Summary
Although lysine acetylation is now recognized as a general protein modification for both histones and non-histone proteins1-3, the mechanisms of acetylation mediated actions are not completely understood. Acetylation of the C-terminal domain (CTD) of p53 was the first example for non-histone protein acetylation4. Yet the precise role of the CTD acetylation remains elusive. Lysine acetylation often creates binding sites for bromodomain-containing “reader” proteins5,6; surprisingly, in a proteomic screen, we identified SET as a major cellular factor whose binding with p53 is totally dependent on the CTD acetylation status. SET profoundly inhibits p53 transcriptional activity in unstressed cells but SET-mediated repression is completely abolished by stress-induced p53 CTD acetylation. Moreover, loss of the interaction with SET activates p53, resulting in tumor regression in mouse xenograft models. Notably, the acidic domain of SET acts as a “reader” for unacetylated CTD of p53 and this mechanism of acetylation-dependent regulation is widespread in nature. For example, p53 acetylation also modulates its interactions with similar acidic domains found in other p53 regulators including VPRBP, DAXX and PELP1 (refs. 7-9), and computational analysis of the proteome identified numerous proteins with the potential to serve as the acidic domain readers and lysine-rich ligands. Unlike bromodomain readers, which preferentially bind the acetylated forms of their cognate ligands, the acidic domain readers specifically recognize the unacetylated forms of their ligands. Finally, the acetylation-dependent regulation of p53 was further validated in vivo by using a knockin mouse model expressing an acetylation-mimicking form of p53. These results reveal that the acidic domain-containing factors act as a new class of acetylation-dependent regulators by targeting p53 and potentially, beyond.
Keywords: p53, acetylation, deacetylation, SET, transcriptional regulation, Acidic domain
Although the physiological consequences of acetylation at K120 and K164 within the DNA-binding domain have been established in the studies of p53 acetylation-defective mutant mice10,11, the in vivo functions of CTD acetylation remain elusive. Interestingly, by examining the mutant mice expressing C-terminal truncated forms of p53, two recent studies have shown that loss of the CTD results in p53 activation12,13, suggesting that the CTD may act as a docking site for negative regulators of p53. Nevertheless, the identity of the negative regulators and the consequences of CTD acetylation remain unclear. To identify proteins that bind p53 in a manner dependent on its CTD acetylation status, we synthesized both unacetylated (Un-Ac) and fully-acetylated (Ac) biotin-conjugated CTD peptides and used the immobilized peptides as affinity columns to purify cellular factors (Fig. 1a). As shown in Fig. 1b, we failed to identify any proteins enriched in the acetylated p53 CTD column. Instead, coomassie blue staining of the bound fractions revealed a major band of ~38 kD from the unacetylated p53 column that was completely absent from the acetylated one. Mass spectrometry analysis of this band revealed 28 unique peptides identical to SET (Fig. 1c and Extended Data Fig. 1a), an oncoprotein that is activated by translocation-associated gene fusions in patients with acute myeloid leukemia14. Although a previous study reported an interaction between p53 and SET15, the impact of CTD acetylation on the functional consequences of this interaction remains unclear.
Figure 1. Identification of SET as a specific co-repressor of C-terminal unacetylated p53.
a, Schematic diagraph of synthesized biotin-conjugated p53 CTD. b, Coomassie Blue staining of the protein complex bound with p53 CTD. c, Schematic diagraph of SET. DD: dimerization domain; ED: earmuff domain; AD: acidic domain. d, In vitro binding assay of p53 CTD and purified SET. e, Western blot analysis of the interaction between p53 and SET in nuclear fraction of H1299 cells. f, EMSA showing SET/p53-DNA complex formation in vitro. g, Luciferase assays of SET-mediated regulation on p53 transactivity in H1299 cells. h, Western blot analysis of the endogenous interaction between p53 and SET upon doxorubicin (Dox) treatment in HCT116 cells. i, ChIP analysis of p53 or SET recruitment on p21 promoter upon Dox treatment in HCT116 cells. j, A model of dynamic promoter-recruitment of SET regulated by p53 CTD acetylation status. Error bars indicate mean ± s.d., n=3 for technical replicates. Data were shown as representative of three experiments. Uncropped blots were shown in Supplementary Fig. 1.
Acetylation-dependent disruption of the p53-SET interaction was confirmed in vitro with purified SET protein (Fig. 1d). Moreover, expression of CBP, the enzyme responsible for CTD acetylation, completely abrogated the formation of SET complex with wildtype p53 (p53WT), but not with CTD acetylation-deficient p53 (p53KR) mutant, validating that CTD acetylation is crucial for the p53-SET interaction in cells (Fig. 1e). Interestingly, other modifications on the CTD lysine residues, including methylation, ubiquitination, sumoylation and neddylation, had no dramatic effect on this binding, underscoring the specificity of acetylation-dependent control of p53-SET interactions (Extended Data Fig. 1b-e).
Next, we tested whether SET acts as a transcriptional cofactor by forming a p53-SET complex on p53 target promoter. As shown in Fig. 1f, although SET alone showed no obvious DNA binding activity, in the presence of both p53 and SET, a slower migrating SET/p53-DNA complex was formed and super-shifted by p53- or SET-antibody. Further binding-domain mapping indicate that the CTD of p53 directly interacts with the acidic domain (AD) of SET (Extended Data Fig. 1f-h). To determine the impact of SET on the transcriptional activity of p53, we measured transactivation of a p53-responsive reporter gene. Indeed, p53-mediated transactivation was abrogated upon co-expression of wildtype SET, but not a SET mutant lacking the acidic domain required for p53 binding (Fig. 1g). Conversely, wildtype SET-mediated repression was abrogated when a p53 mutant lacking the CTD was expressed (Fig. 1g). Notably, the interaction of endogenous p53 and SET was easily detected in unstressed cells; however, upon DNA damage, despite increased p53 levels, the p53-SET interaction was largely diminished, likely due to the induction of CTD acetylation (Fig. 1h). Moreover, chromatin immunoprecipitation (ChIP) assays revealed that the recruitment of SET to the promoter of p53 targets was largely inhibited (Fig. 1i and Extended Data Fig. 1i-k). Together, these data indicate that SET acts as a transcriptional co-repressor of p53 but acetylation of the CTD leads to abrogate the repression through disrupting the p53-SET interactions upon DNA damage (Fig. 1j).
We further examined whether inactivation of SET influences the activities of p53 in human cancer cells. Indeed, RNAi-mediated depletion of SET markedly elevated the expression of p53 targets, such as p21 and PUMA, without affecting the steady-state levels of endogenous p53 in HCT116 colorectal carcinoma cells (Fig. 2a). Similar effects were obtained in other human cancer cell lines that express wildtype p53, including MCF7 (breast carcinoma), U2OS (osteosarcoma), H460 (lung carcinoma) and SU-DHL-5 (B-cell lymphoma) (Fig. 2b). Moreover, this induction of p21 and PUMA expression was completely abrogated in isogenic HCT116 p53−/− cells (Fig. 2c), indicating that the SET-mediated effects are p53-dependent. Further analysis of U2OS and p53-null U2OS cells by SET knockdown identified a number of p53 targets that are upregulated upon inactivation of SET in a p53-dependent manner and SET knockdown induced p53-dependent cell growth repression in those cells (Extended Data Fig. 2a-c and Extended Data Fig. 3a-b). To examine the impact of SET on p53-mediated tumor suppression, we tested whether SET depletion affects cell growth in xenograft tumor models. As shown in Fig. 2d, SET knockdown dramatically suppressed tumor growth of HCT116 cells, but not isogenic HCT116 p53−/− cells. Moreover, such p53-dependent effects were further validated in HCT116 p53 knockout cells generated by CRISPR/Cas9-mediated genome editing technique (Extended Data Fig. 3c-e). These data indicate that the p53-SET interaction is crucial for the tumor growth suppression by p53.
Figure 2. SET negatively regulates p53 transactivity by inhibiting p300/CBP-mediated H3K18 and H3K27 acetylation on p53 target promoter.
a, b, c Western blot analysis of SET knockdown-mediated effect on p53 activity in cells. d, Xenograft analysis of SET-mediated effect on tumor growth. e, ChIP analysis of SET knockdown-mediated effect on histone modifications at p21 promoter in HCT116 cells. f, In vitro acetylation assay of SET effect on p300-mediated H3K18 and H3K27 acetylation. g, ChIP analysis of SET-mediated effect on p53-dependent H3K18 and H3K27 acetylation on p21 promoter in H1299 cells. h, A model of SET-mediated regulation on p53 transactivity. Error bars indicate mean ± s.d., n=3 for technical replicates in (e) and (g); n=5 (p53+/+ group) or n=3 (p53−/− group) for biological replicates in (d). Data were shown as representative of three experiments. Uncropped blots were shown in Supplementary Fig. 1.
Since SET apparently had no dramatic effect on protein stability, DNA binding, or acetylation levels of p53 (Extended Data Fig. 4a-c), we examined whether SET suppressed p53-mediated transactivation by affecting the chromatin modifications at p53 target promoters. ChIP analysis revealed that SET depletion significantly increased the acetylation levels of H3K18 and H3K27 at p21 and PUMA promoter without obviously affecting H3K9, H3K14, H4K16 or pan-H4 acetylation (Fig. 2e and Extended Data Fig. 4d). p300/CBP, which majorly targets H3K18 and H3K27 acetylation in vivo16,17, acts as a key co-activator in p53-mediated transcriptional activation18-20. We then examined whether SET suppresses p300/CBP-mediated acetylation of H3K18 and H3K27 as SET had no obvious effect on the recruitment of p300/CBP (Extended Data Fig. 4e). Indeed, in vitro acetylation assays revealed that SET effectively repressed p300-dependent acetylation of H3K18 and H3K27 (Fig. 2f) and these finds were further verified on p53 target promoters by ChIP analysis (Fig. 2g and Extended Data Fig. 4f). Taken together, these data indicate that SET represses p53-mediated transactivation by inhibiting p300/CBP-dependent acetylation of H3K18 and H3K27 on p53 target promoters (Fig. 2h).
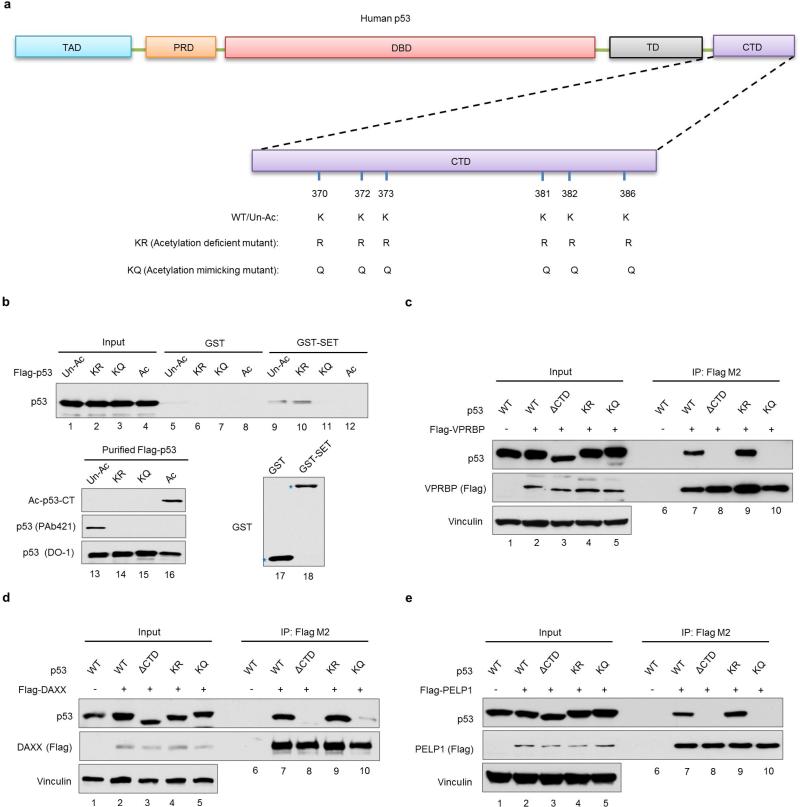
Numerous studies indicate that lysine acetylation often creates docking sites for “reader” proteins that possess bromodomain, a structural motif that forms a recognition surface for acetylated lysine5,6. Our analysis of the p53-SET interaction suggests that the acidic domain of SET serves as a “converse reader” that binds the lysine-rich CTD of p53 in a manner that can be specifically abrogated upon acetylation of these lysine residues. To further evaluate this model, we examined whether p53 interacts with other proteins in a similar manner. Several transcription cofactors known to interact directly with p53, including VPRBP, DAXX and PELP1 (refs. 7-9), also contain acidic domains similar to that of the SET protein (Fig. 3a and Extended Data Fig. 5a). Their acidic domains also readily bound unacetylated, but not acetylated, p53 CTD (Fig. 3b-d). Similar results were also obtained when the full-length proteins of VPRBP, DAXX and PELP1 were tested (Extended Data Fig. 5b). More importantly, their interactions (VPRBP, DAXX and PELP1) with wildtype p53, but not the acetylation-deficient p53KR mutant, were inhibited by CBP-induced acetylation in human cells (Extended Data Fig. 5c-e).
Figure 3. Acidic domain-containing proteins represent a new class of “reader” for their unacetylated ligands.
a, Schematic diagraph of acidic domain (AD)-containing protein SET, VPRBP, DAXX and PELP1. b, c, d, In vitro binding assay of p53 CTD and acidic domain of VPRBP (b), DAXX (c) or PELP1 (d). e, Schematic diagraph of lysine-rich domain (KRD)-containing protein histone H3, KU70 and FOXO1. f, g, h, In vitro binding assay between purified SET acidic domain and lysine-rich domain of H3 (f), KU70 (g) or FOXO1 (h). i, A model of acetylation-dependent regulation of the interactions between lysine-rich domain (KRD)-containing proteins and their acidic domain (AD)-containing “readers”. Uncropped blots were shown in Supplementary Fig. 1.
Previous studies showed that SET also regulates the activities of several other cellular factors, including histone H3, KU70 and FOXO1, through direct interactions21-23. Notably, the binding regions of all three proteins contain a lysine-rich domain (KRD) similar to the CTD of p53 (Fig. 3e). More importantly, those lysine residues have also been reported to be acetylated in vivo24-26. To test whether SET-mediated interactions with these factors are also regulated by acetylation, we performed in vitro binding assays of the acidic domain of SET with unacetylated vs. acetylated lysine-rich domain of H3, KU70 and FOXO1. Indeed, the acidic domain of SET interacted with unacetylated, but not acetylated, lysine-rich domains of H3, KU70 and FOXO1 (Fig. 3f-h). Similar results were also obtained when the full-length SET protein was used in the binding assays (Extended Data Fig. 5f-h), suggesting that the SET interactions with H3, KU70 and FOXO1 are abrogated by acetylation in a manner analogous to the p53-SET binding. Since VPRBP, DAXX and PELP1 are also implicated in transcription regulation, we examined whether these factors interact with H3 in a similar manner. Indeed, VPRBP, DAXX and PELP1 specifically bound unacetylated H3 whereas, as expected, bromodomain proteins such BRD4 and BRD7 recognized only acetylated H3 (Extended Data Fig. 5i-j).
Above data indicate that this mechanism of acetylation-dependent regulation is widespread in nature. Since the positive charge within lysine-rich domain can attract the negative charge of the acidic domain, these lysine clusters form a docking site for acidic domain-containing regulators. However, upon acetylation, the positive charge of lysine sidechains is neutralized, abolishing the docking site for the acidic domain-containing regulators. Conversely, deacetylation of these lysine residues reverses the effects and promotes the recruitment of acidic domain-containing regulators (Fig. 3i). Thus, unlike bromodomain readers, which preferentially bind the acetylated forms of their cognate ligands, the acidic domain readers specifically recognize the unacetylated forms of their ligands.
To corroborate this notion, we compared the SET-binding properties of the acetylation-deficient mutant p53KR and an acetylation-mimicking mutant p53KQ (Extended Data Fig. 6a). As shown in Extended Data Fig. 6b, the p53KR mutant, like unacetylated p53, strongly bound SET; conversely, like acetylated p53, the p53KQ mutant completely abolished the interaction with SET. Similar results were also obtained upon analysis of the acetylation-modulated interactions of p53 with VPRBP, DAXX and PELP1 (Extended Data Fig. 6c-e).
To further determine the physiological significance of these interactions in vivo, we generated p53KQ/KQ mutant mice (Extended Data Fig. 7a-d). While heterozygous p53+/KQ mice displayed normal postnatal development, p53KQ/KQ homozygous mice were neonatal lethal (Extended Data Fig. 7e). All newborn p53KQ/KQ pups were slightly smaller than their p53+/+ littermates (Fig. 4a), lacked milk in their stomach and died within one day of birth, apparently due to dehydration from lack of maternal nourishment. In addition, live p53KQ/KQ mice also displayed uncoordinated movements, consistent with neurological impairments. Indeed, the brains of p53KQ/KQ mice appeared smaller than those of p53+/+ mice (Fig. 4b). Immunohistochemistry analysis of p53KQ/KQ brain sections revealed a marked induction of cleaved Caspase 3 staining without an obvious increase in p53 protein levels (Fig. 4c and Extended Data Fig. 7f), suggesting that the neurological defects of p53KQ/KQ mice may reflect increased apoptosis due to deregulation of the p53KQ protein. In accord with this notion, the major apoptotic transcriptional targets of p53 (Bax and Puma) are significantly up-regulated in p53KQ/KQ brain tissue (Fig. 4d). Indeed, various tissues of p53KQ/KQ mice displayed distinct patterns of induction of the different p53 target genes, suggesting tissue-specific activation of the target genes by p53KQ in vivo (Fig. 4d).
Figure 4. The physiological significance of acetylation-dependent dissociation of p53 from its acidic domain-containing “readers”.
a, The new born of p53+/+ and p53KQ/KQ mice. b, The brains from p53+/+ and p53KQ/KQ mice. c, Immunohistochemistry analysis of brain sections from p53+/+ and p53KQ/KQ mice. d, RT-qPCR analysis of p53 target gene expression in p53+/+ and p53KQ/KQ tissues. e, Western blot analysis of the interaction between p53 and acidic domain-containing proteins in p53+/+ or p53KQ/KQ MEFs treated with proteasome inhibitor Epoxomicin. f, Cell growth analysis of p53+/+ or p53KQ/KQ MEFs (P3). g, Morphological representative of p53+/+ and p53KQ/KQ MEFs from P0 to P4. h, SA-β-gal staining of p53+/+ and p53KQ/KQ MEFs (P3). i, Western blot analysis of p21 and p53 expression in p53+/+ and p53KQ/KQ MEFs. j, Western blot analysis of p53 targets in Set conditional knockout MEFs. Error bars indicate mean ± s.d., n=3 for technical replicates in (d); n=3 for biological replicates in (f). Data were shown as representative of three experiments. Uncropped blots were shown in Supplementary Fig. 1.
Notably, the p53-SET interaction was readily detected in p53+/+, but not p53KQ/KQ, MEFs (Fig. 4e). Similar results were also obtained for the other acidic domain-containing cofactors (VPRBP, DAXX and PELP1), suggesting that the p53KQ mutant recapitulates acetylation-mediated effects on p53 in vivo. Moreover, p53KQ/KQ MEFs displayed a severe proliferation defect (Fig. 4f) and exhibited clear signs of senescence, including a flat and enlarged morphology with large multinucleated nuclei and marked senescence-associated beta-galactosidase (SA-β-Gal) staining (Fig. 4g-h; Extended Data Fig. 7g-h). In addition, Western blot analysis revealed a dramatic increase in the steady-state levels of p21 protein in p53KQ/KQ MEFs (Fig. 4i). To directly address the role of SET in vivo, we generated Set mutant mice (Extended Data Fig. 8a-b). Although the characterization of these mice was not complete (Extended Data Fig. 8c-e), we prepared Setflox/flox MEFs for functional analysis. As shown in Fig. 4j, upon Cre-mediated Set deletion, the expression of p53 target genes, such as p21 and Puma, was significantly induced, indicating SET as a critical regulator of p53 in vivo. Together, these data validate the critical role of CTD acetylation in p53 activation in vivo.
Previous studies showed that a p53KR knockin mutant targeting the same CTD lysine residues does not significantly affect mouse development or p53 activities in mouse tissues or embryonic fibroblasts27,28. Thus, loss of modifiable CTD lysines may neutralize the overall impact on p53 function by abrogating both negative and positive effects of regulation through the different types of CTD modifications. Surprisingly, p53KQ knockin mice die shortly after birth with dramatic p53 activation. Like p53KR, p53KQ also eliminates other types of modifications on these lysine residues; however, p53KQ mimics the acetylated form while p53KR resembles unacetylated p53. Thus, the striking difference between the phenotypes of p53KQ and p53KR mutant mice underscores the role of CTD acetylation in vivo.
The acidic domain-containing proteins in this study are referred to a specific group of proteins that harbor long clustered distribution of acidic amino acids. Searching the Uniprot database with our motif-finding algorithm29, we identified 49 polypeptides with highly acidic domains similar to SET, many of which are involved in transcriptional regulation and chromatin remodeling (Extended Data Table 1). In addition, by using Species-Specific Prediction of lysine (K) Acetylation program (SSPKA)30, we also identified 49 proteins containing a cluster of lysine residues that can potentially bind these acidic domains in an acetylation-modulated manner (Extended Data Table 2). Based on our data, we propose that acetylation-mediated regulation, whereby acetylation of p53 abrogates its association with the acidic domain-containing cofactors, can be expanded to a general mode of post-translational control for protein interactions that involve other acidic domain-containing factors and their acetylatable ligands.
Methods
General Data Reports
There is no statistical method to pre-evaluate the sample size in this study. The experiments (including animal related experiments) were not randomized. The investigators were not blinded to experiments. No samples/data were excluded except the xenograft mice with obvious unhealthy status.
Cell Culture, Plasmid Generation, Transfection and Reagent Treatment
H1299, U2OS, MCF7, H460 and HCT116 cell lines were cultured in DMEM medium with supplementing 10% (vol/vol) FBS. SU-DHL-5 cell line was cultured in IMDM medium with supplementing 10% (vol/vol) FBS. MEFs were cultured in DMEM medium with supplementing 10% (vol/vol) heat-inactivated FBS. All the cell lines were obtained from ATCC and have been proved as negative of mycoplasma contamination. No cell lines used in this work were listed in ICLAC database. The cell lines were freshly thawed from the purchased seed cells and were cultured for no more than 2 months. The morphology of cell lines were checked every week and compared with the ATCC cell line image to avoid cross-contamination or misuse of cell lines. SET stable knockdown cells were generated by lentivirus-based infection of shRNA. SET cDNA was purchased from Addgene (Plasmid# 24998) and the full-length cDNA or the various fragments were sub-cloned into pWG-F-HA, pCMV-Myc or PGEX-2TL vectors. Each p53 plasmid was generated by sub-cloning human p53 cDNA (including full-length or various fragments) into pWG-F-HA, pcDNA3.1 or PGEX-2TL vectors. The point-mutation constructs (including p53-KR and -KQ) were generated by using a site-directed mutagenesis Kit (Stratagene, 200521). Expressing construct and siRNA transfection were performed by Lipofectamine 2000 (Invitrogen, 11668-019) according to the manufacturer's protocol. To transfer oligos into SU-DHL-5 cells, electroporation was used by following Kit manufacture's protocol (Lonza PBC3-00675). DNA damage inducer Doxorubicin was used as 1 μM for 24 hours. Proteasome inhibitor Epoxomicin was used as 100 nM for 6 hours. Cells were treated with TSA (1 μM) and Nicotinamide (5 mM) for 6 hours to inhibit HDAC activity in the assays in which p53 acetylation needed to be maintained. Ad-GFP and Ad-Cre-GFP virus were purchased from Vector Biolabs (Cat. #: 1761 and 1710).
Mouse Model
To generate the knockin mice, W4/129S6 mouse embryonic stem (ES) cells (Taconic, Hudson, NY, USA) were electroporated with a targeting vector containing homologous regions flanking mouse p53 exon 11, in which all 7 lysines were mutated to glutamines (p53-KQ allele). A neomycin resistance gene cassette flanked by two LoxP sites (LNL) was inserted into intron 10 to allow selection of targeted ES cell clones with G418. ES cell clones were screened by Southern blotting with EcoRI-digested genomic DNA, using a probe generated from PCR amplification in the region outside the homologous region in the targeting vector. The correctly targeted ES cell clones containing the K to Q mutations were injected into C57BL/6 blastocysts, which were then implanted into pseudopregnant females to generate chimeras. Germline transmission was accomplished by breeding chimeras with C57BL/6 mice. Subsequently, mice containing the targeted allele were bred with Rosa26-Cre mice to remove the LNL cassette and to generate mice with only the K to Q mutations. To confirm the mutations inserted in p53+/KQ mice, we sequenced p53 cDNA derived from mRNA isolated from p53+/KQ MEFs. All seven K-to-Q mutations were confirmed and no additional mutations were found. The offspring were genotyped by PCR using a primer set (Forward: 5’-GGGAGGATAAACTGATTCTCAGA-3’, Reverse: 5’- GATGGCTTCTACTATGGGTAGGGAT-3’).
To generate a Set conditional knockout mouse, the exon2 of the Set gene is floxed and deletion of the exon2 results in a frameshift and the truncation of the C-terminal domain. The targeting vector of Set contains 10 kb genomic DNA spanning exon2, a neomycin resistance gene cassette and loxP sites are inserted flanking exon2. To increase targeting frequency, a Diphtheria toxin A cassette is inserted at the 3’ end of the targeting vector to reduce random integration of the modified Set genomic DNA. A new Bgl II restriction site is also inserted to facilitate Southern blot screening. Among the 200 mouse ES cell clones screened, eight of them were identified to have integrated floxed exon2 by southern blot using a 5’ probe, which detects a 14-kb band for wild type allele and an 11-kb band for the floxed exon2 allele (Setflox). Two of the clones were then injected into blastocysts to generate Set chimera mice and they were bred to produce germline transmission of the floxed exon2 allele. Setflox/+ mice were intercrossed to generate set homozygote conditional knockout mice (Setflox/flox). Maintenance and experimental procedures of mice were approved by the Institutional Animal Care and Use Committee (IACUC) of Columbia University.
In vitro Binding Assay
For in vitro peptide binding assay: Equal amount of each synthesized biotin-conjugated peptide (made as column or as batch) was incubated with highly concentrated Hela nuclear extract (NE) or purified proteins for 1 hour or overnight at 4 °C. After washing with BC100 buffer (20 mM Tris-HCl pH 7.9, 100 mM NaCl, 10% Glycerol, 0.2 mM EDTA, 0.1% Triton X-100) for three times, the binding components were eluted by high-salt buffer (20 mM Tris-HCl pH 7.9, 1000 mM NaCl, 1% DOC, 10% Glycerol, 0.2 mM EDTA, 0.1% Triton X-100) or by boiling with 1× Laemmli buffer for further analysis. For in vitro GST-fusion protein binding assay: The Escherichia coli containing GST or GST-fusion protein expressing constructs were grew in the shaker at 37 °C until the O.D. 600 was about 0.6. And then 0.1 mM IPTG was added and incubated the Escherichia coli at 25 °C for 4 hours or overnight to induce GST or GST-fusion protein expression. After purification by GST·Bind™ Resin (Novagen, 70541), equal amount of immobilized GST or GST-fusion proteins were incubated with other purified proteins for 1 hour at 4 °C, followed by washing with BC100 buffer for three times. The binding components were eluted by boiling with 1× Laemmli buffer and subjected to western blot analysis.
Co-Immunoprecipitation Assay (Co-IP)
Whole cellular extract (WCE) were prepared by BC100 buffer plus sonication. Nuclear extract (NE) was prepared by sequentially lysing cells with HB buffer (20 mM Tris-HCl pH7.9, 10 mM KCl, 1.5 mM MgCl2, 1 mM PMSF, 1× protease inhibitor (Sigma); for cytosolic fraction) and BC400 buffer (20 mM Tris-HCl pH 7.9, 400 mM NaCl, 10% Glycerol, 0.2 mM EDTA, 0.5% Triton X-100, 1 mM PMSF, 1× protease inhibitor; for nuclear fraction). Carefully adjust the salt concentration of NE to 100 mM. 2 μg of indicated antibody (or 20 μl Flag M2 Affinity Gel (Sigma, A2220)) was added into WCE or NE and incubated overnight at 4 °C, followed by adding 20 μl Protein A/G agarose (Santa Cruz, sc-2003; only for IP by unconjugated antibody mentioned above) for 2 hours. After washing with BC100 buffer for three times, the binding components were eluted by Flag peptide (Sigma, F3290), by 0.1% Trifluoroacetic acid (TFA, Sigma, 302031) or by boiling with 1× Laemmli buffer and subjected to western blot assay.
Purification of Ub-, Sumo- or Nedd-p53 conjugates from cells
To prepare Ub-p53: H1299 cells were co-transfected with p53, Mdm2 and 6×HA-Ub expressing plasmids for 48 hours. The cells were lysed by Flag lysis buffer (50 mM Tris-HCl pH=7.9, 137 mM NaCl, 10 mM NaF, 1 mM Na3VO4, 10% Glycerol, 0.5 mM EDTA, 1% Triton X-100, 0.2% Sarkosyl, 0.5 mM DTT, 1 mM PMSF, 1× protease inhibitor) and total Ub-conjugated proteins were purified by anti-HA-agarose (Sigma, A2095) and eluted by 1×HA peptide (Sigma I2149). To prepare Sumo-p53 or Nedd-p53: H1299 cells were co-transfected with p53, Mdm2 (only for Nedd-p53 preparation) and 6×His-HA-Sumo1 or 6×His-HA-Nedd8 expressing plasmids for 48 hours. The cells were lysed by Guanidine lysis buffer (6 M guanidin-HCl, 0.1 M Na2HPO4, 6.8 mM NaH2PO4, 10 mM Tris-HCl pH=8.0, 0.2% Triton-X100, freshly supplemented with 10 mM β-mercaptoethanol and 5 mM imidazole) with mild sonication. After overnight pull-down by Ni+-NTA agarose (Qiagen 30230), the binding fractions were sequentially washed with Guanidine lysis buffer, Urea buffer I (8 M urea, 0.1 M Na2HPO4, 6.8 mM NaH2PO4, 10 mM Tris-HCl pH=8.0, 0.2% Triton-X100, freshly supplemented with 10 mM β-mercaptoethanol and 5 mM imidazole) and Urea buffer II (8 M urea, 18 mM Na2HPO4, 80 mM NaH2PO4, 10 mM Tris-HCl pH=6.3, 0.2% Triton-X100, freshly supplemented with 10 mM β-mercaptoethanol and 5 mM imidazole). Precipitates were eluted by Elution buffer (0.5 M imidazole, 0.125 M DTT). All purified proteins were dialyzed against BC100 buffer before applying to subsequent pull-down assay. After pull-down assay, the interaction between SET and each p53-conjugate was detected by western blot with anti-p53 (DO-1) antibody.
Mass Spectrometry Assay
The protein complex was separated by SDS-PAGE and stained with GelCode Blue reagent (Pierce, 24592). The visible band was cut and digested with trypsin and then subjected to liquid chromatography (LC) MS/MS analysis.
Luciferase Assay
A firefly reporter (p21-Luci reporter) and a Renilla control reporter were co-transfected with indicated expressing constructs into H1299 cells for 48 hours and the relative luciferase activity was measured by dual-luciferase assay protocol (Promega, E1910).
Electrophoretic Mobility Shift Assay (EMSA)
Highly purified p53 or SET was incubated with a 32p-labelled probe (160 bp) containing p53-binding element of p21 promoter in 1× binding buffer (10 mM Hepes, pH 7.6, 40 mM NaCl, 50 μM EDTA, 6.25% Glycerol, 1 mM MgCl2, 1 mM Spermidine, 1 mM DTT, 50 ng/μl BSA, 5 ng/μl sheared single strand salmon DNA) for 20 minutes at room temperature (RT). For super-shift assay, α-p53 or α-SET antibody was pre-incubated with purified p53 or SET in the reaction system without probe for 30 minutes at RT and then added probe for further 20 minutes incubation. The complex was analyzed by 4% TBE-PAGE and visualized by autoradiography. The probe was obtained by PCR, labelled by T4 kinase (NEB, M0201S) and purified by Bio-Spin column (Bio-Rad, 732-6223).
Chromatin Immunoprecipitation (ChIP) Assay
Cells were fixed by 1% formaldehyde for 10 minutes at RT and lysed with ChIP Lysis Buffer (50 mM Tris-HCl pH 8.0, 5 mM EDTA, 1% SDS, 1× protease inhibitor) for 10 minutes at 4 °C. After sonication, the lysates were centrifuged, and the supernatants were collected and pre-cleaned in Dilution Buffer (20 mM Tris-HCl pH 8.0, 2 mM EDTA, 150 mM NaCl, 1% Triton X-100, 1× protease inhibitor) by salmon sperm DNA saturated protein A agarose (Millipore, 16-157) for 1 hour at 4 °C. The pre-cleaned lysates were aliquot equally and incubated with indicated antibodies overnight at 4 °C. Saturated Protein A agarose was added into each sample and incubated for 2 h at 4 °C. The agarose was washed with TSE I (20 mM Tris-HCl pH 8.0, 2 mM EDTA, 150 mM NaCl, 0.1% SDS, 1% Triton X-100), TSE II (20 mM Tris-HCl pH 8.0, 2 mM EDTA, 500 mM NaCl, 0.1% SDS, 1% Triton X-100), Buffer III (10 mM Tris-HCl pH 8.0, 1 mM EDTA, 0.25 M LiCl, 1% DOC, 1% NP40), and Buffer TE (10 mM Tris-HCl pH 8.0, 1 mM EDTA), sequentially. The binding components were eluted (1% SDS, 0.1 M NaHCO3) and performed reverse cross-link at 65 °C for at least 6 hours. DNA was extracted by PCR purification Kit (Qiagen, 28106). Real-time PCR was performed to detect relative enrichment of each protein or modification on indicated genes.
Cell Growth Assay
Approximate 1×105 cells were seeded into 6-well plate with three replicates. The cell growth was monitored in consecutive days, as indicated, by using Countess™ automated cell counter (Invitrogen) or by staining with 0.1% crystal violet. For quantitative analysis of the crystal violet staining, the crystal violet was extracted from cells by 10% acetic acid and the relative cell number was measured by detecting the absorbance at 590 nm.
Xenograft Model
1×106 cells were mixed with Matrigel (Corning, 354248) as 1:1 ratio for total 200 ul volume. The cell-matrix complex was subcutaneously injected into the nude mice (NU/NU; 8-weeks old; female; strain 088; Charles River). After 3 weeks, the mice were sacrificed and the tumor weight was measured. The experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Columbia University. None of the experiments were limit exceeded for tumor burden (10% total bodyweight or 2 cm in diameter).
RT-qPCR
Total RNA was extracted by TRIzol (Invitrogen, 15596-026) and precipitated by ethanol. 1 μg of total RNA was reversed into cDNA by SuperScript® III First-Strand Synthesis SuperMix (Invitrogen, 11752-50). The relative expression of each target was measured by qPCR and the data were normalized by the relative expression of GAPDH or β-Actin.
Immunohistochemistry (IHC)
FFPE sections of mouse brain tissue samples were stained with indicated antibodies and visualized by DAB exposure.
Protein Purification
The Flag tagged p53 or SET expressing construct was transfected into H1299 cells for 48 hours and the cells were lysed with Flag lysis buffer. After centrifuge, the Flag M2 Affinity Gel was added into supernatant and incubated 1 hour at 4 °C. After intensively washing by Flag Lysis Buffer for six times, the purified proteins were eluted with Flag peptide. For purification of acetylated p53, expressing construct CBP was co-transfected with p53 vector for 48 hours. TSA and Nicotinamide were added into the medium for the last 6 hours and the cells were harvested with Flag Lysis Buffer supplemented with TSA and Nicotinamide. The C-terminal unacetylated p53 was removed by p53-PAb421 antibody and then the acetylated p53 was purified as described above.
In vitro Acetylation Assay
0.5 μg recombinant H3 was incubated with 20 ng purified p300 in 1×HAT buffer (50 mM Tris-HCl, pH 7.9; 1 mM DTT; 10 mM sodium butyrate, 10% glycerol) containing 0.1 mM Ac-CoA for 30 min at 30 °C. After reaction, the products were assayed by western blot with indicated antibodies. To measure the effect of SET on p300-mediated H3 acetylation, pre-incubate H3 and purified SET (1 μg) in 1×HAT buffer for 20 min at RT before adding other components for subsequent in vitro acetylation assay.
Generation of p53 Knockout (p53-KO) Cell Line by CRISPR/Cas9 Technique
Cells were transfected with constructs expressing Cas9-D10A (Nickase) and control sgRNAs or sgRNAs targeting p53 exon3 (Santa Cruz: sc-437281 for control; sc-416469-NIC for targeting p53). After 48 hours of transfection, cells were suspended, diluted and re-seeded to make sure single clone formation. More than 30 clones were picked up and the expression of p53 in each single clone was evaluated by western blot with both α-p53 (DO-1) and α-p53 (FL-393) antibodies. Further verification of the positive clones was done by sequencing the genomic DNA to make sure that the functional genomic editing happened (insertion or deletion-mediated frame-shift of p53 open reading frame (ORF)). Two (U2OS) or three (HCT116) clones were finally selected for subsequent experiments. The p53 knockout-mediated effect was verified to be reproducible in these independent clones. The targeting sequences of p53 loci for the sgRNAs were: 1) TTGCCGTCCCAAGCAATGGA; 2) CCCCGGACGATATTGAACAA.
RNA-Seq
U2OS (CRISPR Ctr or CRISPR p53-KO) cells were transfected with control siRNA or SET-specific siRNA (three oligos) for 4 days. Each sample group has at least two biological replicates. Total RNA was prepared by TRIzol reagent (Invitrogen, 15596-026). The RNA quality was evaluated by Bioanalyzer (Agilent) and confirmed that the RIN > 8. Before performing RNA-seq analysis, a small aliquot of each sample was subjected to RT-qPCR analysis to confirm SET knockdown efficiency. RNA-seq analysis was performed at Columbia Genome Center. Specifically, from total RNA samples, mRNAs were enriched by poly-A pull-down and then preceded for library preparation by using Illumina TruSeq RNA prep kit. Libraries were then sequenced using Illumina HiSeq2000. Samples were multiplexed in each lane and yielded targeted number of single-end 100bp reads for each sample. RTA (Illumina) was used for base calling and bcl2fastq (version 1.8.4) was used for converting BCL to fastq format, coupled with adaptor trimming. Reads were mapped to a reference genome (Human: NCBI/build37.2) using Tophat (version 2.0.4). Relative abundance of genes and splice isoforms were determined using cufflinks (version 2.0.2) with default settings. Differentially expressed genes were tested under various conditions using DEseq, an R package based on a negative binomial distribution that models the number reads from RNA-seq experiments and test for differential expression. To further analyze the differentially expressed genes in a more reliable interval, the following filter strategies were applied: 1) the average of FPKM in either sample group > 0.1; 2) the fold change between CRISPR Ctr/si-Ctr group and CRISPR Ctr/si-SET group >2; 3) p value between CRISPR Ctr/si-Ctr group and CRISPR Ctr/si-SET group <0.01.
To retrieve potentially known p53 target genes which were repressed by SET in a p53-dependent manner, we searched the filtered RNA-Seq result by following strategies: 1) the expression level in CRISPR Ctr/si-SET group was at least 2 fold higher than that in CRISPR Ctr/si-Ctr group; 2) the expression level in CRISPR Ctr/si-SET group was at least 2 fold higher than that in CRISPR p53-KO/si-SET group. The filtered genes which were also clearly verified as p53 target genes by literatures were collected and presented as Heatmap.
Bioinformatic Analysis
For Discovery of Acidic domains in the Human Proteome: Our motif finding algorithm initially searches for sequence motifs with a minimum acidic composition of 76% using a sliding window of 36 residues, as dictated by experimental results. Motifs found to be partially overlapping were merged into single motifs. Lastly, flanking non-acidic residues were cropped-out from the final motif. Motif discovery was carried out using the UniProt database, which contains 20,187 canonical human proteins manually annotated and reviewed. For prediction of proteins binding Acidic domain-containing proteins and regulated by acetylation: We identified proteins that can potentially bind long acidic domains in a similar way to p53: using a K-rich region whose binding properties can be regulated by acetylation. We used the training set assembled in SSPKA, which combines lysine acetylation annotations from multiple resources obtained either experimentally or in the scientific literature. This dataset lists all annotated acetylation sites for a given protein individually. We generated acetylation motifs with multiple acetylation sites by clustering those sites found to within a maximum distance of 11 residues in sequence. Following this, we searched for acetylation motifs with five or more lysines where at least three of them are annotated as acetylation sites.
Statistical Analysis
Results were shown as the means ± s.d.. Difference was determined by using a two-tailed, unpaired Student t test in all figures except those described below. In Fig. 1g, difference was evaluated by one-way ANOVA with Bonferroni post hoc test. In Fig. 2d and g, Extended Data Fig. 2c, Extended Data Fig. 3b and d, Extended Data Fig. 4f and Extended Data Fig. 7h, difference was measured by two-way ANOVA with Bonferroni post hoc test. All statistical analysis was performed by using GraphPad Prism software. p < 0.05 was denoted as statistically significant.
Extended Data
Extended Data Figure 1. Further analysis of p53-SET interaction.
a, A list of SET peptides identified by mass spectrometry. b, In vitro binding assay of methylated p53 CTD and purified SET. c, d, e, In vitro binding assay between SET and purified ubiquitinated, sumoylated or neddylated form of p53. f, g, Western blot analysis of domains of p53 and SET for their interaction. In vitro binding assay was performed by incubating immobilized GST, GST-p53 or GST-SET with each purified SET or p53, as indicated. h, Western blot analysis of the interaction between p53 and SET in cells. H1299 cells were co-transfected with indicated expressing constructs and the nuclear extract was subjected to Co-IP assay. i, j, k, ChIP analysis of p53 or SET recruitment onto PUMA (i), TIGAR (j) or GLS2 (k) promoter. HCT116 cells were treated with or without 1 μM doxorubicin for 24 hours and then the cellular extracts were subjected to ChIP assay by indicated antibodies. Asterisks indicate the specific bands of indicated proteins. Error bars indicate mean ± s.d., n=3 for technical replicates. Data were shown as representative of three experiments. Uncropped blots were shown in Supplementary Fig. 1.
Extended Data Figure 2. RNA-seq analysis to identify genes regulated by p53-SET interplay.
a, Western blot analysis of the expression of p53 in U2OS-derived CRISPR control cells or CRISPR p53-KO cells. b, Heatmap of genes regulated by p53-SET interplay. U2OS (CRISPR Ctr or CRISPR p53-KO) cells were transfected with control siRNA or SET-specific siRNA for 4 days and the total RNA were prepared for RNA-seq analysis with two or three biological replicates, as indicated. Known p53 target genes which were also repressed by SET in a p53-dependent manner were selected and presented as a Heatmap. The relative SET expression was shown in the last row of the Heatmap. c, qPCR validation of the genes regulated by p53-SET interplay. Error bars indicate mean ± s.d., n=3 for technical replicates. Data were shown as representative of three experiments. Uncropped blots were shown in Supplementary Fig. 1.
Extended Data Figure 3. SET-mediated effects on cell proliferation and tumor growth.
a, b, Representative image (a) or quantitative analysis (b) of SET knockdown-mediated effect on cell growth of U2OS-derived CRISPR control cells or CRISPR p53-KO cells. c, Western blot analysis of the expression of p53 in HCT116-derived CRISPR control cells or CRISPR p53-KO cells. d, Xenograft analysis of SET-mediated effect on tumor growth by HCT116-derived CRISPR control cells or CRISPR p53-KO cells. e, Western blot analysis of p53 expression in control or derived HCT116 cell lines, as indicated. Error bars indicate mean ± s.d., n=3 in (b) or n=5 in (d) for biological replicates. Uncropped blots were shown in Supplementary Fig. 1.
Extended Data Figure 4. SET regulates histone modifications on p53 target promoter.
a, Western blot analysis of SET knockdown-mediated effect on p53 C-terminal acetylation in HCT116 cells. Doxorubicin (Dox)-treated cells were also analyzed in parallel as a positive control. b, Western blot analysis of SET-mediated effect on CBP-induced p53 C-terminal acetylation in H1299 cells. c, e, ChIP analysis of promoter-recruitment of p53 (c) or p300/CBP (e) upon SET depletion in HCT116 cells. d, ChIP analysis of SET knockdown-mediated effect on histone modifications on PUMA promoter in HCT116 cells. f, ChIP analysis of SET-mediated effect on p53-dependent H3K18 and H3K27 acetylation on PUMA promoter. Error bars indicate mean ± s.d., n=3 for technical replicates. Data were shown as representative of three experiments. Uncropped blots were shown in Supplementary Fig. 1.
Extended Data Figure 5. Acetylation regulates the interaction between acidic-domain-containing proteins and their acetylatable ligands.
a, A summary table of characteristic features of acidic domain-containing protein SET, VPRBP, DAXX and PELP1. The acidic amino acids were underlined. b, In vitro binding assay of p53 CTD and purified full-length of VPRBP, DAXX or PELP1. c, d, e, Western blot analysis of the interaction between p53 and VPRBP (c), DAXX (d) or PELP1 (e) in nuclear fraction of H1299 cells. f, g, h, In vitro binding assay between purified SET and lysine-rich domain of H3 (f), KU70 (g) or FOXO1 (h). i, In vitro binding assay of H3 lysine-rich domain and purified VPRBP, DAXX or PELP1. j, In vitro binding assay of H3 lysine-rich domain and BRD4 or BRD7 (nuclear extract). Uncropped blots were shown in Supplementary Fig. 1.
Extended Data Figure 6. p53KQ mutant mimics acetylated p53.
a, Schematic diagraph of human unacetylated p53, acetylation-deficient or acetylation-mimicking mutant of p53. b, In vitro binding assay of SET and different types of p53, as indicated. c, d, e, Western blot analysis of the interaction between acidic domain-containing proteins (c, VPRBP; d, DAXX; e, PELP1) and different types of p53 in cells. H1299 cells were co-transfected with indicated expressing constructs, and the nuclear extract was subjected to Co-IP assay. Asterisks indicate the purified proteins. Uncropped blots were shown in Supplementary Fig. 1.
Extended Data Figure 7. Generation of the p53KQ/KQ mice.
a, Schematic diagram of gene targeting strategy to replace p53 C-terminal 7 lysines with 7 glutamine in mouse p53. b, Southern blot screening of ES cells to identify p53+/KQ clones. c, PCR genotyping analysis of wildtype mouse (110 bps), p53+/KQ heterozygous mouse (110 bps and 150 bps), and p53KQ/KQ homozygous mouse (150 bps only). d, Sequencing analysis of the transcripts prepared from p53+/KQ heterozygous mouse spleen. e, A summary table of observed numbers from p53+/KQ heterozygous intercrosses. f, Positive control of p53 staining in IHC assay. The spleen tissue sections of p53+/+ mice treated with or without 6 Gy γ-radiation was stained with p53 (CM-5) antibody. g, h, Representative image (g) or quantitative analysis (h) of SET knockdown-mediated cell growth of p53+/+ or p53KQ/KQ MEFs (P2). Error bars indicate mean ± s.d., n=3 for biological replicates. Uncropped blots were shown in Supplementary Fig. 1.
Extended Data Figure 8. Characterization of Set conditional knockout mice.
a, Schematic diagraph of strategy to generate Set conditional knockout mice. b, Validation of Set knockout in embryos (E8.5) by genotyping and western blot analysis. c, A summary table of observed numbers from Set+/− intercrosses. d, Representative picture of Set+/+ and Set−/− embryos (E10.5). e, qPCR analysis of the expression of p53 target genes in Set+/+ and Set−/− embryos (E10.5). Error bars indicate mean ± s.d., n=3 for technical replicates. Data were shown as representative of three experiments. Uncropped blots were shown in Supplementary Fig. 1.
Extended Data Table 1. A list of human proteins containing acidic domain with a minimum percentage of acidic residues of 76% within a 36 residues-long window.
Proteins are clustered into different categories depending on the biological process they are involved. Each protein is described by its UniProt accession code (1st column), protein name (2nd column) and a list of GO terms (5th column). The corresponding acidic domains are described by their position in sequence (3rd column) and their sequence (4th column).
| UniProtID | Protein Name | Acidic Domain Position | Acidic Domain Sequence | Biological Function (GO) | |
|---|---|---|---|---|---|
| Proteins Involved in Gene Expression Control through DNA Binding, Transcription Regulation and Chromatin Remodeling |
Q8IZL8 Q92688 Q9UL68 Q01538 A1YPR0 Q86V15 Q01105 PODMEO Q7Z6M4 Q6PL18 Q9BTT0 Q7Z6Z7 Q12873-3 Q96KQ7 Q8IX15 Q8WYB5 P19338 Q5H9L4 Q13029 P27797 Q9UER7 Q4LE39 Q9UPS6-2 P39687 P09429 Q9BT43 P17480 Q15911 Q9UK99 Q9Y4B6 |
Proline-, glutamic acid- and
leucine-rich protein 1 Acidic leucine-rich nuclear phosphoprotein 32 family member B Myelin transcription factor 1-like protein Myelin transcription factor 1 Zinc finger and BTB domain-containing protein 7C Zinc finger protein castor Protein SET Protein SETSIP Transcription termination factor 4, mitochondrial ATPase family AAA domain-containing protein 2 Acidic leucine-rich nuclear phosphoprotein 32 family member E E3 ubiquitin-protein ligase HUWE1 Isoform 3 of Chromodomain-helicase-DNA-binding protein 3 Histone-lysine N-methyltransferase EHMT2 Homeobox and leucine zipper protein Homez Histone acetyltransferase KAT6B Nucleolin Transcription initiation factor TFIID subunit 7-like PR domain zinc finger protein 2 Calreticulin DAXX_HUMAN Death domain-associated protein 6 AT-rich interactive domain-containing protein 4B SET1B_HUMAN Isoform 2 of Histone-lysine N-methyltransferase SETD1B AN32A_HUMAN Acidic leucine-rich nuclear phosphoprotein 32 family member A High mobility group protein B1 DNA-directed RNA polymerase III subunit RPC7-like UBF1_HUMAN Nucleolar transcription factor 1 Zinc finger homeobox protein 3 F-box only protein 3 Protein VPRBP |
886 - 963 156 - 232 107 - 169 257 - 315 124 - 178 1671 - 1724 236 - 289 248 - 301 332 - 380 242 - 288 158 - 203 2425 - 2469 6-48 289 - 331 507 - 549 1062 - 1103 233 - 274 326 - 367 261 - 301 368 - 407 433 - 471 528 - 566 1042 - 1079 164 - 201 178 - 214 157 - 192 710 - 745 453 - 488 417 - 451 1395 - 1429 |
DEEEEEEEEEEEEEEEEEEEEEDFEEEEEDEEEYFEEEE EEEEEFEEEFEEEEGELEEEEEEEDEEEEEELEEVEDLE DSDAEVDGVDEEEEDEEGEDEEDEDDEDGEEEEFDEEDD EDEDVEGDEDDDEVSEEEEEFGLDEEDEDEDEDEEEEE EEYSEDNDEPGDEDEEDEEGDREEEEEIEEEDEDDDEDG EDVEDEEEEEEEEEEEEEEEENED EEEDEEEEEEEEEEEEDEEEEEEEEEEEEEEEEEEEEEE EEEEEEEEEEAAPDVIFQED EIMEPGGDGGEEDDKEDDDDDEDDDDEEDEEEEEEEEED DDDDTEDFADQENLPD ESSQEDEEEELELPEEEAEDDEDEDDDEDDDDEDDDEDD DDEDLRTDSEESLPE DMDDEEGEGEEDDDDDEEEEGLEDIDEEGDEDEGEEDED DDEGEEGEEDEGEDD DDEEGGEDDDDDDDDGDEGEEELEDIDEGDEDEGEEDED DDEGEEGEEDEGEDD DDKRASLDEDEDDDDEEDNDEDDNDEDDDDEDDDEAEDN DEDEDDDEEE ESSEEGEDQEHEDDGEDEDDEDDDDDDDDDDDDDDEDDE DEEDGEEE EEEDDEDGDEDDEEEEENEAGPPEGYEEEEEEEEEEDED EDEDEDE EDEDDSQDEEEEEEEDEEDDQEDDEGEEGDEDDDDDGSE MELDED DEEEEEEEMVVSEEEEEEEEEGDEEEEEEVEAADEDDEE DDDE EVEALTEQLSEEEEEEEEEEEEEEEEEEEEEEEEDEESG NQSD EVWCLDEEEEEEEEELPEDDEEEEEEEEEDDDDDDDDV IIQD ELSKESSEEEEEEEDEEEEEEEEEEEEDEEEEEEEEEEE EEE VKE DSRSNNDDDEDEDDEDEDEDEDEDEDEDKEEEEEDCSEE YLE EVHDLGEEEEEEEEEDEEEEEDDDDDELEDEGEEEASMP NE EEEEDKKRKEEEEAEDKEDDEDKDEDEEDEEDKEEDEEE D ETDDEDDEESDEEEEEEEEEEEEEATDSEEEEDLEQMQE DETNKEEDEDDEEAEEEEEEEEEEEDEDDDDNNEEEEFE EEQESTEEEEEAEEEEEEEDDDDDDSDDRDESENDDED EGLDDEEEDEDEEEYDEDAQVVEDEEDEDEEEEGEEED EKSKKKKEEEEDEEDEEDEEEEEDEEDEDEEEDDDDE EEEVTSEEDEEKEEEEEKEEEEEEEYDEEEHEEETD ESSSEDESEDGDENEEDDEDEDDDEDDDEDEDNESE EKVEPAEEEAEEEEEEEEAEEEEEEEEEEEEEEEDE DEYEEMEEEEEEEEEEDEDDDSADMDE5DEDDEEE EDEDEEEDQEEEEQEEEDDDEDDDDTDDLDELDTD |
Chromatin binding, Transcription factor
binding, poly(A) RNA binding Protein binding, Histone binding, RNA polymerase binding Sequence-specific DNA binding, Transcription factor, Zinc binding Sequence-specific transcription factor, Zinc binding Nucleic acid binding, Metal ion binding DNA binding, Metal ion binding DNA binding, Histone binding, Phosphatase inhibitor Chromatin binding DNA binding, RNA binding, Protein binding Histone binding, Chromatin binding, Hydrolase Histone binding, Phophatase inhibitor activity DNA binding, ligase activity, poly(A) RNA binding DNA binding, Helicase activity, poly(A) RNA binding Histone methyltransferase, p53 binding, C2H2 Zinc finger domain DNA binding, Sequence-specific transcription factor, Transcription co-repressor DNA binding, Histone acyltransferase, Transcription factor binding DNA binding, RNA binding, Protein binding Transcription coactivator, Transcription factor binding, Histone acyltransferase binding DNA binding, sequence-specific DNA binding transcription factor, Zinc binding, histone-lysine N-methyltransferase androgen receptor binding, carbohydrate binding, complement component C1q binding Androgen receptor binding, Heat shock protein binding, Histone binding DNA binding, Protein binding, Transcription regulatory region DNA binding Histone-lysine N-methyltransferase, Nuycleotide binding, RNA binding Gene expression, Intracellular signal transduction, nucleocytoplasmic transport DNA binding, Protein binding, Transcription factor binding Gene expression, Innate immune response, RNA polymerase III activity poly(A) RNA binding, RNA pol I CORE element seq-specific DNA binding, RNA pol I upstream control element seq-specific DNA binding DNA binding, sequence-specific DNA binding transcription factor, Protein binding ubiquitin-protein transferase activity Histone kinase, Ser/Thr kinase, Protein binding |
| DNA-related (Replication, Repair) |
P07199 P20962 |
Major centromere autoantigen
B Parathymosin |
403 - 446 504 - 537 38 - 74 |
EGEEEEEEEEEEEEEEGEGEEEEEEGEEEEEEGGEGEEL GEEEE EGGEDSDSDSEEEDDEEEDDEDEDDDDDEEDGDE EEEENGAEEEEEETAEDGEEEDEGEEEDEEEEEEDDE |
Centromeric DNA binding, Chromatin
binding, DNA binding DNA replication, Immune system process |
| RNA-related (Processing, Translation) |
Q96MU7 O60841 P12270 Q6ZU64 Q9NW13 Q9UQ88 P21127 |
YTH domain-containing protein
1 Eukaryotic translation initiation factor 5B Nucleoprotein TPR Coiled-coil domain-containing protein 108 RNA-binding protein 28 Cyclin-dependent kinase 11A Cyclin-dependent kinase 11B |
198 - 264 528 - 566 1948 - 1983 1768 - 1803 223 - 257 291 - 323 303 - 335 |
ENEEEGVEEDVEEDEEVEEDAEEDEEVDEDGEEEEEEEE EEEEEEEEEEEEYEQDERDQKEEGNDYD ENPEEEEEEEEEEEEDEESEEEEEEEGESEGSEGDEEDE DDEEEDDDENDGEHEDYEEDEEDDDDDEDDTGMGDE EEEEEELEEEEEEEEETEEEELGKEEIEEKEEERDE EEEDMEEEENDDDDDDDDEEDGVFDDEDEEEENIE EEEEEEEEEEEEEGSTSEESEEEEEEEEEEEEE EEEEEEEEEEEEEGSTSEESEEEEEEEEEEEEE |
poly(A) RNA binding, RNA
binding GTPase activity, poly(A) RNA binding, GTP binding chromatin binding, heat shock protein binding, mRNA binding poly(A)RNA binding nucleotide binding, poly(A) RNA binding ATP binding, cyclin-dependent protein ser/thr kinase ATP binding, cyclin-dependent protein ser/thr kinase, poly(A) RNA binding |
| Other |
Q5TCY1 P46060 Q5JTC6 O60721 P21817 O43847 |
Tau-tubulin kinase 1 Ran GTPase-activating protein 1 APC membrane recruitment protein 1 Sodium/potassium/calcium exchanger 1 RYR1_HUMAN Ryanodine receptor 1 NRDC_HUMAN Nardilysin |
732 - 779 358 - 404 369 - 410 854 - 894 1872 - 1911 141 - 179 |
EEEEEEEEDEEEEEEDEEEEEEEEEEEEEEEEEEEEEEE AAAAVALGE DDEDEEEEEEGEEEEEEAEEEEEEDEEEEEEEEEEEEEE PQQRGQGE EEMALPDDDDEEEEEEEEVELEEEEEEVKEEEEDDDLEY LWE DGGDSEEEEEEEEEQEEEEEEEEQEEEEEEEEEEEEKGN EE EEEEEEDEEEEGEEEDEEEKEEDEEETAQEKEDEEKEEE E DDEEEEEVEEEEDDDEDSGAEIEDDDEEGFDDEDEFDDE |
ATP binding, protein serine/threonine
kinase activity GTPase activator activity beta-catenin binding, phosphatidylinositol-4,5-bisphosphate binding calcium, potassium:sodium antiporter activity, symporter activity Calcium ion channel, Calmoduling binding Epidermal growth factor binding, Metalloendopeptidase, Zinc ion binding |
| Function not clear |
Q86TY3 Q7L0X2 Q8TC90 P0C7V8 |
Uncharacterized protein
C14orf37 Glutamate-rich protein 6 Coiled-coil domain-containing glutamate-rich protein 1 DDB1 - and CUL4-associated factor 8-like protein 2 |
604 - 651 16 - 63 301 - 344 107 - 146 |
DQLESEEGQEDEDEEDEEDEDEEEEDEEEDEEDKDADSL DEGLDGDTE DQKESEEELEEEEEEEEVEEEEEEVEEEEEEVEEEEEEV VEEELVGEE EEEEEVEDEEEEVEDEEEEEVEEAEYVEEGEEELEEEEL EEEEE EEETEREEEDEEIQEEGGEEEEEEEEEEEEEEEEEEEEE E |
Membrane NA NA NA |
Extended Data Table 2. A List of human proteins containing lysine-rich domain with at least five lysines where three or more lysines are annotated as acetylation sites in the SSPKA database.
Each protein is described by its UniProt accession code and their protein name (1st and 2nd column, respectively). Acetylated motifs are described by the position of their annotated acetylation sites contained and their sequence (3rd and 4th column, respectively).
| UniProt ID | Protein Name | Acetylated Lysines | Sequence of Lysine-rich Domain | |
|---|---|---|---|---|
| Transcription Factor | 015525 | Transcription factor MafG | 53, 60, 71, 76 | EEIVQLKQRRRTLKNRGYAASCRVKRVTQKEELEKQ |
| P18146 | Early growth response protein 1 | 422, 424, 425 | KIHLRQKDKKADKSW | |
| P52630 | Signal transducer and activator of transcription 2 | 182, 184, 194, 197 | RYKIQAKGKTPSLDPHQTKEQKILQETL | |
| Q16236 | Nuclear factor erythroid 2-related factor 2 | 533, 536, 538, 541, 543, 548, 554, 555 | QDLDHLKDEKEKLLKEKGENDKSLHLLKKQLSTLY | |
| Q9Y2Y9 | Krueppel-like factor 13 | 166, 168, 180 | LESPQRKHKCHYAGCEKVYGKSSHLKA | |
| P04150 | Glucocorticoid receptor | 480, 492, 494, 495 | PACRYRKCLQAGMNLEARKTKKKIKGIQ | |
| P43694 | Transcription factor GATA-4 | 312, 319, 321, 323 | RPLAMRKEGIQTRKRKPKNLNKSK | |
| P06733* | Alpha-enolase | 60, 71, 80, 89 | KTRYMGKGVSKAVEHINKTIAPALVSKKLNVTEQEKIDKLMI | |
| P23769 | Endothelial transcription factor GATA-2 | 389, 390, 399, 403, 405, 406, 408, 409 | NRPLTMKKEGIQTRNRKMSNKSKKSKKGAECFE | |
| Transcriptional Regulation (Except Transcription Factor), Chromatin Remodeling | 060563 | Cyclin-T1 | 380, 386, 390 | SQKQNSKSVPSAKVSLKEYRAKH |
| P04406* | Glyceraldehyde-3-phosphate dehydrogenase | 251, 254, 259, 260 | LTCRLEKPAKYDDIKKWKQAS | |
| P06748* | Nucleophosmin | 141, 150, 154, 155 | LLSISGKRSAPGGGSKVPQKKVKLAAD | |
| 250, 257, 267, 273 | VEDIKAKMQASIEKGGSLPKVEAKFINYVKNCFRMT | |||
| P09874 | Poly [ADP-ribose] polymerase 1 | 498, 505, 508 | WAPRGKSGAALSKKSKGQVKEE | |
| P19338 | Nucleolin | 70, 79, 87 102, 109, 116, 124, 132 |
VWSPTKKVAVATPAKKAAVTPGKKAAATP KTVTPAKAVTTPGKKGATPGKALVATPGKKGAAIPAKGAKNGK |
|
| P51531 | Probable global transcription activator SNF2L2 | 996, 997, 999, 1003 1547, 1551, 1553, 1555, 1556 |
DGSEKDKKGKGGAKTLMNTI LNKKDDKGRDKGKGKKRPNRGK |
|
| Q00987 | E3 ubiquitin-protein ligase Mdm2 | 466, 467, 469, 470 | ACFTCAKKLKKRNKPCP | |
| Q13547 | Histone deacetylase 1 | 432, 438, 439, 441 | EGEGGRKNSSNFKKAKRVKTED | |
| Q92793 | CREB-binding protein | 1797, 1806, 1809 1583, 1586, 1587, 1588, 1591, 1592, 1595, 1597 |
SLPSCQKMKRWQHTKGCKRKTNGG G3QGDSKNAKKKNNKKTNKNKSSISRA |
|
| Q92831 | Histone acetyltransferase KAT2B | 416, 428, 430, 441, 442 | SSSPACKASSGLEANPGEKRKMTDSHVLEEAKKPRVMGD | |
| P27695* | DNA-(apurinic or apyrimidinic site) lyase | 24, 27, 31, 32, 35 | RTEPEAKKSKTAAKKNDKEAAGEG | |
| P62805 | Histone H4 | 6, 9, 13, 17, 21, 32 | MSGRGKGGKGLGKGGAKRHRKVLRDNIQGITKPAIRRL | |
| Q92922 | SWI/SNF complex subunit SMARCC1 | 345, 346, 354, 359 | SRKKSGKKGQASLYGKRRSQKEEDEQE | |
| P26358 | DNA (cytosine-5)-methyltransferase 1 | 1111, 1113, 1115, 1117, 1119, 1121 | SPGNKGKGKGKGKGKPKSQACEP | |
| Q13569 | G/T mismatch-specific thymine DNA glycosylase | 83, 84, 87 | KKPVESKKSGKSAKSKE | |
| Q8TEK3 | Histone-lysine N-methyltransferase, H3 lysine-79 specific | 397, 398, 401 | PSKARKKKLNKKGRKMA | |
| Q92841 | Probable ATP-dependent RNA helicase DDX17 | 108, 109, 121, 129 | GGGLPPKKFGNPGERLRKKKWDLSELPKFEKNEY | |
| P68431 | Histone H3.1 | 5, 10, 15, 19, 24, 28, 37, 38 | MARTKQTARKSTGGKAPRKQLATKAARKSAPATGGVKKPHRYRP | |
| Q92522 | Histone H1x | 179, 182, 185 | KKGAGAKKDKGGKAKKTAA | |
| P46100 | Transcriptional regulator ATRX | 1933, 1935, 1936, 1939 | YTKKKKKGKKGKKDSSSSG | |
| Q6DN03 | Putative histone H2B type 2-C | 13, 16, 17, 21, 24 | FAPAPKKGSKKAVTKAQKKDGKKR | |
| P05114 | Non-histone chromosomal protein HMG-14 | 3, 5, 14, 18, 27, 31, 38, 42, 48, 53, 55, 59, 61 | MPKRKVSSAEGAAKEEPKRRSARLSAKPPAKVEAKPKKAAAKDKSSDKKVQTKGKRGAK GKQAEVAN |
|
| DNA Repair and Integrity | P12956 | X-ray repair cross-complementing protein 6 | 539, 542, 544, 553, 556 | DYNPEGKVTKRKHDNEGSGSKRPKVEYSEE |
| Q9UQE7 | Structural maintenance of chromosomes protein 3 | 105, 106, 113, 114 | RRVIGAKKDQYFLDKKMVTKND | |
| P27695* | DNA-(apurinic or apyrimidinic site) lyase | 24, 27, 31, 32, 35 | RTEPEAKKSKTAAKKNDKEAAGEG | |
| Other DNA Related Function | 094761 | ATP-dependent DNA helicase Q4 | 376, 380, 382, 385, 386 | RSRLLRKQAWKQKWRKKGECFGG |
| Ribosome Biogenesis | P06748* | Nucleophosmin | 141, 150, 154, 155 250, 257, 267, 273 |
LLSISGKRSAPGGGSKVPQKKVKLAAD VEDIKAKMQASIEKGGSLPKVEAKFINYVKNCFRMT |
| Specific Molecular/Biological Function Uncertain | P81534 | Beta-defensin 103 | 48, 54, 61, 66, 67 | VLSCLPKEEQIGKCSTRGRKCCRRKK |
| Q3BBV0 | Neuroblastoma breakpoint family member 1 | 1101, 1103, 1105, 1106 | VGEIEKKGKGKKRRGRRS | |
| Q8N7X0 | Androglobin | 337, 340, 343 | KDGKEVKDVKEFKPESSLT | |
| Q6ZQR2 | Uncharacterized protein C9orf171 | 237, 240, 246 | EQKATQKAIKLEKKQKWLGKL | |
| Others | P04406* | Glyceraldehyde-3-phosphate dehydrogenase | 251, 254, 259, 260 | LTCRLEKPAKYDDIKKWKQAS |
| P09622 | Dihydrolipoyl dehydrogenase, mitochondrial | 267, 271, 273, 277 | FQRILQKQGFKFKLNTKVTGATK | |
| P40939 | Trifunctional enzyme subunit alpha, mitochondrial | 350, 353, 359 | HGQVLCKKNKFGAPQKDVKHLA | |
| Q9NP61 | ADP-ribosylation factor GTPase-activating protein 3 | 223, 228, 229 | KPNQAKKGLGAKKGSLGAQ | |
| Q9Y6F6 | Protein MRVI1 | 398, 402, 405 | EKRFAGKAGGKLAKAPGLKD | |
| 205, 214, 223, 229, 236 | AACLLPKLDE LRDEGKASSAKQRLKCASLQKFGERAFKAWAVAR | |||
| P02768 | Serum albumin | 543, 548, 560, 565, 569, 581, 584, 588, 597, 598 | ICTLSEKERQIKKQTALVELVKHKPKATKEQLKAVMDDFAAFVEKCCKADDKETCFAEE GKKLVAASQ |
|
| P62328 | Thymosin beta-4 | 4, 12, 15 | MSDKPDMAEIEKFDKSKLKKT | |
| Q13576 | Ras GTPase-activating-like protein IQGAP2 | 1467, 1471, 1474 | SIKLDGKGEPKGAKRAKPVK | |
| Q15283 | Ras GTPase-activating protein 2 | 208, 209, 211 | PSRNDQKKTKVKKKTS | |
| Q99075 | Proheparin-binding EGF-like growth factor | 96, 97, 99, 104 | EHGKRKKKGKGLGKKRDPCLR | |
| P06733* | Alpha-enolase | 60, 71, 80, 89 | KTRYMGKGVSKAVEHINKTLAPALVSKKLNVTEQEKIDKLMI | |
| P15692 | Vascular endothelial growth factor A | 142, 147, 149, 152 | RARQEKKSVRGKGKGQKRKRKKS | |
| P10636 | Microtubule-associated protein tau | 571, 574, 576, 584, 591, 597, 598, 607, 615 | VPMPDLKNVKSKIGSTENLKHQPGGGKVQIINKKLDLSNVQSKCGSKDNIKHVPGGG |
Supplementary Material
Acknowledgements
We thank Dr. F Giancotti, Dr. X Yang, Dr. R K Vadlamudi and Dr. W An to provide the reagents for this work. We also thank Dr. Richard Baer for discussion and suggestions. This work was supported by the National Cancer Institute of the National Institutes of Health under Award 5R01CA193890, 5RO1CA190477, 5RO1CA085533 and 2P01CA080058 to W.G. and GM030518 and CA121852 to B.H. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Author Contributions The experiments were conceived and designed by D.W., N.K., G.L. and W.G.. The experiments were performed mainly by D.W. and N.K.. Bioinformatic analysis was performed by G.L.. Mass spectrometry analysis was performed by W.L. Xenograft assay was performed by D.W. and L.J.. Data were analyzed and interpreted by D.W., N.K., G.L., W-G.Z., J.Q., B.H. and W.G.. The manuscript was written by D.W., N.K., G.L. and W.G..
Author Information RNA-seq data is available through NCBI Gene Expression Omnibus (GEO) database with the access number GSE83635.
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
Online Content Methods, Extended Data display items and Source Data are available in the online version of the paper.
Supplementary Information is available in the online version of the paper.
References
- 1.Zhao S, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. doi:10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. doi:10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 3.Kim SC, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Molecular cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. doi:10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 4.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 5.Dhalluin C, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. doi:10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 6.Marmorstein R, Zhou MM. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harbor perspectives in biology. 2014;6:a018762. doi: 10.1101/cshperspect.a018762. doi:10.1101/cshperspect.a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim K, et al. Vpr-binding protein antagonizes p53-mediated transcription via direct interaction with H3 tail. Molecular and cellular biology. 2012;32:783–796. doi: 10.1128/MCB.06037-11. doi:10.1128/MCB.06037-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao LY, et al. Negative regulation of p53 functions by Daxx and the involvement of MDM2. The Journal of biological chemistry. 2004;279:50566–50579. doi: 10.1074/jbc.M406743200. doi:10.1074/jbc.M406743200. [DOI] [PubMed] [Google Scholar]
- 9.Nair BC, et al. Proline, glutamic acid and leucine-rich protein-1 is essential for optimal p53-mediated DNA damage response. Cell death and differentiation. 2014;21:1409–1418. doi: 10.1038/cdd.2014.55. doi:10.1038/cdd.2014.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li T, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–1283. doi: 10.1016/j.cell.2012.04.026. doi:10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang L, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. doi:10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simeonova I, et al. Mutant mice lacking the p53 C-terminal domain model telomere syndromes. Cell reports. 2013;3:2046–2058. doi: 10.1016/j.celrep.2013.05.028. doi:10.1016/j.celrep.2013.05.028. [DOI] [PubMed] [Google Scholar]
- 13.Hamard PJ, et al. The C terminus of p53 regulates gene expression by multiple mechanisms in a target- and tissue-specific manner in vivo. Genes & development. 2013;27:1868–1885. doi: 10.1101/gad.224386.113. doi:10.1101/gad.224386.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Lindern M, et al. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′ half to different genes: characterization of the set gene. Molecular and cellular biology. 1992;12:3346–3355. doi: 10.1128/mcb.12.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JY, et al. Inhibition of p53 acetylation by INHAT subunit SET/TAF-Ibeta represses p53 activity. Nucleic acids research. 2012;40:75–87. doi: 10.1093/nar/gkr614. doi:10.1093/nar/gkr614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Z, et al. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell. 2013;154:297–310. doi: 10.1016/j.cell.2013.06.027. doi:10.1016/j.cell.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin Q, et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. The EMBO journal. 2011;30:249–262. doi: 10.1038/emboj.2010.318. doi:10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. doi:10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. doi:10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 20.Berger SL. Keeping p53 in check: a high-stakes balancing act. Cell. 2010;142:17–19. doi: 10.1016/j.cell.2010.06.026. doi:10.1016/j.cell.2010.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsumoto K, Nagata K, Okuwaki M, Tsujimoto M. Histone- and chromatin-binding activity of template activating factor-I. FEBS letters. 1999;463:285–288. doi: 10.1016/s0014-5793(99)01632-4. [DOI] [PubMed] [Google Scholar]
- 22.Kim KB, et al. Inhibition of Ku70 acetylation by INHAT subunit SET/TAF-Ibeta regulates Ku70-mediated DNA damage response. Cellular and molecular life sciences : CMLS. 2014;71:2731–2745. doi: 10.1007/s00018-013-1525-8. doi:10.1007/s00018-013-1525-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chae YC, et al. Inhibition of FoxO1 acetylation by INHAT subunit SET/TAF-Ibeta induces p21 transcription. FEBS letters. 2014;588:2867–2873. doi: 10.1016/j.febslet.2014.06.053. doi:10.1016/j.febslet.2014.06.053. [DOI] [PubMed] [Google Scholar]
- 24.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. doi:10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Cohen HY, et al. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Molecular cell. 2004;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 26.Daitoku H, et al. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. doi:10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krummel KA, Lee CJ, Toledo F, Wahl GM. The C-terminal lysines fine-tune P53 stress responses in a mouse model but are not required for stability control or transactivation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10188–10193. doi: 10.1073/pnas.0503068102. doi:10.1073/pnas.0503068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng L, Lin T, Uranishi H, Gu W, Xu Y. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Molecular and cellular biology. 2005;25:5389–5395. doi: 10.1128/MCB.25.13.5389-5395.2005. doi:10.1128/MCB.25.13.5389-5395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.UniProt C. UniProt: a hub for protein information. Nucleic acids research. 2015;43:D204–212. doi: 10.1093/nar/gku989. doi:10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, et al. Accurate in silico identification of species-specific acetylation sites by integrating protein sequence-derived and functional features. Scientific reports. 2014;4:5765. doi: 10.1038/srep05765. doi:10.1038/srep05765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.