Abstract
IMPORTANCE
Developmental brain disorders are a group of clinically and genetically heterogeneous disorders characterized by high heritability. Specific highly penetrant genetic causes can often be shared by a subset of individuals with different phenotypic features, and recent advances in genome sequencing have allowed the rapid and cost-effective identification of many of these pathogenic variants.
OBJECTIVES
To identify novel candidate genes for developmental brain disorders and provide additional evidence of previously implicated genes.
DATA SOURCES
The PubMed database was searched for studies published from March 28,2003, through May 7,2015, with large cohorts of individuals with developmental brain disorders.
DATA EXTRACTION AND SYNTHESIS
A tiered, multilevel data-integration approach was used, which intersects (1) whole-genome data from structural and sequence pathogenic loss-of-function (pLOF) variants, (2) phenotype data from 6 apparently distinct disorders (intellectual disability, autism, attention-deficit/hyperactivity disorder, schizophrenia, bipolar disorder, and epilepsy), and (3) additional data from largescale studies, smaller cohorts, and case reports focusing on specific candidate genes. All candidate genes were ranked into 4 tiers based on the strength of evidence as follows: tier 1, genes with 3 or more de novo pathogenic loss-of-function variants; tier 2, genes with 2 de novo pathogenic loss-of-function variants; tier 3, genes with 1 de novo pathogenic loss-of-function variant; and tier 4, genes with only inherited (or unknown inheritance) pathogenic loss-of-function variants.
MAIN OUTCOMES AND MEASURES
Development of a comprehensive knowledge base of candidate genes related to developmental brain disorders. Genes were prioritized based on the inheritance pattern and total number of pathogenic loss-of-function variants identified amongst unrelated individuals with any one of six developmental brain disorders.
STUDY SELECTION
A combination of phenotype-based and genotype-based literature review yielded 384 studies that used whole-genome or exome sequencing, chromosomal microarray analysis, and/or targeted sequencing to evaluate 1960 individuals with developmental brain disorders.
RESULTS
Our initial phenotype-based literature review yielded 1911 individuals with pLOF variants involving 1034 genes from 118 studies. Filtering our results to genes with 2 or more pLOF variants identified in at least 2 unrelated individuals resulted in 241 genes from 1110 individuals. Of the 241 genes involved in brain disorders, 7 were novel high-confidence genes and 10 were novel putative candidate genes. Fifty-nine genes were ranked in tier 1,44 in tier 2,68 in tier 3, and 70 in tier 4. By transcending clinical diagnostic boundaries, the evidence level for 18 additional genes that were ranked 1 tier higher because of this cross-disorder approach was increased.
CONCLUSIONS AND RELEVANCE
This approach increased the yield of gene discovery over what would be obtained if each disorder, type of genomic variant, and study design were analyzed independently. These results provide further support for shared genomic causes among apparently different disorders and demonstrate the clinical and genetic heterogeneity of developmental brain disorders.
Developmental brain disorders (DBDs) are a group of heterogeneous conditions characterized by deficits that affect multiple functional domains, such as cognition, behavior, communication, and motor skills.1 Developmental brain disorders include intellectual disability or developmental delay (ID/DD), autism spectrum disorder (ASD), attention-deficit/hyperactivity disorder (ADHD), schizophrenia, bipolar disorder (BD), and epilepsy. The clinical presentation of these disorders is highly variable, ranging from mild to severe impairments across the major areas of development.2 Clinical and epidemiologic studies1–3 reveal that DBDs frequently cooccur with other developmental and/or neuropsychiatric disorders and share common and overlapping signs and symptoms.3
Developmental brain disorders are considered distinct clinical entities by the DSM-54 and the International Classification of Diseases, 10th Revision.5 However, previous studies1,6–8 provide strong evidence of common underlying molecular pathways and shared genetic causes, such as copy number and single-nucleotide variants, among apparently different DBDs.
For example, structural and sequence variants in NRXN1 (HGNC8008) are associated with Pitt-Hopkins–like syndrome, ID/DD, ASD, ADHD, schizophrenia, BD, and epilepsy.9 Variants in ADNP (HGNC15766), CHD8 (HGNC20153), SCN2A (HGNC10588), and PTCHD1 (HGNC26392) have similarly been reported in individuals with a range of DBDs.10–13 Likewise, recurrent copy number variants (CNVs), including deletions at 1q21.1, 16p11.2, 17q12, and 22q11.2, have been identified in individuals with ID/DD, ASD, schizophrenia, and epilepsy.14–17
Because of the phenotypic and genetic heterogeneity of DBDs, the identification of causative genetic variants has been challenging. However, recent advances in copy number and next-generation sequencing technologies, such as whole-exome sequencing (WES) and whole-genome sequencing (WGS), have revolutionized the diagnostic approach to DBDs, revealing causative genetic mutations in approximately 25% to50% of individuals with a DBD.18–21 These technologies have also been successfully used in the research setting for gene discovery in large cohorts of individuals with ASD, ID/DD, epilepsy, and schizophrenia, generating vast amounts of publicly available genomic data.22–24
Other large-scale studies25–27 of individuals with DBDs have found that identifying independent de novo pathogenic loss-of-function (pLOF) variants (ie, nonsense, frameshift, or splice site) in the same gene among unrelated individuals is a powerful statistical approach to reliably identifying disease causative genes. Sanders et al25 studied 928 individuals with ASD and applied a permutation test to determine that 2 or more de novo pLOF variants were highly unlikely to occur by chance and provide significant evidence for ASD association (P = .008; false discovery rate [q] = 0.005). Willsey et al26 similarly assessed 1043 probands with ASD and found that observing 2 and 3 de novo pLOF variants in the same gene in unrelated individuals identifies an ASD gene with a 97.8% and greater than 99.9%chance of being a true ASD gene, respectively (q = 0.02 and 0.0002, respectively), whereas genes with a single de novo pLOF variant are more likely than not to be true ASD genes (54.7% chance, q = 0.45). Likewise, Dong et al27 studied 787 families with ASD (2963 individuals) and concluded that genes with a single de novo pLOF variant have a 50.4% (q = 0.496) probability of being associated with ASD, whereas genes with at least 2 de novo pLOF variants have a 97.6% probability of being associated with ASD (q = 0.024).
This robust statistical approach has been used to identify disease risk genes in large cohorts of individuals with DBDs, such as ASD, yielding a steadily increasing number of genes associated with ASD. However, rare pLOF variants identified in a single individual are usually not discussed in the results of these publications and often remain as isolated findings relegated to the supplemental data. Genomic data from smaller cohorts and case reports are similarly not routinely pooled with data from larger studies, thus precluding the identification of potential individuals who may represent the second or third de novo pLOF variant, moving the gene over the threshold for being considered disease causative. Moreover, previous studies25–27 using this approach have been restricted to cohorts of individuals ascertained based on a single categorical diagnosis; therefore, genomic data from individuals with apparently distinct DBDs are not being jointly analyzed in search of independent pLOF variants in the same gene among unrelated individuals.
In this study, we expanded the aforementioned method and developed a multilevel data-integration approach, which capitalizes on 3 genotype-phenotype data sources: (1) genomic data from structural and sequence pLOF variants (de novo and inherited), (2) phenotype data from 6 apparently distinct DBDs (cross-disorder), and (3) data from large-scale studies, smaller cohorts, and case reports (cross-study). We used this method to identify and categorize DBD candidate genes and shared genetic causes and molecular pathways that transcend clinical diagnostic boundaries.
Methods
Development of a Comprehensive Cross-Disorder Genotype-Phenotype Knowledge Base
We conducted a tiered approach to develop a comprehensive database of DBD candidate genes. Our first step comprised an exhaustive phenotype-based literature review of studies using WGS, WES, CNV analysis, and/or targeted sequencing to evaluate large cohorts of individuals with ID/DD, ASD, ADHD, schizophrenia, BD, and/or epilepsy. We selected these 6 diagnoses because more studies with publicly available genomic data on these disorders are available relative to other DBDs. We searched the PubMed database for articles published from March 28, 2003, through May 7, 2015, for our search algorithm, limited to studies in humans and written in the English language. The MeSH terms used were intellectual disability, developmental delay, mental retardation, autism, epilepsy, seizures, schizophrenia, bipolar disorder, ADHD, attention-deficit/hyperactivity disorder, whole-exome sequencing, whole-genome sequencing, targeted sequencing, chromosomal microarray analysis, single-gene deletions, microdeletions, and exonic deletions. Reference lists of retrieved articles were also searched for other relevant studies. We reviewed all available genotype and phenotype data included in the results section and supplemental data of each article. Institutional review board approval and informed consent were not required because all the data included were pooled from publicly available studies.
We included all cases with pLOF variants, which we defined as LOF sequence variants and single-gene deletions that include one or more exons. We excluded individuals with intronic, missense, or silent sequence variants, individuals with duplications or deletions involving more than one coding gene, and individuals with more than one pLOF variant. For each individual, we documented the presence or absence of each of the 6 DBDs, the type of genomic variant and inheritance pattern, and the type of genomic diagnostic platform used in the study. The phenotype information was used only as part of the inclusion criteria.
We subsequently selected those genes with 2 or more pLOF variants identified in 2 or more unrelated individuals and conducted a second comprehensive genotype-based literature review focused on each of these genes to identify additional pLOF variants and/or clinical manifestations from smaller cohort studies and case reports (eFigure and eTable 1 in the Supplement). To avoid double counting individuals and variants, we carefully annotated the cohort name, individual identification number, phenotype, and all available variant information and removed duplicate entries. Two investigators (A.J.G-.M. and A.M.-D-.L.) independently reviewed all the data points included in the knowledge base. Although our study design resembles a meta-analysis in the comprehensive literature review, it does not follow the formal statistical approach used in meta-analyses. To provide an easily accessible resource for researchers, physicians, and other health care professionals, we created a comprehensive online database with all our genotype and phenotype findings, which can be accessed at http://geisingeradmi.org/dbdgenes.
DBD Candidate Gene Prioritization
To classify and prioritize our findings, we ranked all DBD candidate genes into 4 tiers based on the strength of evidence as follows: tier 1, genes with 3 or more de novo pLOF variants; tier 2, genes with 2 de novo pLOF variants; tier 3, genes with 1 de novo pLOF variant; and tier 4, genes with only inherited (or unknown inheritance) pLOF variants. Genes from tiers 1 and 2 are considered high-confidence DBD causative genes, whereas those from tiers 3 and 4 are categorized as emerging DBD causative genes. In addition to the de novo events, genes from all tiers could have any number of inherited (or unknown inheritance) variants.
Results
Identification and Prioritization of DBD Candidate Genes
Our initial phenotype-based literature review yielded 1911 individuals with pLOF variants involving 1034 genes from 118 studies. Filtering our results to genes with 2 or more pLOF variants identified in at least 2 unrelated individuals resulted in 241 genes from 1110 individuals. The genotype-based literature review identified 850 additional individuals with pLOF variants in 85 of the 241 initially identified genes, from 266 new studies, for a total of 1960 individuals with pLOF variants in 241 genes (eTable 2 in the Supplement).
The pLOF sequence variants were identified in 1516 individuals (77.3%) and were detected using targeted sequencing (820 [54.1%]), WES (660 [43.5%]), and WGS (36 [2.4%]). Singlegene exonic deletions were observed in 444 individuals (22.7%), with most variants from studies using genome-wide chromosomal microarray analysis(CMA) (413 [93.0%]) and the remaining 31 (7.0%) being identified with targeted CNV analysis. Overall, 1109 variants (56.6%) were identified using an unbiased, genome-wide diagnostic platform (WGS, WES, or CMA), whereas 851 (43.4%) came from studies using targeted genomic analyses. Information regarding the inheritance pattern was not available for 671 individuals (34.2%). Of the 1289 with available inheritance data, 810 (62.8%) were de novo and 479 (37.2%) were inherited (Table 1).
Table 1.
Summary of Inheritance Pattern and Type of Diagnostic Platform Used to Identify 1960 pLOF Variants in 241 DBD Candidate Genes
| Inheritance Pattern |
No. of Variants | |||||
|---|---|---|---|---|---|---|
| pLOF Variants |
WGS | WES | CMA | Targeted CNV |
Targeted Sequencing |
|
| De novo | 810 | 20 | 367 | 92 | 8 | 323 |
| Inherited | 479 | 16 | 135 | 183 | 8 | 137 |
| Unknown | 671 | 0 | 158 | 138 | 15 | 360 |
| Total | 1960 | 36 | 660 | 413 | 31 | 820 |
Abbreviations: CMA, genome-wide chromosomal microarray analysis; CNV, copy number variant; DBD, developmental brain disorder; pLOF, pathogenic loss-of-function; WES, whole exome sequencing; WGS, whole genome sequencing.
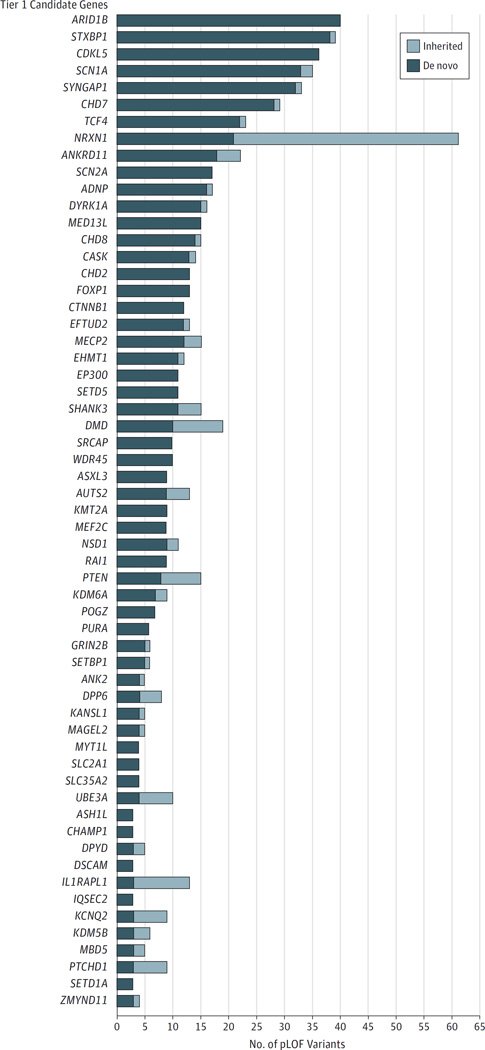
We ranked all DBD candidate genes into 4 tiers based on the strength of evidence (Table 2). Tier 1 includes 59 genes with 3 or more de novo pLOF variants. Tier 2 comprises 44 genes with 2 de novo pLOF variants. Tier 3 includes 68 genes with 1 de novo pLOF variant, and tier 4 contains 70 genes with only inherited or unknown inheritance pLOF variants. Genes with the strongest level of evidence from tier 1 can be further subdivided based on the number and inheritance pattern of all pLOF variants observed (Figure 1). Six genes have more than 25 de novo pLOF variants each(ARID1B[HGNC18040],STXBP1 [HGNC 11444], CDKL5 [HGNC 11411], SCN1A [HGNC 10585], SYNGAP1 [HGNC 11497], and CHD7 [HGNC 20626]), 21 genes have 10 to 22 de novo pLOF variants, and 32 genes harbor 3 to 9 de novo pLOF variants.
Table 2.
Prioritization of Candidate Genes for Developmental Brain Disorders Based on the Number of De Novo pLOF Variants
| Gene Rank |
No. of De Novo pLOF Variants |
No. of Genes |
Genesa |
|---|---|---|---|
| Tier 1 | ≥3 | 59 |
ADNP, ANK2, ANKRD11, ARID1B, ASH1L, ASXL3, AUTS2, CASK, CDKL5, CHAMP1, CHD2, CHD7, CHD8, CTNNB1, DMD, DPP6, DPYD, b DSCAM, DYRK1A, EFTUD2, EHMT1, EP300, FOXP1, GRIN2B, IL1RAPL1,b IQSEC2, KANSL1, KCNQ2,b KDM5B, KDM6A, KMT2A, MAGEL2, MBD5, MECP2, MED13L, MEF2C, MYT1L, NRXN1, NSD1, POGZ, PTCHD1, PTEN, PURA, RAI1, SCN1A, SCN2A, SETBP1, SETD1A, SETD5, SHANK3, SLC2A1, SLC35A2, SRCAP, STXBP1, SYNGAP1, TCF4, UBE3A, WDR45, ZMYND11b |
| Tier 2 | 2 | 44 |
CBL, CSMD1,b
CTNNA3, CUL3, DDX3X, DIP2A, DLG2,b
ELAVL2, FHIT, GATAD2BHIST1H1E, HIVEP3, ITSN1, KATNAL2, KDM5C,b KDM6B, KMT2C, LAMA2, LRP2,b MOV10, MYH10, NBEA, NCKAP1, NEDD9, NFIA, PHF2, PHF21A, RALGAPB, RIMS1, SETD2,b SLC6A8,b SMC1A, SPAST, TAF13, TBR1, TCF7L2, TNRC6B, TRIO,b TTN, UPF3B,b WAC, WDFY3, YTHDC1, ZWILCH |
| Tier 3 | 1 | 68 |
APH1A, ARHGAP24, ATRX, AXLBAIAP2, BIRC6, BRWD1, CACNA2D3, CCDC91, CD163L1, CDC42BPB, CHRNA7, CSDE1, CSTF2T, CTTNBP2, DDHD2,b DNM3, DPP3, DST, EDA2R, EP400, ERBB4, FAM190A, FCRL6, FLG, GABRB3, GALNTL4, GGNBP2, GSDMC, HNRNPU, JUP, KAL1, KDM3A, L1CAM,b LEO1, LINGO2,b LPP, MACROD2,b MCPH1, MSRA, MYOC, NAA15, NCKAP5, NR3C2, OR2T10, PARK2, PCDH15,b PDE11A, PLCB1, PPM1D, PSD3, PTPRD, QRICH1, RAB2A, RAB39B, RANBP17, RBFOX1, SCARA3, SLC16A2, SLC4A10,b SLC6A1, SLC9A6, STXBP5, SUCLG2, TGM1, THSD7A, TSPAN17, WHSC1 |
| Tier 4 | Only inherited, or unknown, variants |
70 |
ACACA, AIFM3, ANKS1B, AP1S2, ARHGEF38, BRAT1, CACNA1C, CACNA2D2, CACNA2D4, CALCR, CAPN12, CARKD, CDH13, CERS4, CHD1L, CNTN4, CNTN6, CNTNAP2, COBL, CUL4B, CYFIP1, CYLC2, DDX53, DISC1, DNAH10, EYS, GJB6, GRIP1, IMMP2L, INADL, IQGAP2, KANK1, KIAA0100, KRT34, KYNU, LAMC3, LRBA, MAPT, MCC, MIB1, MTMR12, NAALADL2, NLGN3, NRG3, NRXN3, OR52M1, PAH, PAK3, PCOLCE, PHF15, PLA1A, PLA2G4F, PTPRM, RELN, RNASET2, SDK1, SGSM3, SLCO1B1, SLCO1B3, SPARCL1, TRPM1, TRPM3, TTLL3, UTP6, VIL1, VPS13B, WWOX, ZBBX, ZNF559, ZNF774 |
Abbreviation: pLOF, pathogenic loss of function.
Genes set in boldface type are genes that would have been missed with a categorical diagnostic approach (evidence required to be included in the database came from individuals with different developmental brain disorders). Underlined genes are novel developmental brain disorder candidate genes.
Genes that rank one tier higher (increased evidence level) because of our cross-disorder approach.
Figure 1.
Pathogenic Loss-of-Function (pLOF) Variants With Known Inheritance Pattern in Tier 1 Candidate Genes for Developmental Brain Disorders
Variants with unknown inheritance patterns are not included in the figure. Tier 1 refers to genes with 3 or more de novo pLOF variants.
Increased Yield of Risk Allele Identification and Discovery of Novel DBD Candidate Genes
Compared with the use of our multilevel data-integration approach, had we used a classic categorical diagnostic approach for gene discovery and only considered genes with 2 or more pLOF variants identified in unrelated individuals with the same DBD, the total number of unique disease-associated genes would be 208. By implementing a cross-disorder approach to DBDs and integrating genotype and phenotype data from individuals with 6 apparently distinct disorders, we provided the required evidence to include 33 additional genes in the knowledge base (12 high-confidence genes from tiers 1 and 2 and 21 emerging candidate genes from tiers 3 and 4), which would have been missed with a categorical diagnostic approach (Table 2). Furthermore, by transcending clinical diagnostic boundaries, we increased the evidence level for 18 additional genes, which were ranked one tier higher because of our cross-disorder approach (Table 2).
Moreover, our multilevel data-integration approach identified 7 novel high-confidence DBD candidate genes (tier 2) and provided evidence of 10 novel putative candidate genes (tiers 3 and 4), which were not previously considered to act as mendelian genes with high penetrance and large effect size in any brain disorder (Table 3). Evidence of 2 of the novel candidate genes (ELAVL2 [HGNC 3313] and OR52M1 [HGNC 15225]) came from individuals with different disorders with different types of genomic variants (structural and sequence) reported by different studies, whereas 10 additional genes (HIVEP3 [HGNC 13561], MOV10 [HGNC 7200], NEDD9 [HGNC 7733], RALGAPB [HGNC 29221], YTHDC1 [HGNC 30626], CCDC91 [HGNC 24855], EDA2R [HGNC 17756], MSRA [HGNC 7377], ARHGEF38 [HGNC 25968],and EYS [HGNC 21555]) were identified by the cross-disorder and cross-study approach and 5 (ZWILCH [HGNC 25468], LPP [HGNC 6679], OR2T10 [HGNC 19573], CYLC2 [HGNC 2583], and LRBA [HGNC 1742]) exclusively by the cross-study approach (unrelated individuals with variants in each gene had the same DBD). All the pLOF variants identified for these genes came from the supplemental data of different studies, which did not discuss such findings in the main report. For example, 2 unrelated individuals from different cohort studies have de novo pLOF variants in NEDD9: one with ID/DD and ASD from the Simons Simplex Collection reported by Iossifov et al28 and another with epilepsy reported by the EuroEPINOMICS-RES Consortium et al.29 Mutations in this gene were previously reported in individuals with different types of cancer, including melanoma, glioblastoma, and gastric cancer, but not in association with any developmental or neuropsychiatric disorder.30
Table 3.
Novel Candidate Genes Identified Using a Multilevel Data-Integration Approach
| Gene | No. of Cases |
DBDa | Inheritance | Platform | Tier | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID/DD | ASD | ADHD | SCZ | BD | EP | De Novo | Inherited | NA | WGS | WES | CMA | |||
| ELAVL2 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | 2 |
| HIVEP3 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 2 |
| MOV10 | 2 | 1 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 2 |
| NEDD9 | 2 | 1 | 1 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 2 | 0 | 2 |
| RALGAPB | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 2 | 0 | 2 |
| YTHDC1 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 2 |
| ZWILCH | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 1 | 0 | 2 |
| CCDC91 | 2 | 1 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 3 |
| EDA2R | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 3 |
| LPP | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 3 |
| MSRA | 2 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 2 | 3 |
| OR2T10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | 3 |
| ARHGEF38 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 4 |
| CYLC2 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 4 |
| EYS | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 4 |
| LRBA | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 4 |
| OR52M1 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 1 | 4 |
Abbreviations: ADHD, attention-deficit/hyperactivity disorder; ASD, autism spectrum disorder; BD, bipolar disorder; CMA, chromosomal microarray analysis; DD, developmental delay; EP, epilepsy; ID, intellectual disability; NA, not available; SCZ, schizophrenia; WES, whole-exome sequencing; WGS, whole-genome sequencing.
More than one developmental brain disorder may be reported in the same individual.
Variable Expressivity of DBD Genes
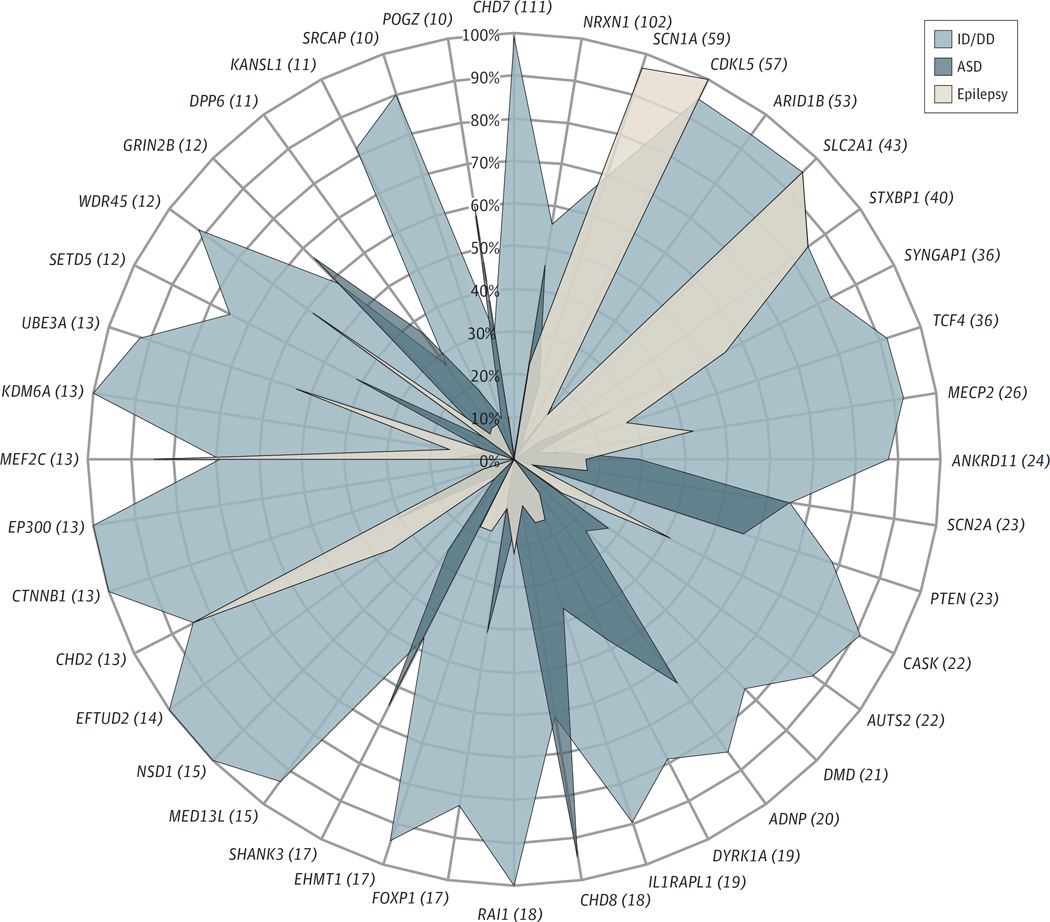
Of the 241 DBD candidate genes, 75 (31.1%) were associated with a single disorder (ASD, 63; ID/DD, 6; schizophrenia, 5; and ADHD, 1), 79 (32.8%) with 2 disorders, 49 (20.3%) with 3 disorders, 25 (10.4%) with 4 disorders, and 11 (4.6%) with 5 disorders. Two genes, NRXN1 and PARK2 (HGNC 8607), were associated with all 6 DBDs. Although there is significant phenotypic heterogeneity and diagnostic overlap of apparently different disorders associated with most of the DBD candidate genes, the prevalence of reported disorders varies among genes, and a specific DBD association profile can be observed for each gene. Furthermore, certain genes appear to be enriched for specific disorders more than others. For example, as shown in Figure 2, the frequency of epilepsy is greater than that of ID/DD in individuals with pLOF variants in SCN1A, which in turn is greater than the frequency of ASD. Conversely, individuals with pLOF variants in CHD8 have a higher frequency of ASD than ID/DD and epilepsy, whereas probands with variants in ARID1B are more likely to be diagnosed as having ID/DD than ASD or epilepsy. However, the extent and detail of the phenotype data included in our study are restricted to the clinical information reported by multiple articles, which is frequently scarce and incomplete. For example, large cohort studies often only report the categorical diagnosis used as inclusion criteria for the study (eg, epilepsy) but omit important neurodevelopmental comorbidities (eg, ID/DD, ASD). In addition, many individuals recruited to study cohorts based on certain categorical diagnoses, such as ID/DD and ASD, are young and have not passed through the age of risk to develop other disorders, including schizophrenia and epilepsy. By integrating apparently different categorical diagnoses and multiple study designs, we likely decrease the burden of some phenotyping biases.
Figure 2.
Frequency of Intellectual Disability or Developmental Delay (ID/DD), Autism Spectrum Disorder (ASD), and Epilepsy in Individuals With Pathogenic Loss-of-Function (pLOF) Variants in Tier 1 Candidate Genes for Developmental Brain Disorders
The figure shows 40 tier 1 genes selected based on the total number of cases (≥10). The total number of pLOF variants (de novo, inherited, and unknown inheritance) is shown in parentheses for each gene. We only included 3 of the 6 developmental brain disorders because these were the most common phenotypes reported by the individual studies. Tier 1 refers to genes with 3 or more de novo pLOF variants.
Discussion
In this study, we developed and implemented a genomicdriven, tiered approach to DBD candidate gene identification, which capitalizes on the vast and rapidly increasing amount of publicly available genotype and phenotype data on multiple individuals with a broad range of brain disorders. We identified 241 candidate genes for DBD and prioritized them based on the strength of evidence. Our multilevel data-integration approach increased the yield of DBD candidate gene discovery over what would be obtained if each disorder, type of genomic variant, and study design were analyzed independently.
The power of our approach comes from genotype and phenotype data integration at multiple levels. This is the first study, to our knowledge, to integrate (1) genomic data from structural and sequence pLOF variants identified using different diagnostic platforms (WGS, WES, CMA, and targeted genomic approaches), (2) phenotype data from any 1 of 6 apparently distinct brain disorders (ID/DD, ASD, ADHD, schizophrenia, BD, and epilepsy), and (3) data from different types of study designs (large-scale cohorts, case series, and case reports). Moreover, we included de novo and inherited pLOF variants to capture a broader spectrum of genomic variation. We focused our analyses on pLOF variants to provide the highest level of confidence on the deleteriousness of the observed variants. Al-though recent studies22,31 have begun to include de novo missense variants predicted to be damaging by a combination of in silico tools (developed to model the effects of amino acid substitutions on protein structure and function), we excluded missense events, which account for most events identified by large-scale sequencing studies, to avoid ambiguity in the interpretation of their functional consequences. This approach results in higher specificity and reduced sensitivity.
Previous studies25–27 have found that identifying independent de novo pLOF variants in the same gene among unrelated individuals is a powerful statistical approach to reliably identifying disease-causative genes. In our study, we identified 59 genes with at least 3 and 44 genes with 2 independent de novo pLOF variants, thus providing strong evidence to consider this group of 103 genes as high-confidence DBD causative genes. In addition, previous studies26,27 have also found that genes with a single de novo pLOF variant are more likely than not to be true risk alleles with a 50.4% to 54.7% chance of truly being disease associated. We identified 68 genes with 1 de novo variant plus at least another pLOF event that was inherited or of unknown inheritance and 70 genes with 2 or more pLOF inherited (or of unknown inheritance) variants. Although the strength of evidence (based on de novo variant counts) is not as robust as that of genes within tiers 1 and 2, we consider these genes as emerging DBD causative genes. We hypothesize that as we continue our ongoing approach to DBD gene discovery, a subgroup of genes included in tiers 3 and 4 will soon move up to tiers 1 and 2 because of the rapidly increasing number of studies reporting a wealth of genomic and phenotypic data from individuals with a variety of brain disorders. If pLOF variants were to be identified in DBD candidate genes among unaffected individuals (properly screened for neuropsychiatric disorders), the pathogenicity tier of such genes would be reduced.
This type of study has certain limitations. First, the amount and quality of the phenotype data used for our multilevel data integration approach rely on the clinical information provided by each individual study, which is often limited. Second, we excluded missense variants, deletions involving more than 1 coding gene, and individuals with more than 1 pLOF variant, which decreases the sensitivity of our method. However, the use of such strict inclusion and exclusion criteria limits the number of potential confounding factors and provides the strongest and least ambiguous evidence for the candidate genes identified. Third, we based part of our approach on a method that was developed based on de novo variants exclusively, and our study also includes inherited pLOF variants. To circumvent this fact, we created a tiered system for gene prioritization, which ranks genes based on the number of de novo variants. With this approach, genes within tiers 1 and 2, which have at least 2 independent de novo pLOF variants, are considered high confidence, and genes with in tiers 3 and 4 (1 de novo and only inherited variants, respectively) are considered emerging candidate genes. Fourth, because our approach relies on published data, it is subject to publication bias.
A previous study31 took a similar cross-disorder approach to identify DBD candidate genes. Li et al31 pooled WES and WGS data from 3555 trios with any 1 of 4 disorders (ID, ASD, epilepsy, or schizophrenia) from 36 studies and used the Transmission and De Novo Association program31 to identify a higher prevalence of de novo variants (mainly missense variants) in probands relative to their unaffected control siblings. On the basis of annotation of de novo exonic variants, they prioritized 764 potential candidate genes, of which 53 were associated with more than 1 disorder and 1, SCN2A, with all 4 DBDs. The main differences with our study include our exclusion of missense variants to avoid ambiguous pathogenicity interpretations and our inclusion of (1) 2 additional brain disorders (ADHD and BD), (2) structural pLOF variation in addition to sequence variants, (3) inherited (and unknown inheritance) pLOF variants, and (4) a larger number and different types of studies (351 publications in addition to the 36 studies reviewed by Li et al31).
Our results are consistent with the concept of developmental brain dysfunction, which our group recently proposed to encompass a continuum of developmental disabilities and neuropsychiatric disorders.1 This model emphasizes atypical brain development as a common denominat or that can manifest as cognitive, behavioral, and/or motor impairments and arise from a genetic anomaly or an insult to the developing brain. Furthermore, the specific profile of impairments (type and severity) resulting from the underlying brain dysfunction can be used to determine an individual’s categorical clinical diagnosis and guide treatment.
Conclusions
The continued efforts of the scientific community to establish and expand large data-sharing international consortia dedicated to study the underpinnings of brain disorders will further increase our knowledge of genomic causes shared by apparently distinct disorders. The creation of a comprehensive database, updated in real time, of all potentially causative variants associated with all medical disorders will allow for continuous gene discovery. By sharing genotype and phenotype data on individuals with DBDs with fellow researchers, physicians, and other health care providers, common underlying molecular pathways that may be amenable to potential therapeutic interventions are likely to emerge.
Acknowledgments
Funding/Support: This study was supported by grant RO1MH074090 from the National Institute of Mental Health (Drs Martin and Ledbetter).
Role of the Funder/Sponsor: The funding source had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Author Contributions: Drs Gonzalez-Mantilla and Moreno-De-Luca contributed equally to this work. Dr Martin had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: All authors.
Acquisition, analysis, or interpretation of data: Gonzalez-Mantilla, Moreno-De-Luca, Martin.
Drafting of the manuscript: Gonzalez-Mantilla, Moreno-De-Luca, Martin.
Critical revision of the manuscript for important intellectual content: All authors.
Obtained funding: Ledbetter, Martin.
Administrative, technical, or material support: Moreno-De-Luca, Ledbetter, Martin.
Study supervision: Moreno-De-Luca, Ledbetter, Martin.
Conflict of Interest Disclosures: Dr Ledbetter reported working as a consultant for Natera Inc. However, there was no financial support from this company for research or preparation of the article. No other disclosures were reported.
Additional Contributions: Daniel Moreno-De-Luca, MD, critically reviewed the manuscript and Tristan Nelson, BA, developed the website with our DBD candidate gene knowledge base.
Dr Moreno-De-Luca was not compensated for his review. Mr Nelson is employed by Geisinger Health System and receives compensation for his work, which included the development of the DBD candidate gene website.
REFERENCES
- 1.Moreno-De-Luca A, Myers SM, Challman TD, Moreno-De-Luca D, Evans DW, Ledbetter DH. Developmental brain dysfunction: revival and expansion of old concepts based on new genetic evidence. Lancet Neurol. 2013;12(4):406–414. doi: 10.1016/S1474-4422(13)70011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reiss AL. Childhood developmental disorders: an academic and clinical convergence point for psychiatry, neurology, psychology and pediatrics. J Child Psychol Psychiatry. 2009;50(1–2):87–98. doi: 10.1111/j.1469-7610.2008.02046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeargin-Allsopp M, Boyle C, van Naarden-Braun K, Trevathan E. The epidemiology of developmental disabilities. In: Accardo PJ, editor. Capute and Accardo’s Neurodevelopmental Disabilities in Infancy and Childhood. Vol I. Neurodevelopmental Diagnosis and Treatment. 3rd. Baltimore, MD: Paul H. Brookes Publishing Co; 2008. pp. 61–104. [Google Scholar]
- 4.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 5th. Arlington, VA: American Psychiatric Publishing; 2013. [Google Scholar]
- 5.World Health Organization. The ICD-10 Classification of Mental and Behavioural Disorders: Clinical Descriptions and Diagnostic Guidelines. Geneva, Switzerland: World Health Organization; 2010. [Google Scholar]
- 6.Martin J, Cooper M, Hamshere ML, et al. Biological overlap of attention-deficit/hyperactivity disorder and autism spectrum disorder: evidence from copy number variants. J Am Acad Child Adolesc Psychiatry. 2014;53(7):761.e26–770.e26. e726. doi: 10.1016/j.jaac.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCarthy SE, Gillis J, Kramer M, et al. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry. 2014;19(6):652–658. doi: 10.1038/mp.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreno-De-Luca D, Moreno-De-Luca A, Cubells JF, Sanders SJ. Cross-disorder comparison of four neuropsychiatric CNV loci. Curr Genet Med Rep. 2014;2(3):151–161. [Google Scholar]
- 9.Béna F, Bruno DL, Eriksson M, et al. Molecular and clinical characterization of 25 individuals with exonic deletions of NRXN1 and comprehensive review of the literature. Am J Med Genet B Neuropsychiatr Genet. 2013;162B(4):388–403. doi: 10.1002/ajmg.b.32148. [DOI] [PubMed] [Google Scholar]
- 10.Helsmoortel C, Vulto-van Silfhout AT, Coe BP, et al. A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat Genet. 2014;46(4):380–384. doi: 10.1038/ng.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernier R, Golzio C, Xiong B, et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158(2):263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim YS, State MW. Recent challenges to the psychiatric diagnostic nosology: a focus on the genetics and genomics of neurodevelopmental disorders. Int J Epidemiol. 2014;43(2):465–475. doi: 10.1093/ije/dyu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaudhry A, Noor A, Degagne B, et al. DDD Study. Phenotypic spectrum associated with PTCHD1 deletions and truncating mutations includes intellectual disability and autism spectrum disorder. Clin Genet. 2015;88(3):224–233. doi: 10.1111/cge.12482. [DOI] [PubMed] [Google Scholar]
- 14.Cancrini C, Puliafito P, Digilio MC, et al. Italian Network for Primary Immunodeficiencies. Clinical features and follow-up in patients with 22q11.2 deletion syndrome. J Pediatr. 2014;164(6):1475.e2–1480.e2. e1472. doi: 10.1016/j.jpeds.2014.01.056. [DOI] [PubMed] [Google Scholar]
- 15.Moreno-De-Luca A, Evans DW, Boomer KB, et al. The role of parental cognitive, behavioral, and motor profiles in clinical variability in individuals with chromosome 16p11.2 deletions. JAMA Psychiatry. 2015;72(2):119–126. doi: 10.1001/jamapsychiatry.2014.2147. [DOI] [PubMed] [Google Scholar]
- 16.Haldeman-Englert C, Jewett T. 1q21.1 microdeletion. In: Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews(R) Seattle: University of Washington; 1993. [Google Scholar]
- 17.Moreno-De-Luca D, Mulle JG, Kaminsky EB, et al. SGENE Consortium; Simons Simplex Collection Genetics Consortium; GeneSTAR. Deletion 17q12 is a recurrent copy number variant that confers high risk of autism and schizophrenia. Am J Hum Genet. 2010;87(5):618–630. doi: 10.1016/j.ajhg.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Need AC, Shashi V, Hitomi Y, et al. Clinical application of exome sequencing in undiagnosed genetic conditions. J Med Genet. 2012;49(6):353–361. doi: 10.1136/jmedgenet-2012-100819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312(18):1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee H, Deignan JL, Dorrani N, et al. Clinical exome sequencing for genetic identification of rare mendelian disorders. JAMA. 2014;312(18):1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srivastava S, Cohen JS, Vernon H, et al. Clinical whole exome sequencing in child neurology practice. Ann Neurol. 2014;76(4):473–483. doi: 10.1002/ana.24251. [DOI] [PubMed] [Google Scholar]
- 22.De Rubeis S, He X, Goldberg AP, et al. DDD Study. Homozygosity Mapping Collaborative for Autism; UK10K Consortium. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515(7526):209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell SM, Moran JL, Fromer M, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506(7487):185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deciphering Developmental Disorders S; Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519(7542):223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanders SJ, Murtha MT, Gupta AR, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485(7397):237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willsey AJ, Sanders SJ, Li M, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155(5):997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong S, Walker MF, Carriero NJ, et al. De novo insertions and deletions of predominantly paternal origin are associated with autism spectrum disorder. Cell Rep. 2014;9(1):16–23. doi: 10.1016/j.celrep.2014.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iossifov I, O’Roak BJ, Sanders SJ, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515(7526):216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.EuroEPINOMICS-RES Consortium; Epilepsy Phenome/Genome Project; Epi4K Consortium. De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am J Hum Genet. 2014;95(4):360–370. doi: 10.1016/j.ajhg.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beck TN, Nicolas E, Kopp MC, Golemis EA. Adaptors for disorders of the brain? the cancer signaling proteins NEDD9, CASS4, and PTK2B in Alzheimer’s disease. Oncoscience. 2014;1(7):486–503. doi: 10.18632/oncoscience.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Cai T, Jiang Y, et al. Genes with de novo mutations are shared by four neuropsychiatric disorders discovered from NPdenovo database [published online May 5, 2015] Mol Psychiatry. doi: 10.1038/mp.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]