Abstract
We report the main characteristics of ‘Bacillus dakarensis’ P3515T sp. nov., ‘Bacillus sinesaloumensis’ P3516T sp. nov., ‘Gracilibacillus timonensis’ P2481T sp. nov., ‘Halobacillus massiliensis’ P3554T sp. nov., ‘Lentibacillus massiliensis’ P3089T sp. nov., ‘Oceanobacillus senegalensis’ P3587T sp. nov., ‘Oceanobacillus timonensis’ P3532T sp. nov., ‘Virgibacillus dakarensis’ P3469T sp. nov. and ‘Virgibacillus marseillensis’ P3610T sp. nov., that were isolated in 2016 from salty stool samples (≥1.7% NaCl) from healthy Senegalese living at Dielmo and N’diop, two villages in Senegal.
Keywords: Culturomics, halophilic species, human gut microbiota, new species, taxonogenomics
Culturomics has allowed to culture 247 new bacterial species, greatly increasing our understanding of the human gut repertoire thanks to high-throughput culture conditions with a rapid identification method of grown colonies using matrix-assisted desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) [1]. As a part of the culturomics approach, we isolated in 2016 from stool samples from healthy patients from Senegal nine bacteria that could not be identified by our MALDI-TOF MS screening on a Microflex spectrometer (Bruker Daltonics, Bremen, Germany) [2]. The study was approved by the ethics committee of the Institut Hospitalo-Universitaire Méditerranée Infection under number 2016-011, and all patients provided written informed consent.
The salinity of the stool specimens was measured by a salinity refractometer (Thermo Scientific, Villebon-sur-Yvette, France). One gram of each stool specimen was diluted in 10 mL of distilled water and centrifuged for 10 minutes at 5000g. Then 100 μL of supernatant was deposited in the refractometer, and the result were directly displayed on the screen (in ‰) and then reported (in % NaCl).
Stool samples were inoculated in an aerobic blood culture bottles (Becton Dickinson, Le Pont-de-Claix, France) including an halophilic medium prepared in modifying a Columbia broth medium (Sigma-Aldrich, Saint-Quentin-Fallavier, France) by adding (per litre: 1% (w/v) MgSO4, 0.1% (w/v) MgCl2, 0.4% (w/v) KCl, 0.1% (w/v) CaCl2, 0.05% (w/v) NaHCO3, 0.2% (w/v) of glucose, 0.5% (w/v) of yeast extract (Becton Dickinson), and from 10 to 15% (w/v) NaCl according to the required salinity with a pH adjusted to 7.5 and incubated for 3 days in aerobic atmosphere at 37°C. The amount of solutes per litre was given in weight volume (w/v) percentage concentration and calculated by the following formula: Concentration in % (w/v) = 100 × [(mass solute in g)/(volume solution in mL)].
The initial agar-grown colonies were obtained after 24 to 48 hours of incubation at 37°C. The 16S rRNA gene was sequenced using fD1-rP2 primers as previously described, using a 3130-XL sequencer (Applied Biosciences, Saint-Aubin, France). Because all strains exhibited a 16S rRNA sequence homology of <98.7% with its phylogenetically closest species, we propose the creation of nine new species according to the nomenclature [3].
Strain Marseille-P3515T was isolated in a stool sample (4% NaCl) from a 17-year-old adolescent boy living in N’diop. The strain was first isolated in a halophilic medium with 15% (w/v) NaCl. Growth occurred in culture media with NaCl concentration ranging from 0 to 15% (w/v) NaCl (optimum, 7.5% (w/v) NaCl). Agar-grown colonies are beige, circular and shiny with a mean diameter of 1.2 mm. Bacterial cells were Gram positive, rod shaped and polymorphic. Strain Marseille-P3515T was motile and spore forming with a positive catalase and oxidase reaction. Strain Marseille-P3515T exhibited a 97.96% sequence identity with Bacillus circulans strain NBRC 13626 (GenBank accession no. NR_112632 (Fig. 1). We propose to classify strain Marseille-P3515T as a new species of the genus Bacillus within the family Bacillaceae in the phylum Firmicutes. Strain Marseille-P3515T is the type strain of ‘Bacillus dakarensis’ (da.ka.ren′sis, N.L. masc. adj. dakarensis, the name of the capital of Senegal where the stool sample was collected).
Fig. 1.
Phylogenetic tree showing position of ‘Bacillus dakarensis’ Marseille-P3515T and ‘Bacillus sinesaloumensis’ Marseille-P3516T relative to other phylogenetically close neighbours. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using maximum-likelihood method within MEGA software. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 500 times to generate majority consensus tree. Only bootstrap scores of at least 75 were retained. Scale bar indicates 1% nucleotide sequence divergence.
Strain Marseille-P3516T was isolated in a stool sample (3.8% NaCl) from a 10-year-old girl from N’diop in the same halophilic medium as for strain Marseille-P3515T, but this strain could grow in media ranging from 0 to 10% (w/v) NaCl (optimum, 5% (w/v) NaCl). Colonies are beige, circular and shiny with a mean diameter of 1.2 mm. Bacterial cells were Gram positive, rod shaped and polymorphic. Strain Marseille-P3516T was catalase and oxidase positive and exhibited a 98.5% sequence identity with Bacillus humi strain LMG 22167 (GenBank accession no. NR_025626.1) (Fig. 1) [4], which allowed us to classify it as a member of the genus Bacillus within the family Bacillaceae in the phylum Firmicutes. Strain Marseille-P3516T is the type strain of the new species ‘Bacillus sinesaloumensis’ (si.ne.sa.lou.men′sis, N.L. masc. adj. sinesaloumensis, a vast agropastoral area in Senegal where the village of N’diop, where the stool sample was obtained, is situated).
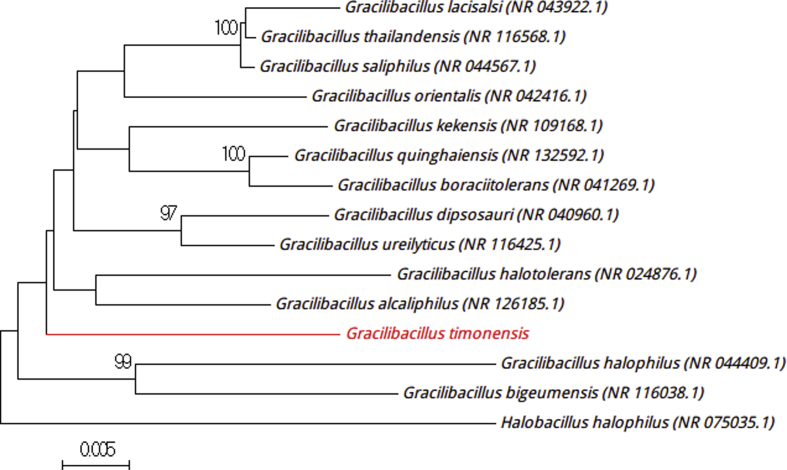
Strain Marseille-P2481T is a Gram-positive, motile, spore-forming organism isolated from a stool sample (1.7% NaCl) from a 10-year-old boy from N’diop. Catalase activity is positive and oxidase negative. The first isolation was realized in a halophilic medium with 10% (w/v) NaCl, but the strain could tolerate up to 20% (w/v) NaCl. The growing colonies are orange and circular, with a mean diameter of 2 mm. Individual cells exhibit a diameter of 1.9 × 0.5 μm and have a slightly curved form with a flagellum under electron microscopy. Strain P2481T exhibited a 16S rRNA gene sequence identity of 97% with Gracilibacillus alcaliphilus strain SG103 T (GenBank accession NR_126185) (Fig. 2) [5]. We proposed the creation of a new species within the Gracilibacillus genus in the family Bacillaceae in the phylum Firmicutes. Strain Marseille-P2481T is the type strain of the new species ‘Gracilibacillus timonensis’ (ti.mo.n.en′sis, N.L. fem. adj. timonensis, from the name Hôpital de la Timone, the hospital of Marseille, France).
Fig. 2.
Phylogenetic tree showing position of ‘Gracilibacillus timonensis’ Marseille-P2481T relative to other phylogenetically close neighbours. Sequences alignment and phylogenetic inferences were realized as explained for Fig. 1. Scale bar indicates 0.5% nucleotide sequence divergence.
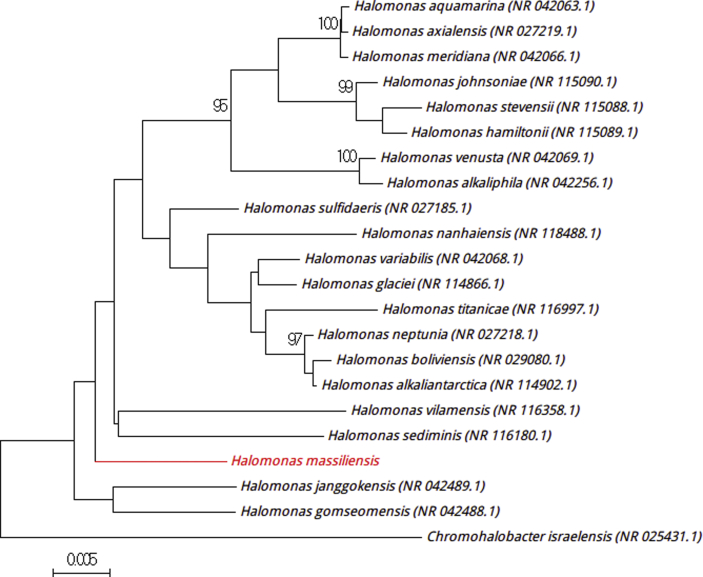
Strain Marseille-P3554T was isolated in a stool sample (2.1%) from a 12-year-old boy living in N’diop. The strain Marseille-P3554T is Gram positive, catalase negative and oxidase positive. The first isolation was realized in a halophilic medium with 10% (w/v) NaCl, but the strain was able to grow with 5 to 20% (w/v) NaCl, with optimum growth at 7.5% (w/v) NaCl. The agar colonies are grey, circular, smooth and convex, with a mean diameter of 2 mm. Individual cells exhibit a diameter of 0.3–0.6 × 2.0–4.0 μm. Strain Marseille-P3554T exhibited a 16S rRNA sequence divergence of >1.3% with Halobacillus yeomjeoni strain MSS-402 (GenBank accession no. NR_043251) (Fig. 3) [6]. On the basis of this finding, we propose to classify ‘Halobacillus massiliensis’ as a new representative of the Halobacillus genus belonging to the family Bacillaceae in the phylum Firmicutes. Strain Marseille-P3554T is the type strain of ‘Halobacillus massiliensis’ (mas.si.li.en′sis, L. masc. adj. massiliensis, ‘of Massilia,’ the old Roman name for Marseille, where the strain was isolated).
Fig. 3.
Phylogenetic tree showing position ‘Halobacillus massiliensis’ Marseille-P3554T relative to other phylogenetically close neighbours. Sequences alignment and phylogenetic inferences were realized as explained for Fig. 1. Scale bar indicates 0.5% nucleotide sequence divergence.
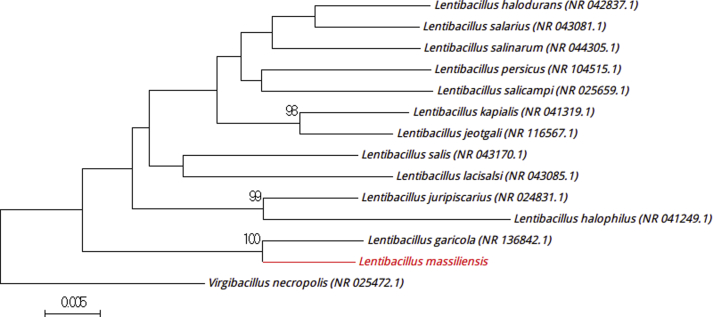
Strain Marseille-P3089T is also a Gram-positive, motile, spore-forming, catalase-negative and oxidase-positive bacterium. This strain was isolated from a stool sample (2.1% NaCl) from a 19-year-old man from N’diop. Strain Marseille-P3089T required salinity ranging from 0.5 to 20% (w/v) NaCl (optimum, 10% (w/v) NaCl). The colonies are yellow and circular, and entire, smooth and convex, with a 1.5 mm diameter. Individual cells exhibit a diameter of 0.3–0.6 × 2.0–4.0 μm. Strain Marseille-P3089T exhibited a 16S rRNA sequence similarity of 98.3% with Lentibacillus garicola (strain SL-MJ1, GenBank accession NR_136842), its closest related species with standing in nomenclature (Fig. 4) [7]. On the basis of this difference, we propose the creation of ‘Lentibacillus massiliensis,’ a member of the genus Lentibacillus, family Bacillaceae and phylum Firmicutes. Strain Marseille-P3089T is the type strain of ‘Lentibacillus massiliensis’ (mas.si.li.en′sis, L. masc. adj. massiliensis, ‘of Massilia,’ the old Roman name for Marseille, where the strain was isolated).
Fig. 4.
Phylogenetic tree showing position ‘Lentibacillus massiliensis’ Marseille-P3089T relative to other phylogenetically close neighbours. Sequences alignment and phylogenetic inferences were realized as explained for Fig. 1. Scale bar indicates 0.5% nucleotide sequence divergence.
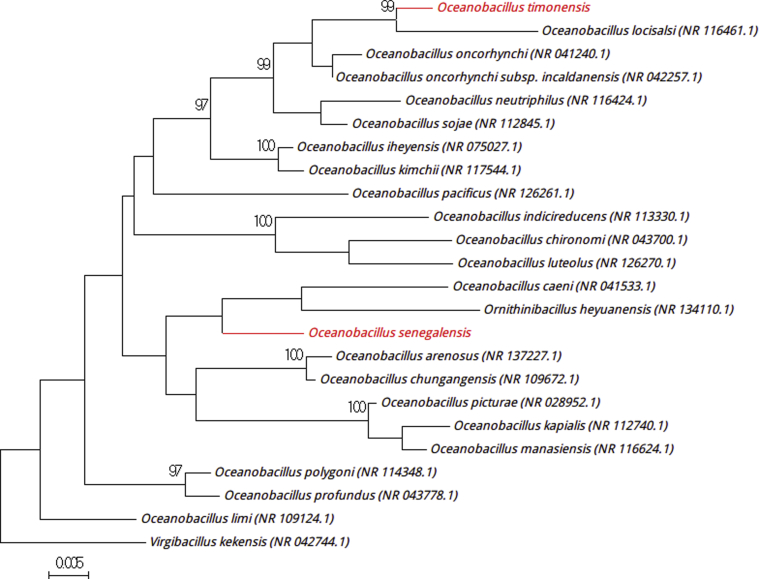
Strain Marseille-P3587T was isolated from a stool sample (2.1% NaCl) from a 7-year-old boy living in N’diop. Growth was first obtained in 15% (w/v) NaCl but occurs at 5 to 20% (w/v) NaCl, with an optimum at 7.5% (w/v) NaCl. Colonies are smooth, circular, yellowish and shiny with a mean diameter of 2.5 mm. Bacterial cells are Gram positive, rod shaped and polymorphic, and the strain Marseille-3587T is catalase and oxidase positive. The 16S rRNA gene showed 96.6% sequence identity with Oceanobacillus polygoni strain SA9 (GenBank accession no. NR_114348.1), the phylogenetically closest species (Fig. 5) [8]. Considering the sequence similarity of <98.7%, we proposed the creation of a new species, ‘Oceanobacillus senegalensis,’ within the phylum Firmicutes. Strain Marseille-P3587T is the type strain of the new species ‘Oceanobacillus senegalensis’ (se.ne.ga.len′sis, L. masc. adj. senegalensis, ‘related to Senegal,’ the name of the country where the stool sample was collected).
Fig. 5.
Phylogenetic tree showing position of ‘Oceanobacillus senegalensis’ Marseille-P3587T and ‘Oceanobacillus timonensis’ Marseille-P3532T relative to other phylogenetically close neighbours. Sequences alignment and phylogenetic inferences were realized as explained for Fig. 1. Scale bar indicates 0.5% nucleotide sequence divergence.
Strain Marseille-P3532T was isolated from the same stool sample as for strain Marseille-P3516T. The first isolation was obtained in a halophilic medium with 15% (w/v) NaCl. However, the strain was able to grow with 0.5 to 20% (w/v) NaCl, with optimum growth at 7.5% (w/v) NaCl. Grown colonies are beige, circular and shiny with a mean diameter of 2 mm. Bacterial cells are Gram positive, rod shaped and polymorphic. Strain Marseille-3532T is catalase and oxidase positive and exhibited a 98.2% sequence identity with Oceanobacillus oncorhynchi strain R-2 (GenBank accession no. NR_041240.1) (Fig. 5) [9], the phylogenetically closest species with standing in nomenclature. Therefore, we proposed to classify the species ‘Oceanobacillus timonensis’ as a member of the genus Oceanobacillus within the phylum Firmicutes. Strain Marseille-P3532T is the type strain of the new species ‘Oceanobacillus timonensis’ (ti.mo.nen′sis, L. fem. adj. timonensis, ‘related to Timone,’ the name of the main university hospital in Marseille, France, where the strain was isolated).
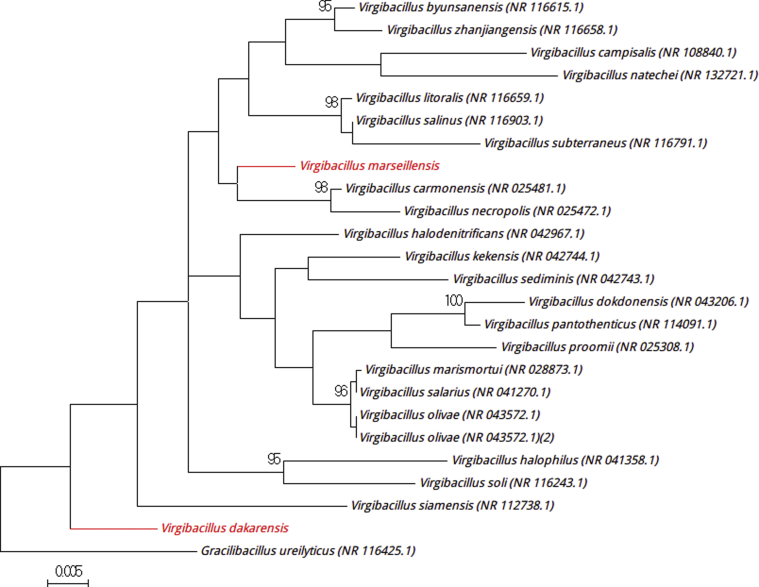
Strain Marseille-P3469T was isolated from a stool sample (2% NaCl) from a 15-year-old adolescent boy in the village of Dielmo. The first isolation was obtained in a halophilic medium with 10% (w/v) NaCl. However, the strain was able to grow between 0 and 20% (w/v) NaCl, with optimum growth observed at 10% (w/v) NaCl. Agar-grown colonies are cream coloured, circular and shiny, with a mean diameter of 4 mm. Bacterial cells are Gram positive, rod shaped and polymorphic. Strain Marseille-3469T is catalase and oxidase positive. Strain Marseille 3469T exhibited a 96.6% sequence identity with Virgibacillus byunsanensis strain ISL-24 (GenBank accession no. NR_116615.1) (Fig. 6) [10], the phylogenetically closest species, which led us to classify it as a member of the genus Virgibacillus within the family Bacillaceae in the phylum Firmicutes. Strain Marseille-P3469T is the type strain of the proposed new species ‘Virgibacillus dakarensis’ (da.ka.ren′sis, N.L. fem. adj. dakarensis, ‘related to Dakar,’ the capital of Senegal, where the stool sample was collected).
Fig. 6.
Phylogenetic tree showing position of ‘Virgibacillus dakarensis’ Marseille-P3469T and ‘Virgibacillus marseillensis’ Marseille-P3610T relative to other phylogenetically close neighbours. Sequences alignment and phylogenetic inferences were realized as explained for Fig. 1. Scale bar indicates 0.5% nucleotide sequence divergence.
Strain Marseille-P3610T was isolated in a stool sample (2.6% NaCl) from a 7-year-old boy living in N’diop. The strain was first isolated in a halophilic medium with 10% (w/v) NaCl but requires 2 to 20% (w/v) NaCl, with optimum growth at 10% (w/v) NaCl. Colonies are cream, circular and shiny with a mean diameter of 4 mm. Bacterial cells are Gram positive, rod shaped and polymorphic. Strain Marseille-3610T is catalase and oxidase positive. Strain 3610T exhibited a sequence similarity of 98% with Virgibacillus carmonensis strain LMG-20964 (GenBank accession no. NR_025481), the phylogenetically closest species (Fig. 6) [4]. Therefore, we propose the classification of ‘Virgibacillus marseillensis’ as a member of the family Bacillaceae in the phylum Firmicutes. Strain Marseille-P3610T is the type strain of the new species ‘Virgibacillus marseillensis’ (mar.sei.llens′s, N.L. fem. adj. marseillensis, ‘related to Marseille,’ the name of the region in France where the laboratory of La Timone hospital, where the strain was isolated, is situated).
MALDI-TOF MS spectra
MALDI-TOF MS spectra of strains are available online (http://www.mediterranee-infection.com/article.php?laref=256&titre=urms-database).
Nucleotide sequence accession numbers
The 16S r RNA gene sequences were deposited in GenBank under the following accession numbers: ‘Bacillus dakarensis’ Marseille-P3515T (LT671589), ‘Bacillus sinesaloumensis’ Marseille-P3516T (LT671591), ‘Gracilibacillus timonensis’ Marseille-P2481 (LT223702), ‘Halobacillus massiliensis’ Marseille-P3554T (LT671593), ’Lentibacillus massiliensis’ Marseille-P3089T (LT598549), ‘Oceanobacillus senegalensis’ Marseille-P3587T (LT714172) ‘Oceanobacillus timonensis’ Marseille-P3532T (LT671597), ‘Virgibacillus dakarensis’ Marseille-P3469T (LT631544) and ‘Virgibacillus marseillensis’ Marseille-P3610T (LT714170).
Deposit in a culture collection
The strains were deposited in the Collection de Souches de l’Unité des Rickettsies (CSUR, WDCM 875) under numbers ‘Bacillus dakarensis’ Marseille-P3515T (P3515), ‘Bacillus sinesaloumensis’ Marseille-P3516T (P3516), ‘Gracilibacillus timonensis’ Marseille-P2481T (P2248), ‘Halobacillus massiliensis’ P3554T (P3354), ‘Lentibacillus massiliensis’ Marseille-P3089T (P3089), ‘Oceanobacillus senegalensis’ Marseille-P3587T (P3587), ‘Oceanobacillus timonensis’ Marseille-P3532T (P3532), ‘Virgibacillus dakarensis’ Marseille-P3469T (P3469) and ‘Virgibacillus marseillensis’ Marseille-P3610T (P3610).
Acknowledgement
This study was funded by the Fondation Méditerranée Infection.
Conflict of Interest
None declared.
References
- 1.Lagier J.C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PubMed] [Google Scholar]
- 2.Lo C.I., Fall B., Sambe-Ba B., Diawara S., Gueye M.W., Mediannikov O. MALDI-TOF mass spectrometry: a powerful tool for clinical microbiology at Hôpital Principal de Dakar, Senegal (West Africa) PLoS One. 2015;10:e0145889. doi: 10.1371/journal.pone.0145889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramasamy D., Mishra A.K., Lagier J.C., Padhmanabhan R., Rossi M., Sentausa E. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol. 2014;64:384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 4.Heyrman J. Virgibacillus carmonensis sp. nov., Virgibacillus necropolis sp. nov. and Virgibacillus picturae sp. nov., three novel species isolated from deteriorated mural paintings, transfer of the species of the genus Salibacillus to Virgibacillus, as Virgibacillus marismortui comb. nov. and Virgibacillus salexigens comb. nov., and emended description of the genus Virgibacillus. Int J Syst Evol Microbiol. 2003;53:501–511. doi: 10.1099/ijs.0.02371-0. [DOI] [PubMed] [Google Scholar]
- 5.Hirota K., Hanaoka Y., Nodasaka Y., Yumoto I. Gracilibacillus alcaliphilus sp. nov., a facultative alkaliphile isolated from indigo fermentation liquor for dyeing. Int J Syst Evol Microbiol. 2014;64:3174–3180. doi: 10.1099/ijs.0.060871-0. [DOI] [PubMed] [Google Scholar]
- 6.Yoon J.H., Kang S.J., Lee C.H., Oh H.W., Oh T.K. Halobacillus yeomjeoni sp. nov., isolated from a marine solar saltern in Korea. Int J Syst Evol Microbiol. 2005;55:2413–2417. doi: 10.1099/ijs.0.63801-0. [DOI] [PubMed] [Google Scholar]
- 7.Jung W.Y., Lee S.H., Jin H.M., Jeon C.O. Lentibacillus garicola sp. nov., isolated from myeolchi-aekjeot, a Korean fermented anchovy sauce. Antonie Van Leeuwenhoek. 2015;107:1569–1576. doi: 10.1007/s10482-015-0450-2. [DOI] [PubMed] [Google Scholar]
- 8.Hirota K., Hanaoka Y., Nodasaka Y., Yumoto I. Oceanobacillus polygoni sp. nov., a facultatively alkaliphile isolated from indigo fermentation fluid. Int J Syst Evol Microbiol. 2013;63:3307–3312. doi: 10.1099/ijs.0.048595-0. [DOI] [PubMed] [Google Scholar]
- 9.Yumoto I. Oceanobacillus oncorhynchi sp. nov., a halotolerant obligate alkaliphile isolated from the skin of a rainbow trout (Oncorhynchus mykiss), and emended description of the genus Oceanobacillus. Int J Syst Evol Microbiol. 2005;55:1521–1524. doi: 10.1099/ijs.0.63483-0. [DOI] [PubMed] [Google Scholar]
- 10.Yoon J.H., Kang S.J., Jung Y.T., Lee K.C., Oh H.W., Oh T.K. Virgibacillus byunsanensis sp. nov., isolated from a marine solar saltern. Int J Syst Evol Microbiol. 2010;60:291–295. doi: 10.1099/ijs.0.009837-0. [DOI] [PubMed] [Google Scholar]