 43]. Subsequently, an evaluation for the effectiveness of the sanitizers used in the food processing environment, and food contact surfaces is crucial for understanding effective pathogen control. Both Profoam and Kwiksan are Quaternary Ammonium Compound (QACs) cationic surfactants that are widely used in clinical and industrial settings. Similarly, Topactive DES. which has peracetic acid as its active ingredient is used in the food industry and for disinfection of medical supplies. The observed MICs of E. coli O26 of different pathotypes to a range of QACs (Profoam and Kwiksan) and Topactive DES demonstrated that the required MICs for inhibiting the growth of E. coli O26 carrying EHEC virulence markers (EHEC) and pathotypes lacking EHEC virulence factors (aEPEC, pEHEC and NTEC) are similar to that for E. coli O157 Sakai strain used as a control in our study. This suggests that the manufacturers’ recommended concentrations for the tested disinfectants validated for O157 strain are effective for the control of E. coli O26 of various pathotypes and are may indeed be effective for most E. coli regardless of pathotypes or serogroup.
When challenged for their capability to survive disinfectants with sodium hypochlorite as the main component, both human and cattle E. coli O26 isolates of different pathotypes achieved MICs that exceeded the application concentrations. Consequently all forty isolates regardless of their pathotype were deemed to be non-susceptible to the recommended concentrations of Maxifoam and Dairy Chlor 12.5%. Pathogens that survive recommended concentrations of tested disinfectants pose a greater risk of spreading into the food supply chain and subsequently could contribute to the incidence of human disease thereby reinforcing the importance of continued evaluation of disinfectants. Škaloud et al.,[31] reported that the MIC of sodium hypochlorite for both STEC O157 and O26 was 0.5% which is lower than the effective concentrations for disinfection of E. coli O26 in this study. Although these data may suggest variations among E. coli strains in response to disinfectant stress comparison of the current results with others is difficult since the chosen susceptibility method is different. Previous studies raised concerns about the use of disinfectants and developing resistance to antimicrobial agents [32, 36, 44]. These studies suggested that the use of disinfectants may impose selective pressure giving rise to the emergence of cross-resistance and co-resistance for widely used disinfectants and antimicrobial agents. In our study, the percentage of resistance to a range of disinfectants was similar among O26 isolates regardless of their AMR status and no association between the use of disinfectants and development of resistance to antimicrobial agents was found suggesting that the presence of either resistance has not resulted in selection for the other.
Organic acids have been used in foods as preservatives to enhance microbial safety. In addition, acids may be used as interventions in the beef industry to reduce bacterial contamination. Results presented here show that the majority of isolates (87%), regardless of their source and pathotypes, exhibited elevated MICs (≥1024 μg/ml) to the tested acids. A previous study on the influence of organic acids on E. coli O157:H7 demonstrated elevated MICs for those acids as well [32]. In addition, the MICs for the E. coli O26 isolates in this study occurred at low pH (4.55-3.31). Molina et al., [45] have shown that STEC O26 and other STEC serotypes (O91:H2, O111:H-, O145: H-, and O157:H7) did not grow when they were exposed to citric acid and acetic acid at a pH of 4.5. Others reported that treatment with lactic acid at a concentration of 4%, reduced non-O157 including E. coli O26 by 2.3 log [30]. Findings from the current and previous studies suggest that E. coli O26 isolates of different pathotypes utilize a number of acid resistance mechanisms to prevent the lethal effect of acidic stresses. The capacity of E. coli strains to withstand acidic environments during passage and growth of these bacteria in the intestinal tracts of cattle and human and in acidic food is an important factor that influence their ability to survive and subsequently cause disease [46].
Conclusion
In conclusion, E. coli O26 isolated in Australia are a genetically diverse group of organisms that belong to a range of pathotypes. The low level of resistance and the absence of AMR to clinically relevant antimicrobials in Australian cattle bacterial isolates are reflective of the comprehensive controls over the use of antimicrobials in food-production animals in Australia. However, the tolerance of EHEC and stx-negative E. coli O26 pathotypes (i.e., aEPEC, pEHEC and NTEC) to three of the tested disinfectants (Maxifoam, Dairy Chlor 12.5% and Envirosan) and the elevated MICsμg/ml to the acids examined in this study might contribute to bacterial colonisation of food contact surfaces, which may result in product contamination and subsequently foodborne illness. The ability of E. coli O26 isolates to survive a stress intervention was not related to a specific pathotype as isolates lacking EHEC associated markers such as stx or eae persisted at the same proportion as EHEC strains suggesting that other factors affect persistence of E. coli O26 strains. Knowledge of the virulence factors and genetic relatedness of E. coli O26 may improve our understanding of the capability of E. coli O26 to survive stress and subsequently cause human illness. Continuous evaluation of disinfectants and acids for their efficacy in reducing E. coli O26 should be conducted by food industries to assist in ensuring E. coli O26 is limited in its capacity to persist in food processing environments and contribute to foodborne disease.
Acknowledgments
Funding
The authors are gratefully acknowledge funding from Commonwealth Scientific and Industrial Research Organization (CSIRO) and the Department of Environment and Primary industries Victoria (DEPI-V). Additional funding was provided by the National Board of Technical and Vocational Education, Libya/Department of Laboratory Medicine Derna, Libya. Grant No. 2013-202-2.
Availability of data and materials
All data generated or analysed during this study are included in this article.
Authors’ contributions
SL, RB and JB conceived and designed the experiments; SL performed the experiments, acquired the data and drafted the manuscript. RB interpreted the data and critically revised the manuscript. All authors read and approved the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study used existing, cattle and de-identified human isolates from the CSIRO culture collection. New samples or isolates were not collected as part of the study and based on this no ethics approval was required for this study as defined by Griffith University's research ethics arrangements and the National Statement on Ethical Conduct in Human Research. CSIRO own these samples and gave permission for use of these samples.
Abbreviations
- aEPEC
Enteropathogenic E. coli
- AMR
Antimicrobial resistance
- ATCC
American type culture collection
- bfp
Bundle forming pilus
- CDC
Centers for disease control
- CLSI
Clinical and Laboratory Standards Institute
- eae
E. coli attachment and effacing gene
- ecf
eae positive conserved fragments
- EHEC
Enterohaemorrhagic Escherichia coli
- ehx
Enterohemolysin
- EUCAST
European Committee on Antimicrobial Susceptibility Testing
- HC
Hemolytic colitis
- HUS
Hemolytic uremic-syndrome
- MICs
Minimum inhibitory concentrations
- NARMS
National antimicrobial resistance monitoring system
- NCTC
National collection of type cultures
- NTEC
Non-toxigenic E. coli
- PCR
Polymerase chain reaction
- pEHEC
Potential enterohaemorrhagic E. coli
- PFGE
Pulse-field gel electrophoresis
- QACs
Quaternary ammonium compound
- RFLP
Restriction fragment length polymorphism
- SNP
Single nucleotide polymorphism
- TSA
Tryptone soya agar
- USA
United State of America
Contributor Information
Salma A. Lajhar, Email: salmaaibsais.lajhar@griffithuni.edu.au
Jeremy Brownlie, Email: j.brownlie@griffith.edu.au.
Robert Barlow, Email: Robert.Barlow@csiro.au.
References
- 1.Vally H, Hall G, Dyda A, Raupach J, Knope K, Combs B, Desmarchelier P. Epidemiology of Shiga toxin producing Escherichia coli in Australia, 2000-2010. BMC Public Health. 2012;12:63. doi: 10.1186/1471-2458-12-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guth BE, Lopes de Souza R, Vaz TM, Irino K. First Shiga toxin-producing Escherichia coli isolate from a patient with hemolytic uremic syndrome, Brazil. Emerg Infect Dis. 2002;8:535–536. doi: 10.3201/eid0805.010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brooks JT, Sowers EG, Wells JG, Greene KD, Griffin PM, Hoekstra RM, Strockbine NA. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983-2002. J Infect Dis. 2005;192:1422–1429. doi: 10.1086/466536. [DOI] [PubMed] [Google Scholar]
- 4.Paton AW, Paton JC. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfb O111, and rfb O157. J Clin Microbiol. 1998;36:598–602. doi: 10.1128/jcm.36.2.598-602.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delannoy S, Beutin L, Fach P. Towards a molecular definition of enterohemorrhagic Escherichia coli (EHEC): detection of genes located on O island 57 as markers to distinguish EHEC from closely related enteropathogenic E. coli strains. J Clin Microbiol. 2013;51:1083–1088. doi: 10.1128/JCM.02864-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerber A, Karch H, Allerberger F, Verweyen HM, Zimmerhackl LB. Clinical course and the role of Shiga toxin-producing Escherichia coli infection in the hemolytic-uremic syndrome in pediatric patients, 1997-2000, in Germany and Austria: a prospective study. J Infect Dis. 2002;186:493–500. doi: 10.1086/341940. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Cheng Y, Xiong Y, Ye C, Zheng H, Sun H, Zhao H, Ren Z, Xu J. Enterohemorrhagic Escherichia coli specific enterohemolysin induced IL-1beta in human macrophages and EHEC-induced IL-1beta required activation of NLRP3 inflammasome. PloS One. 2012;7:e50288. doi: 10.1371/journal.pone.0050288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van de Kar NC, Monnens LA, Karmali MA, van Hinsbergh VW. Tumor necrosis factor and interleukin-1 induce expression of the verocytotoxin receptor globotriaosylceramide on human endothelial cells: implications for the pathogenesis of the hemolytic uremic syndrome. Blood. 1992;80:2755–2764. [PubMed] [Google Scholar]
- 9.Schmidt H, Beutin L, Karch H. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect Immun. 1995;63:1055–1061. doi: 10.1128/iai.63.3.1055-1061.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jerse AE, Yu J, Tall BD, Kaper JB. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci USA. 1990;87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lynch MJ, Fox EM, O'Connor L, Jordan K, Murphy M. Surveillance of verocytotoxigenic Escherichia coli in Irish bovine dairy herds. Zoonoses Public Health. 2012;59:264–271. doi: 10.1111/j.1863-2378.2011.01443.x. [DOI] [PubMed] [Google Scholar]
- 12.Barlow RS, Mellor GE. Prevalence of enterohemorrhagic Escherichia coli serotypes in Australian beef cattle. Foodborne Pathog Dis. 2010;7:1239–1245. doi: 10.1089/fpd.2010.0574. [DOI] [PubMed] [Google Scholar]
- 13.Blanco J, Blanco M, Blanco JE, Mora A, Gonzalez EA, Bernardez MI, Alonso MP, Coira A, Rodriguez A, Rey J, Alonso JM, Usera MA. Verotoxin-producing Escherichia coli in Spain: prevalence, serotypes, and virulence genes of O157:H7 and non-O157 VTEC in ruminants, raw beef products, and humans. Exp Biol Med. 2003;228:345–351. doi: 10.1177/153537020322800403. [DOI] [PubMed] [Google Scholar]
- 14.Foodborne Outbreak Online Database.CDC. Colorado. 2007. http://wwwn.cdc.gov/foodborneoutbreaks/ accessed on 5 May 2014.
- 15.Foodborne Outbreak Online Database.CDC. Massachusetts. 2006. http://wwwn.cdc.gov/foodborneoutbreaks/. accessed on 1 June 2014.
- 16.Foodborne Outbreak Online Database. CDC. Multistate outbreak. 2010. http://wwwn.cdc.gov/foodborneoutbreaks/ accessed on 8 June 2014.
- 17.CDC. Multistate Outbreak of Shiga Toxin-producing Escherichia coli O26 Infections Linked to Raw Clover Sprouts at Jimmy John's Restaurants. 2012. http://www.cdc.gov/ecoli/2012/o26-02-12/index.html accessed on 24 Sep 2015.
- 18.Ethelberg S, Smith B, Torpdahl M, Lisby M, Boel J, Jensen T, Nielsen EM, Molbak K. Outbreak of non-O157 Shiga toxin-producing Escherichia coli infection from consumption of beef sausage. Clin Infect Dis. 2009;48:e78–81. doi: 10.1086/597502. [DOI] [PubMed] [Google Scholar]
- 19.Hiruta N, Murase T, Okamura N. An outbreak of diarrhoea due to multiple antimicrobial-resistant Shiga toxin-producing Escherichia coli O26:H11 in a nursery. Epidemiol Infect. 2001;127:221–227. doi: 10.1017/S0950268801006069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoshina K, Itagaki A, Seki R, Yamamoto K, Masuda S, Muku T, Okada N. Enterohemorrhagic Escherichia coli O26 outbreak caused by contaminated natural water supplied by facility owned by local community. Jpn J Infect Dis. 2001;54:247–248. [PubMed] [Google Scholar]
- 21.Allerberger F, Friedrich AW, Grif K, Dierich MP, Dornbusch HJ, Mache CJ, Nachbaur E, Freilinger M, Rieck P, Wagner M, Caprioli A, Karch H, Zimmerhackl LB. Hemolytic-uremic syndrome associated with enterohemorrhagic Escherichia coli O26:H infection and consumption of unpasteurized cow’s milk. Int J Infect Dis. 2003;7:42–45. doi: 10.1016/S1201-9712(03)90041-5. [DOI] [PubMed] [Google Scholar]
- 22.Murphy M, Buckley JF, Whyte P, O'Mahony M, Anderson W, Wall PG, Fanning S. Surveillance of dairy production holdings supplying raw milk to the farmhouse cheese sector for Escherichia coli O157, O26 and O111. Zoonoses Public Health. 2007;54:358–365. doi: 10.1111/j.1863-2378.2007.01073.x. [DOI] [PubMed] [Google Scholar]
- 23.De Schrijver K, Buvens G, Posse B, Van den Branden D, Oosterlynck O, De Zutter L, Eilers K, Pierard D, Dierick K, Van Damme-Lombaerts R, Lauwers C, Jacobs R. Outbreak of verocytotoxin-producing E. coli O145 and O26 infections associated with the consumption of ice cream produced at a farm, Belgium, 2007. Euro Surveill. 2008;13:7. doi: 10.2807/ese.13.07.08041-en. [DOI] [PubMed] [Google Scholar]
- 24.Buvens G, Posse B, De Schrijver K, De Zutter L, Lauwers S, Pierard D. Virulence profiling and quantification of verocytotoxin-producing Escherichia coli O145:H28 and O26:H11 isolated during an ice cream-related hemolytic uremic syndrome outbreak. Foodborne Pathog Dis. 2011;8:421–426. doi: 10.1089/fpd.2010.0693. [DOI] [PubMed] [Google Scholar]
- 25.Madic J, Vingadassalon N, de Garam CP, Marault M, Scheutz F, Brugere H, Jamet E, Auvray F. Detection of Shiga toxin-producing Escherichia coli serotypes O26:H11, O103:H2, O111:H8, O145:H28, and O157:H7 in raw-milk cheeses by using multiplex real-time PCR. Appl Environ Microbiol. 2011;77:2035–2041. doi: 10.1128/AEM.02089-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.FSIS. https://www.fsis.usda.gov/wps/portal/fsis/topics/recalls-and-public-health-alerts/recall-summaries. Accessed 11 Jan 2014. 2010–2014.
- 27.Espié E, Mariani-Kurkdjian P, Grimont F, Pihier N, Vaillant V, Francart S, Capek I, de Valk H, Vernozy-Rozand C. Shiga-toxin producing Escherichia coli O26 infection and unpasteurised cows cheese, France, 2005. In: Sofronidis J, editor. Progr Abstr 6th Int Symp Shiga Toxin (Verocytoxin)-Producing Escherichia coli Infect; Melbourne, Australia. West Leederville: Cambridge Publishing; 2006. [Google Scholar]
- 28.CDC. Multistate Outbreaks of Shiga toxin-producing Escherichia coli O26 Infections Linked to Chipotle Mexican Grill Restaurants. 2015. http://www.cdc.gov/ecoli/2015/o26-11-15/index.html accessed on 24 Sep 2016.
- 29.Fukushima H, Hoshina K, Gomyoda M. Long-term survival of shiga toxin-producing Escherichia coli O26, O111, and O157 in bovine feces. Appl Environ Microbiol. 1999;65:5177–5181. doi: 10.1128/aem.65.11.5177-5181.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalchayanand N, Arthur TM, Bosilevac JM, Schmidt JW, Wang R, Shackelford SD, Wheeler TL. Evaluation of commonly used antimicrobial interventions for fresh beef inoculated with Shiga toxin-producing Escherichia coli serotypes O26, O45, O103, O111, O121, O145, and O157:H7. J Food Prot. 2012;75:1207–1212. doi: 10.4315/0362-028X.JFP-11-531. [DOI] [PubMed] [Google Scholar]
- 31.Škaloud J, Pokludová L, Novotná R, Čížek A. Evaluation by conductance assay of Shiga Toxin-producing Escherichia coli (STEC) O157 and O26 and their sensitivity to selected disinfectants. Acta Veterinaria Brno. 2003;72:101. doi: 10.2754/avb200372010101. [DOI] [Google Scholar]
- 32.Beier RC, Poole TL, Brichta-Harhay DM, Anderson RC, Bischoff KM, Hernandez CA, Bono JL, Arthur TM, Nagaraja TG, Crippen TL, Sheffield CL, Nisbet DJ. Disinfectant and antibiotic susceptibility profiles of Escherichia coli O157:H7 strains from cattle carcasses, feces, and hides and ground beef from the United States. J Food Prot. 2013;76:6–17. doi: 10.4315/0362-028X.JFP-12-253. [DOI] [PubMed] [Google Scholar]
- 33.Norman KN, Strockbine NA, Bono JL. Association of nucleotide polymorphisms within the O-antigen gene cluster of Escherichia coli O26, O45, O103, O111, O121, and O145 with serogroups and genetic subtypes. Appl Environ Microbiol. 2012;78:6689–6703. doi: 10.1128/AEM.01259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunter SB, Vauterin P, Lambert-Fair MA, Van Duyne MS, Kubota K, Graves L, Wrigley D, Barrett T, Ribot E. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J Clin Microbiol. 2005;43:1045–1050. doi: 10.1128/JCM.43.3.1045-1050.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.CDC: One-day (24-28 h) standardized laboratory protocol for molecular subtyping of Escherichia coli O157:H7, non-typhoidal Salmonella serotypes, and Shigella sonnei by pulsed field gel electrophoresis (PFGE). Centers for Disease Control and Prevention; 2004.
- 36.Beier RC, Bischoff KM, Ziprin RL, Poole TL, Nisbet DJ. Chlorhexidine susceptibility, virulence factors, and antibiotic resistance of beta-hemolytic Escherichia coli isolated from neonatal swine with diarrhea. Bull Environ Contam Toxicol. 2005;75:835–844. doi: 10.1007/s00128-005-0826-5. [DOI] [PubMed] [Google Scholar]
- 37.Beier RC, Duke SE, Ziprin RL, Harvey RB, Hume ME, Poole TL, Scott HM, Highfield LD, Alali WQ, Andrews K, Anderson RC, Nisbet DJ. Antibiotic and disinfectant susceptibility profiles of vancomycin-resistant Enterococcus faecium (VRE) isolated from community wastewater in Texas. Bull Environ Contam Toxicol. 2008;80:188–194. doi: 10.1007/s00128-007-9342-0. [DOI] [PubMed] [Google Scholar]
- 38.Bielaszewska M, Prager R, Kock R, Mellmann A, Zhang W, Tschape H, Tarr PI, Karch H. Shiga toxin gene loss and transfer in vitro and in vivo during enterohemorrhagic Escherichia coli O26 infection in humans. Appl Environ Microbiol. 2007;73:3144–3150. doi: 10.1128/AEM.02937-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barlow RS, Gobius KS. Pilot survey for antimicrobial resistant (AMR) bacteria in Australian food. Cannon Hill: CSIRO; 2008. [Google Scholar]
- 40.Barlow RS, McMillan KE, Duffy LL, Fegan N, Jordan D, Mellor GE. Prevalence and antimicrobial resistance of Salmonella and Escherichia coli from Australian cattle populations at slaughter. J Food Prot. 2015;78:912–920. doi: 10.4315/0362-028X.JFP-14-476. [DOI] [PubMed] [Google Scholar]
- 41.Bugarel M, Martin A, Fach P, Beutin L. Virulence gene profiling of enterohemorrhagic (EHEC) and enteropathogenic (EPEC) Escherichia coli strains: a basis for molecular risk assessment of typical and atypical EPEC strains. BMC Microbiol. 2011;11:142. doi: 10.1186/1471-2180-11-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delannoy S, Beutin L, Fach P. Discrimination of enterohemorrhagic Escherichia coli (EHEC) from non-EHEC strains based on detection of various combinations of type III effector genes. J Clin Microbiol. 2013;51:3257–3262. doi: 10.1128/JCM.01471-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wells JG, Shipman LD, Greene KD, Sowers EG, Green JH, Cameron DN, Downes FP, Martin ML, Griffin PM, Ostroff SM, Potter ME, Tauxe RV, Wachsmuth IK. Isolation of Escherichia coli serotype O157:H7 and other Shiga-like-toxin-producing E. coli from dairy cattle. J Clin Microbiol. 1991;29:985–989. doi: 10.1128/jcm.29.5.985-989.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sidhu MS, Heir E, Leegaard T, Wiger K, Holck A. Frequency of disinfectant resistance genes and genetic linkage with beta-lactamase transposon Tn552 among clinical staphylococci. Antimicrob Agents Chemother. 2002;46:2797–2803. doi: 10.1128/AAC.46.9.2797-2803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Molina PM, Sanz ME, Lucchesi PMA, Padola NL, Parma AE. Effects of acidic broth and juices on the growth and survival of verotoxin-producing Escherichia coli (VTEC) Food Microbiol. 2005;22:469–473. doi: 10.1016/j.fm.2004.10.002. [DOI] [Google Scholar]
- 46.Arnold CN, McElhanon J, Lee A, Leonhart R, Siegele DA. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J Bacteriol. 2001;183:2178–2186. doi: 10.1128/JB.183.7.2178-2186.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perelle S, Dilasser F, Grout J, Fach P. Detection by 5'-nuclease PCR of Shiga-toxin producing Escherichia coli O26, O55, O91, O103, O111, O113, O145 and O157:H7, associated with the world's most frequent clinical cases. Mol Cell Probes. 2004;18:185–192. doi: 10.1016/j.mcp.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 48.Luedtke BE, Bono JL, Bosilevac JM. Evaluation of real time PCR assays for the detection and enumeration of enterohemorrhagic Escherichia coli directly from cattle feces. J Microbiol Methods. 2014;105:72–79. doi: 10.1016/j.mimet.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 49.Taniuchi M, Walters CC, Gratz J, Maro A, Kumburu H, Serichantalergs O, Sethabutr O, Bodhidatta L, Kibiki G, Toney DM, Berkeley L, Nataro JP, Houpt ER. Development of a multiplex polymerase chain reaction assay for diarrheagenic Escherichia coli and Shigella spp. and its evaluation on colonies, culture broths, and stool. Diagn Microbiol Infect Dis. 2012;73:121–128. doi: 10.1016/j.diagmicrobio.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this article.


