Abstract
The formation of lipid electrophile-protein adducts is associated with many disorders that involve perturbations of cellular redox status. The identities of adducted proteins and the effects of adduction on protein function are mostly unknown and an increased understanding of these factors may help to define the pathogenesis of various human disorders involving oxidative stress. 7-Dehydrocholesterol (7-DHC), the immediate biosynthetic precursor to cholesterol, is highly oxidizable and gives electrophilic oxysterols that adduct proteins readily, a sequence of events proposed to occur in Smith-Lemli-Opitz syndrome (SLOS), a human disorder resulting from an error in cholesterol biosynthesis. Alkynyl lanosterol (a-Lan) was synthesized and studied in Neuro2a cells, Dhcr7-deficient Neuro2a cells and human fibroblasts. When incubated in control Neuro2a cells and control human fibroblasts, a-Lan completed the sequence of steps involved in cholesterol biosynthesis and alkynyl-cholesterol (a-Chol) was the major product formed. In Dhcr7-deficient Neuro2a cells or fibroblasts from SLOS patients, the biosynthetic transformation was interrupted at the penultimate step and alkynyl-7-DHC (a-7-DHC) was the major product formed. When a-Lan was incubated in Dhcr7-deficient Neuro2a cells and the alkynyl tag was used to ligate a biotin group to alkyne-containing products, protein-sterol adducts were isolated and identified. In parallel experiments with a-Lan and a-7-DHC in Dhcr7-deficient Neuro2a cells, a-7-DHC was found to adduct to a larger set of proteins (799) than a-Lan (457) with most of the a-Lan protein adducts (423) being common to the larger a-7-DHC set. Of the 423 proteins found common to both experiments, those formed from a-7-DHC were more highly enriched compared to a DMSO control than were those derived from a-Lan. The 423 common proteins were ranked according to the enrichment determined for each protein in the a-Lan and a-7-DHC experiments and there was a very strong correlation of protein ranks for the adducts formed in the parallel experiments.
Abbreviations: 7-DHC, 7-dehydrocholesterol; Lath, lathosterol; Lan, lanosterol; Chol, cholesterol; SLOS, Smith-Lemli-Opitz syndrome; DHCR7, 7-dehydrocholesterol reductase; MS, mass spectrometry; APCI, atmospheric pressure chemical ionization; HPLC-, high pressure liquid chromatography; MeOH, methanol; NMR, nuclear magnetic resonance; FBS, fetal bovine serum; DMEM, Dulbecco's Modified Eagle Medium; PBS, phosphate buffered saline; DMSO, dimethylsulfoxide
Keywords: Alkynyl sterols, 7-dehydrocholesterol, Cholesterol, HPLC-MS, GC-MS, DHCR7, Smith-Lemli-Opitz Syndrome
Graphical abstract
1. Introduction
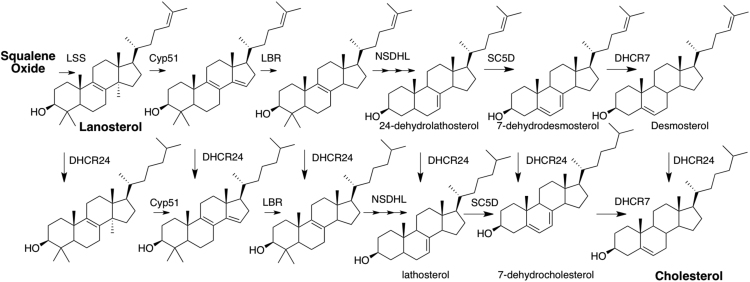
The post-lanosterol pathway for the biosynthesis of cholesterol involves a complex series of chemical transformations [1]. Over 30 discrete compounds are intermediates between lanosterol and cholesterol, with the Bloch and Kandutsch-Russell parallel pathways differing only by unsaturation in the tail of the molecule. An abbreviated Scheme showing a simplified pathway is described in Fig. 1. A number of human disorders have been linked to deficiencies in the enzymes that catalyze the various demethylations, oxidations and reductions in the biosynthetic pathway [2], [3], [4]. Desmosterolosis, for example, derives from mutations in the enzyme involved in the conversion of desmosterol to cholesterol [5] while a more common disorder, Smith-Lemli-Opitz syndrome (SLOS), results from errors in the gene that encodes 7-dehydrocholesterol reductase, the enzyme that promotes the conversion of 7-dehydrocholesterol (7-DHC) to cholesterol. SLOS is an autosomal recessive disorder with a DHCR7 mutation carrier frequency of about 1% [6], [7], [8], [9], [10]. The phenotype is broad, with severe cases suffering pre-term demise and mild cases having minor physical findings with associated learning and behavioral problems [11]. A plasma sterol analysis showing an elevated dehydrocholesterol level and reduced cholesterol level forms the primary biochemical basis of a positive SLOS diagnosis.
Fig. 1.
Partial Scheme for Cholesterol Biosynthesis. Bloch pathway, Top Row of Sterols; Kandutsch-Russell Pathway, Bottom Row.
Plasma and tissue levels of 7-DHC found in SLOS patients can be over 10,000-fold higher than those found in control individuals and recent evidence suggests that the pathology associated with the syndrome may be due in large part to the elevated levels of this sterol [12], [13]. 7-DHC has the highest rate constant for the propagation of free radical peroxidation of any lipid studied to date [14], [15] and the oxysterols formed in the process are toxic to primary cortical neuronal and glial cells in vitro and accelerate differentiation and arborization of cortical neurons [16], [17]. The result of a mutation in DHCR7 then, is a sequence of events that includes: 1. An increase of 7-DHC levels in tissues and fluids. 2. Oxidation of this vulnerable sterol to give a set of oxysterols, some of which are highly electrophilic. 3. Adduction of the reactive 7-DHC-derived electrophiles to target proteins.
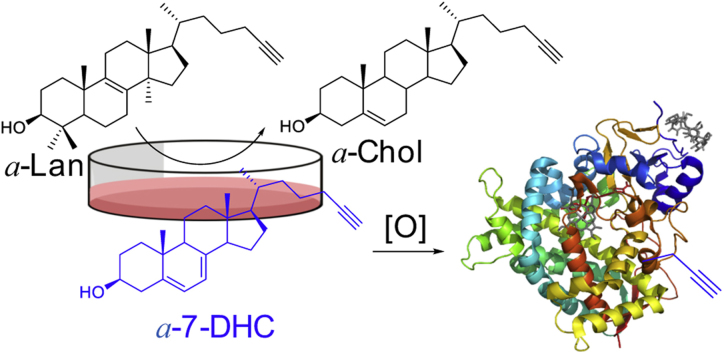
To enquire if 7-DHC-derived electrophiles interact with the cellular proteome, cholesterol precursors bearing attached alkynyl groups were prepared and these compounds were evaluated as sterol surrogates in cell culture models and then used to identify lipid-adducted proteins. Protein adduction by reactive electrophiles likely contributes to chemical toxicities and oxidative stress, but the functional impact of adduction across proteomes is poorly understood [18]. Indeed, the isolation and identification of protein adducts of lipid electrophiles is a not a trivial exercise and various strategies have been developed to aid in this effort [19]. In this regard, we and others have utilized various lipid analogs that are modified with an alkyne functional group which serves as a “tag” that can be used to visualize the sterols in cells or to pull-down lipid-protein adducts by “click” methods [20], [21], [22], [23]. In our hands, this approach has permitted the identification of adduct proteomes of various reactive species derived from fatty acid peroxidation [18], [19], and it has provided evidence that proteins are modified by reactive electrophiles derived from 7-DHC. We now report that an alkyne-modified lanosterol can serve as a viable surrogate in cholesterol biosynthesis, completing the sequence of eighteen enzymatically promoted steps required for its conversion to alkynyl cholesterol in Neuro2a cells and in human fibroblasts. This permits an in situ analysis of the proteome adducted by sterol electrophiles formed in a Neuro2a cellular model of SLOS.
2. Results
2.1. Alkynyl sterols
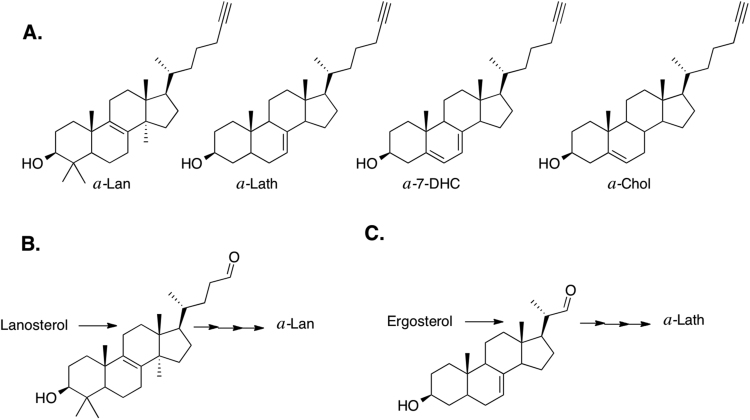
The four alkynyl sterols shown in Fig. 2 were available for use in our studies. Alkynyl-cholesterol (a-Chol) and alkynyl-7-dehydrocholesterol (a-7-DHC) had been previously reported [24], the starting material for the synthesis of both being 3β-hydroxy-5-cholenic acid. The starting material for preparation of alkynyl-lanosterol (a-Lan) was lanosterol itself, while alkynyl-lathosterol (a-Lath) was prepared starting from ergosterol. In both of these syntheses, the double bond in the tail of the starting material was oxidatively cleaved and the aldehyde so generated was subjected to coupling with an appropriate Grignard reagent, which was then reduced with lithium aluminum hydride, see Fig. 2B and C. Details of the syntheses of a-Lan (four steps) and a-Lath (eight steps) are provided in Supporting Information (Appendix A, Appendix A).
Fig. 2.
Alkynyl sterols and their synthetic precursors. A. The four alkynyl sterols synthesized. B. Partial scheme for preparation of a-Lan. C. Partial scheme for preparation of a Lath.
2.2. a-Lathosterol and a-Lanosterol in Neuro2a and Dhcr7-deficient Neuro2a
Cells were grown in the presence or absence of alkynyl sterols for 24–48 h and lipids isolated as described in Materials and Methods. Alkynyl sterols and endogenous sterols were analyzed by reverse-phase HPLC-MS, the alkynyl analogs eluting earlier than the natural compounds. Like their natural counterparts, a-Lath and a-Chol did not separate under any of the HPLC chromatography conditions attempted. These two sterols could be separated by GC however, and a combination of the HPLC-MS and GC was required for a complete analysis of a-sterol and natural sterol product mixtures. The HPLC retention times for the a-sterols under the conditions described in Materials and Methods were a-7-DHC, 3.8 min; (a-Lath and a-Chol), 4.1 min; and a-Lan, 4.8 min. For comparison, natural cholesterol elutes at 9.1 min under the same chromatography conditions.
In Neuro2a cells, both a-Lath and a-Lan undergo bio-conversion to a-Chol as the major product when incubated with the cells for 24 h at concentrations <10 μM. Our initial experiments were carried out with a-Lath at concentrations of 5 μM and 10 μM in Neuro2a cells, the a-sterol profiles found were as follows: For 5 μM a-Lath incubation for 24 h, mole fractions of a-Lath remaining=0.16; a-7-DHC=0.10; and a-Chol=0.74. For 10 μM a-Lath incubation for 24 h, mole fractions of a-Lath remaining=0.32; a-7-DHC=0.21; and a-Chol=0.47. The natural sterol levels found after 24 h incubations with a-Lath were somewhat suppressed compared to levels found in controls, the total sterol levels (a-sterols+natural sterols) totaled 70–80% of the natural sterols found in control cells. Higher concentrations of a-Lath had a larger effect on reducing the total sterol levels. The alkynyl sterols made up 25% and 40% of the total sterol levels at a-Lath concentrations of 5 and 10 µM, respectively.
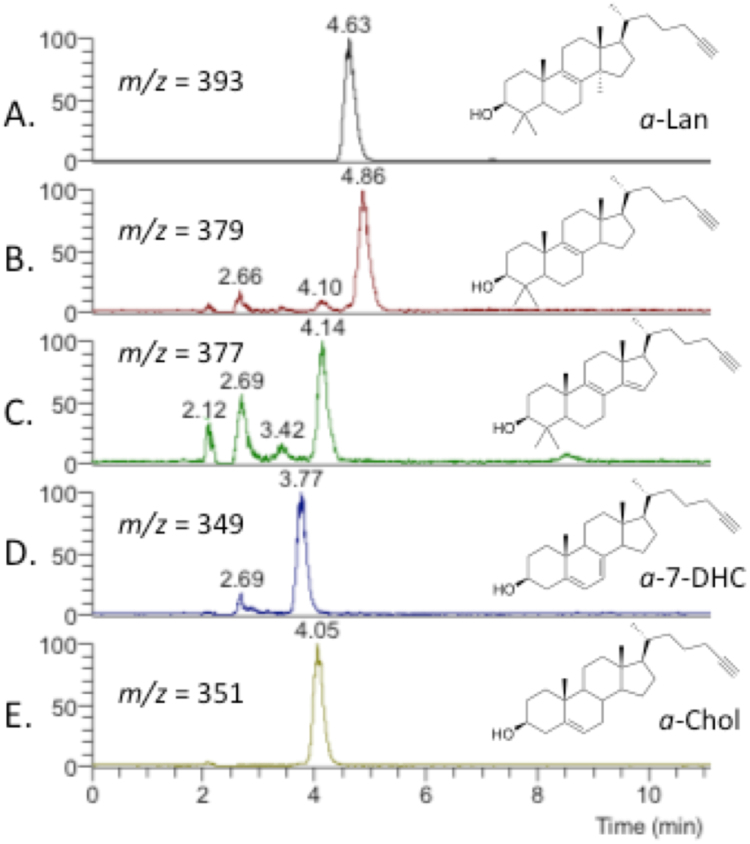
After the exploratory experiments with a-Lath in Neuro2a cells, subsequent studies were carried out with a-Lan at 0.1, 1.0, 5.0 and 10 μM in both Neuro2a and Dhcr7-deficient Neuro2a cells. Fig. 3 presents a typical HPLC-MS chromatogram for a product mixture derived from an experiment of 10 μM of a-Lan incubated in Neuro2a for 24 h. Under these conditions a-Chol is the major alkynyl sterol found in the product mixture, panel E in Fig. 3. The starting material in the experiment, a-Lan (panel A, Fig. 3), is always detected in the mix of alkynyl sterols found as is a-7-DHC, (panel D in Fig. 3). Only trace amounts of other alkynyl sterols, including a-Lath, were found in product mixtures but the two alkynyl compounds shown in panels B and C in Fig. 3 were always detected. The structures assigned to these products are tentative, they have not been independently synthesized, but the diene shown in panel C is the biosynthetic product of CYP51A1 demethylation of a-Lan and this diene is the immediate biosynthetic precursor to the alkene shown in panel B of the Fig. 3.
Fig. 3.
Alkynyl Sterols Found in 24 h Incubations ofa-Lan in Neuro2a Cells. Structures of compounds shown in Panels B at 4.86 min and C at 4.14 min are tentative based on m/z values and the known biosynthetic pathway.
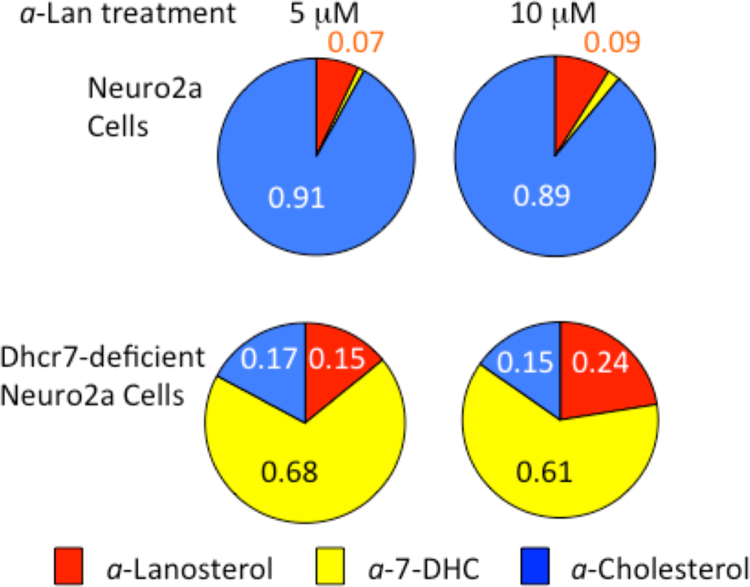
The major alkynyl sterols found in control or Dhcr7-deficient Neuro2a cells after 24 h incubation with a-Lan are a-Lan, a-7-DHC and a-Chol. Fig. 4 shows the alkynyl sterol mole fractions determined from incubations at 5 and 10 μM a-Lan in both cell types. Incubations at 0.1 and 1 μM give similar qualitative results to the incubations carried out at higher concentrations but with more complete consumption of a-Lan at the lower concentrations, the mole fraction at 24 h of a-Chol in control Neuro2a cells approaching 1.0. In a similar way, incubation with 0.1 μM a-Lan in Dhcr7-deficient Neuro2a cells for 24 h gave an alkynyl sterol distribution with a-7-DHC close to 1.0. Total sterol levels (alkynyl + endogenous) in the experiments with a-Lan dropped to about 90% of the endogenous levels in control cells.
Fig. 4.
Mole fractions of alkynyl Sterols Found in 24 h Incubations of a-Lan in Neuro2a and Dhcr7-deficient Neuro2a Cells. Data presented is from three independent experiments with σ<5%.
2.3. a-Lanosterol in human control, SLOS and NPC fibroblasts
To determine the utility of alkyne sterol surrogates in a variety of cell culture models, we cultured human fibroblasts with a-Lan. a-Chol biosynthesis from a-Lan was compared in control fibroblasts and fibroblasts from two different cholesterol disorders, SLOS and Niemann-Pick type C (NPC). NPC is an autosomal recessive disorder characterized by cellular accumulation of unesterified cholesterol and other lipids. Table 1 shows the alkynyl and natural sterol distribution in human fibroblasts incubated with a-Lan for 6 days. The results from experiments with control fibroblasts are similar to those described for the a-Lan in Neuro2a cells. For control fibroblasts as with control Neuro2a cells most of the alkynyl sterol was found as a-Chol (85%) with only fractional a-Lan (8%) and a-7-DHC (7%) found after 6 days of incubation. In contrast, in SLOS fibroblasts, most of the alkynyl sterol (83%) was found in the a-7-DHC fraction after 6 days. We conclude that the results of studies in human fibroblasts parallel those found in control and Dhcr7-deficient Neuro2a cells. a-Lan is converted into a-7-DHC in SLOS fibroblasts while in control and NPC cells, a-Chol is the major product formed. Total cholesterol accumulation in NPC is significantly increased compared to control fibroblasts, a result that accurately reflects the defect described for this disorder.
Table 1.
Sterol distribution in cultured fibroblasts after 6 days incubation with 1 μM a-Lan.a
| Alkynyl sterols |
Natural sterols |
|||||||
|---|---|---|---|---|---|---|---|---|
| a-Lan | a-Lath | a-7-DHC | a-Chol | Lan | Lath | 7-DHC | Chol | |
| Control | 0.055±0.013 | n.d. | 0.051±0.017 | 0.660±0.085 | 0.021±0.003 | n.d. | 0.042±0.008 | 12.7±0.8 |
| fibroblast | ||||||||
| SLOS | 0.143±0.021 | n.d. | 0.657±0.096 | n.d. | 0.015±0.001 | 0.167±0.145 | 3.47±0.51 | 8.1±1.7 |
| fibroblast | ||||||||
| NPC | 0.090±0.05 | n.d. | 0.011±0.011 | 1.308±0.22 | 0.017±0.002 | 0.051±0.089 | 0.049±0.016 | 33.9±6.9 |
| fibroblast | ||||||||
nmol of sterol determined by HPLC-MS and GC per 106 cells. Data presented are averages determined from triplicate analyses. n.d., not detected.
2.4. Adduct protein identification
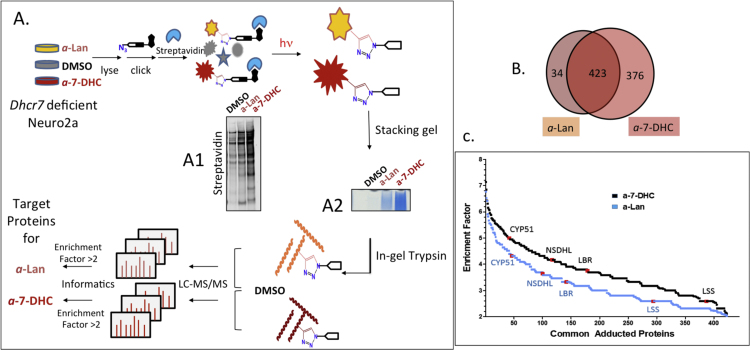
The alkyne functional group serves as a tag that can be used to concentrate covalent sterol-protein adducts formed from the alkynyl sterol precursor. This strategy, which is presented in Fig. 5A, involves incubation of Dhcr7-deficient Neuro2a cells with a-Lan or a-7-DHC for 24 h followed by treatment of the cell lysate with a biotin azide under Huisgen-Sharpless click conditions. The click reaction links any sterol alkyne-protein covalent adducts formed with biotin, which can then be visualized by the use of streptavidin immunoblotting [24]. In the experiments described in Fig. 5A, a photo-cleavable biotin azide [20] (see Supporting information, Fig. S1 for the structure) was used in the click-reaction step and the protein mixture treated with streptavidin beads. This was followed by a pre-wash of the beads to remove un-adducted proteins and the covalent a-sterol protein adducts were then subsequently released by photolysis of the streptavidin beads with a small hand-held UV lamp. As shown in Fig. 5B, a total of 833 proteins were identified as being significantly enriched in the photorelease fractions for the treatment of Dhcr7-deficient Neuro2a cells with 5 μM a-7-DHC or 10 μM a-Lan, 423 of those adducted proteins were found in the incubations of both alkynyl sterols.
Fig. 5.
Protein Adduction inDhcr7-deficient Neuro2a Cells. A. Workflow to identify lipid-adducted proteins. Intensity oown in A. C. Enrichment Ff Streptavidin indicates the degree of protein adduction (A1). Photo-eluted proteins were visualized in short stacking SDS Page gel shown in A2 followed by in-gel trypsin digestion for LC-MS/MS analysis. 10 and 5 μM of a-Lan and a-7-DHC were supplemented respectively. B. Number of total proteins identified from experiments shactor for the 423 common proteins adducted by both a-DHC and a-Lan. Enrichment factor for a-7-DHC (in black) vs. treatment with DMSO and a-Lan (in blue) vs. treatment with DMSO. The enrichment factor is defined as log2 of [spectral count for a protein found for alkyne treatment divided by spectral count of the same protein for DMSO treatment]. Four proteins involved in cholesterol biosynthesis were identified in both the a-7-DHC and a-Lan experiments and are highlighted in red in the Figure. Additional replica data of Streptavidin western and stacking gels are included in Supporting Information (Fig. S2). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.5. Ranking and analysis of adductomes
The alkyne sterol tag used in these studies permitted the identification of adducted proteins and allowed for a preliminary ranking and classification of those proteins. The enrichment factor (log2 [protein spectral counts for treated cells/protein spectral counts for control cells]) was used to compare the enrichment of the 423 common proteins adducted in the a-7-DHC and a-Lan experiments. Inspection of the enrichment factor data shown in Fig. 5C suggested that the adducts identified from a-7-DHC were more highly enriched than the same adducts found in the a-Lan study, even though the concentration of a-Lan used was 10 μM, twice that used with a-7-DHC. Enrichment data of selected proteins involved in cholesterol biosynthesis, (LSS, LBR, NSDHL and CYP51), for example, showed them to be more highly adducted in the a-7-DHC experiment than was found in experiments with a-Lan, see Fig. 1 for the protein function and Fig. 5C for relative enrichment. The enrichment factors for the 423 proteins found common to the a-7-DHC and a-Lan experiments and the ranking of each protein based on enrichment are presented in Table S1 in Supporting information. A comparison of a-7-DHC and a-Lan protein ranks in various protein classes is presented in the Discussion Section.
3. Discussion
3.1. Lipid peroxidation, lipid electrophiles and protein adduction
The free radical chain oxidation of lipids has been the focus of extensive investigation. The primary products of polyunsaturated lipid peroxidation are compounds that readily undergo metabolism or decomposition to give secondary compounds that have important biological activities. 4-Hydroxy-2-nonenal (4-HNE) for example, is a cytotoxic end-product of decomposition of hydroperoxides formed from polyunsaturated lipid peroxidation and 4-HNE is an electrophile that undergoes Michael addition with biological protein nucleophiles [18]. Protein thiols and lysine primary amines, for example, react with 4-HNE to form protein adducts of this lipid-derived electrophile.
Peroxidation has also recently been associated with a lipid intermediate in cholesterol biosynthesis, 7-DHC, and the human disorder (SLOS) has been linked to the reactivity of this sterol [13], [14], [25]. Indeed, 7-DHC is highly susceptible to free radical peroxidation, the B-ring of this compound is some 36 times more reactive than linoleic acid [15] and over 200 times that of cholesterol [26], [27]. Cytotoxic oxysterols are products of 7-DHC peroxidation and evidence has been presented that some of these products are reactive electrophiles [24]. The initial evidence suggesting that 7-DHC electrophile formation and protein adduction was associated with SLOS came from studies with a-7-DHC in control Neuro2a and human fibroblasts as well as with Dhcr7-deficient Neuro2a and SLOS fibroblasts. Thus, a-7-DHC and click cycloaddition provided a means to biotinylate protein-sterol adducts which could then be visualized by immunoblot with a streptavidin-modified fluorophore. In this way, evidence of protein adduction was found when a-7-DHC was incubated with Dhcr7-deficient Neuro2a and SLOS fibroblasts while only background protein adduction was observed in control cell incubations [24].
3.2. Alkynyl sterol metabolism
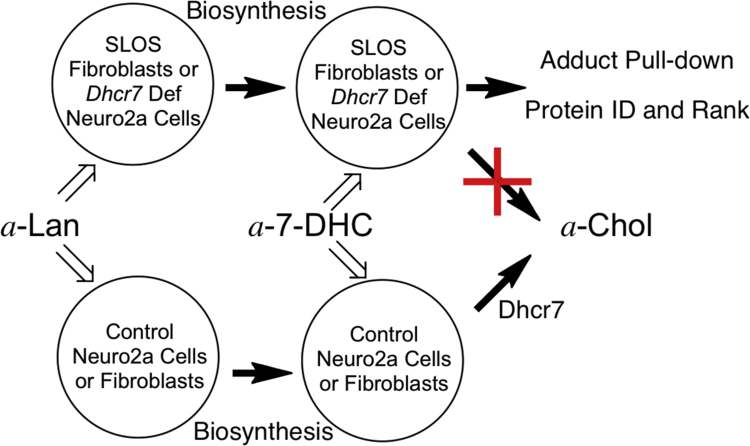
The use of the alkynyl sterols described here in conjunction with the two Neuro2a cell types and control and SLOS fibroblasts provided a means for presentation of alkynyl tagged 7-DHC to the cells in a metabolically meaningful way, see Fig. 6. Thus, introduction of the sterol tag with a-Lan requires a minimum of eighteen enzyme-mediated steps for its conversion to a-Chol. The experiments in both control Neuro2a and human fibroblasts demonstrated that the transformation occurred with efficiency, see Fig. 4 and Table 1.
Fig. 6.
Strategy for alkynyl sterol incubations. In control cells a-Lan and a-7-DHC undergo conversion to a-Chol. In Dhcr7-deficient Neuro2a or SLOS fibroblasts a-7-DHC accumulates and derived oxysterols adduct to proteins.
It is of some interest that the cholesterol biosynthetic apparatus accepts alkyne substitution on the sterol tail of all of the alkynyl sterols, from a-Lan to a-Chol. The mammalian enzymes do in fact tolerate some substrate structural variability, as seems obvious from the fact that parallel pathways exist for cholesterol biosynthesis [1]. Thus, sterols substituted with or without a double bond at C24-C25 are viable substrates for the biosynthetic machinery and modification of the natural sterol with a linear 2-carbon alkyne appears to be no more restrictive than the presence or absence of a C24-C25 double bond in the sterol structure. With the exception of DHCR24, the enzyme-mediated transformations in the post-lanosterol pathways operate on the sterol ring substructure, and the tail of the molecule does not apparently play a decisive role in the transformations.
In contrast to the experiments in control cells, metabolic transformations of a-Lan were prematurely interrupted in Dhcr7-deficient Neuro2a cells and in SLOS fibroblasts. In both cells, the biosynthetic transformation was terminated at a-7-DHC, the penultimate step in the sequence, due to the absence of the Dhcr7 enzyme in Neuro2a cells or the presence of alleles bearing mutations that reduce the enzyme efficacy in SLOS fibroblasts. In Dhcr7-deficient Neuro2a cells, a-7-DHC was the major sterol detected although smaller amounts of a-Chol were also found. In SLOS fibroblasts, a-Chol could not be detected in the cell lysates and again a-7-DHC was the major metabolite found along with unreacted a-Lan.
3.3. Alkynyl sterol adducts
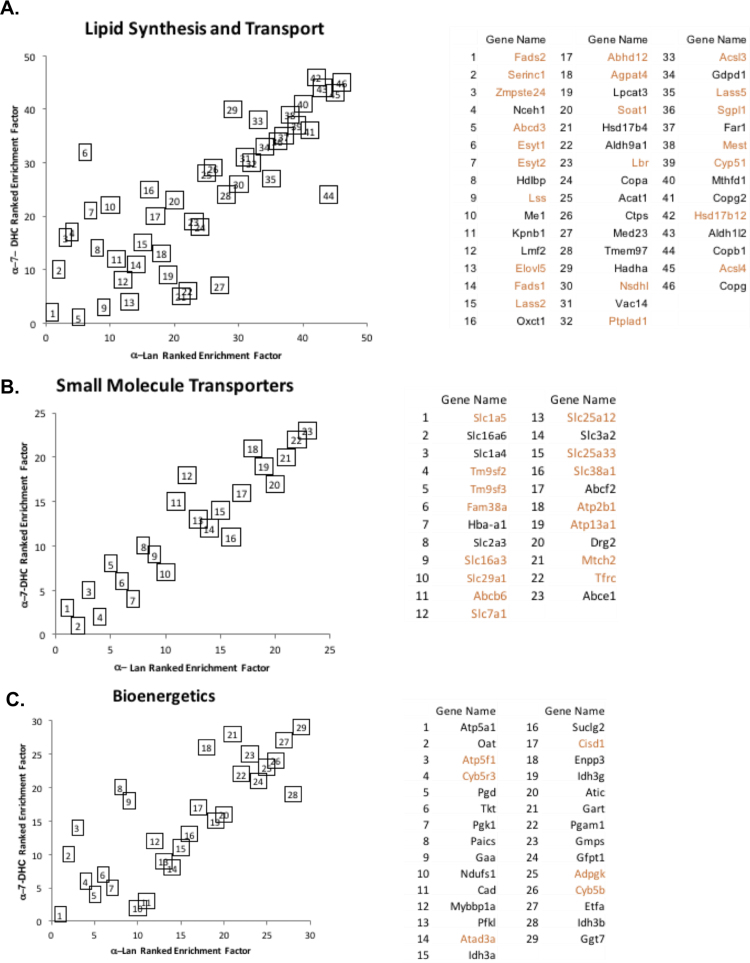
One interpretation of the adductome enrichment data shown in Fig. 5 is that a-7-DHC, or more reasonably an a-7-DHC-derived oxysterol or set of oxysterols, are the proximate electrophiles that form protein adducts. a-7-DHC forms adducts with more proteins than a-Lan and the adduct enrichment is generally higher for a-7-DHC than is the case for a-Lan even though the concentration of a-Lan used (10 μM) was twice that used for a–7-DHC. Metabolism of a-Lan is required in order to generate the same set of adduct-forming electrophiles that derive from a-7-DHC. A more detailed analysis of the 423 adductome common to the a-7-DHC and a-Lan experiments showed a significant correlation of protein enrichment by protein class in the a-7-DHC and a-Lan experiments. Thus, ranking protein adducts within a particular class showed statistically significant Pearson correlations [in brackets] of protein rank for a-7-DHC adduction vs. the rank for the same protein in a-Lan adduction in the following classes: Very strong; small molecule transport [0.93], lipid synthesis and transport [0.81], and bioenergetics [0.80]. Strong; protein synthesis and transport [0.78], cell cycle [0.72], cytoskeleton [0.66], transcription [0.66] and inflammatory response [0.62]. Moderate; signaling [0.54]. Weak; stress response [0.34]. The adduction of neurodevelopment proteins was also found to be strongly correlated in the a-7-DHC and a-Lan experiments but only seven proteins were found in that category while significantly more of the 423 adducted proteins [shown in brackets] were identified in other protein classes; protein synthesis and transport [110], cell cycle [57], signaling [50], lipids [45], transcription [39], bioenergetics [29], cytoskeleton [27], and small molecule transport [23].
Fig. 7 presents the a-7-DHC rank vs a-Lan rank plots of three protein classes (lipids, small molecule transport and bioenergetics) that have Pearson coefficients of 0.80 or greater. It is of particular interest that of the adducted proteins identified in our study, many have been shown to directly interact with cholesterol or oxysterols, including Hsd17b12 [28], the COPI-associated proteins (Copg and Copb1)[29] and the ABC transporters [30]. Additionally, the proteins that function within cellular bioenergetics identified in this study also met our requirements for a strong correlation (Fig. 7C), which could be insightful since recent findings have demonstrated SLOS impacts mitochondrial function [31]. It should also be noted that many of these proteins in bioenergetics, have already been linked to the formation of lipids (Cyb5b)[32] or are known targets of lipid electrophiles (Idh3b) [33].
Fig. 7.
Adduct Enrichment Ranking by Protein Class for thea-7-DHC vs.a-Lan incubations inDhcr7-deficient Neuro2a cells. A. In the lipid synthesis protein class. B. In the small molecule transporter protein class. C. In the bioenergetics protein class. Red names indicate proteins that form non-covalent cholesterol complexes, see reference 23. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Of further interest is the fact that 88 proteins in our 423 adductome list were also reported to interact with cholesterol in a recent study by Hulce et al. [23] In that study, synthetic photoaffinity cholesterol surrogates were used in competitions with cholesterol to identify proteins that have non-covalent sterol binding sites [23]. Our approach identifies covalent sterol-protein adducts, presumably by endogenously formed electrophiles in a different cell line, by a completely different strategy and using a different proteomics MS protocol than the one employed by Hulce et al. The overlap of the 88 proteins is particularly evident in the “lipid synthesis and transport” and the “small molecule transport” classes of proteins shown in Fig. 7A and B. Thus, of the 46 “lipid synthesis and transport” protein adducts found in this study, 23 shown in red in the key to Fig. 7A, were listed in the non-covalent binding study of Hulce et al. Prominent among those proteins in the “lipid synthesis and transport” class are the cholesterol biosynthesis enzymes, Lss, Cyp51, Nsdhl and Lbr. In a similar way, 15 of the 23 covalent adducts we identified in the “small molecule transport group” were also reported in the list of the proteins that form non-covalent complexes with cholesterol.
These findings lend credence to the use of our alkynyl sterol surrogate probes and are in alignment with the biology and biochemistry of the proteins found to be common to the two studies. Lipid synthesis proteins assemble hydrophobic binding pockets for their substrates and transport proteins for both lipids and small molecules make use of bilayer-spanning hydrophobic aggregates to facilitate their function. Sterols and oxysterols are thus suitably disposed to form complexes with these proteins, a process driven by thermodynamics. Protein covalent adduction requires an additional kinetic event, however, and there is no assurance that complex formation will lead to a sterol-protein adduct. A suitable protein nucleophile is required for covalent bond formation and non-covalent association is no guarantee of reaction. In contrast to the lipid and small molecule proteins, far fewer proteins in other classes were found to be common to the non-covalent protein set described by Hulce et al. This is illustrated in the “bioenergetics” proteins shown in Fig. 7C, only 6 of the 29 proteins found as covalent sterol adducts were also listed in the non-covalent set of proteins [23]. Proteins in other categories identified as forming non-covalent complexes with cholesterol were even less represented in the common set with our proteins, i.e. 4 of 59 “cell cycle” and 1 of 41 “transcription” proteins.
The adduct protein set reported here is substantially different from proteins found to be differentially expressed in SLOS mice in an earlier proteomics study [34]. That study compared proteins found in the brain of a SLOS rodent model with the proteome of a control animal. In contrast, the protein adduct set described here results from a cellular treatment with a viable sterol precursor that differs only from the natural cholesterol precursor by alkynyl substitution in the sterol tail.
3.4. Mechanism of sterol adduction
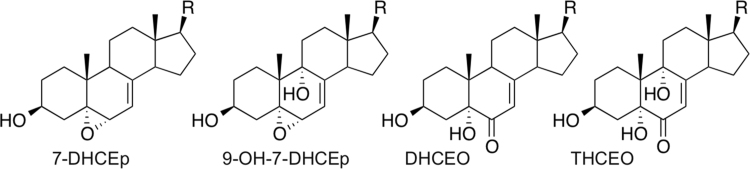
The correlation of adduct ranks in the a-7-DHC vs. a-Lan treatments provided strong evidence that the two alkynyl sterols are adducting cellular proteins by the same mechanism and this, along with the demonstration that a-7-DHC generally gives rise to higher adduct enrichment than a-Lan, lends support to the notion that 7-DHC, and more particularly oxysterols derived from 7-DHC, are the sterol electrophiles that react to form protein adducts. Peroxidation of 7-DHC yields over a dozen different oxysterols, several of which may be reactive electrophiles, see Fig. 8. The 5,6-α epoxide of 7-DHC (7-DHCEp) is one of the oxysterols formed in the peroxidation of 7-DHC and this allylic epoxide is an extremely reactive electrophile, forming covalent adducts with nucleophiles immediately upon mixing the reactants [24]. We expect that the 9-α-hydroxy-5,6-α epoxide (9-OH-7-DHCEp) shown in Fig. 8, a major product of 7-DHC peroxidation, would also be a highly reactive electrophile. The B-ring α,β–unsaturated ketones DHCEO and THCEO, also shown in Fig. 8, would likely have modest electrophilic reactivity as well.
Fig. 8.
Potential 7-DHC Oxysterol Electrophiles. Four primary peroxidation products of 7-DHC that are potentially reactive electrophiles. The 5,6-α allylic epoxide and the α, β unsaturated 6-keto sterols are possible electrophilic candidates derived from 7-DHC.
The studies reported here do not identify the reactive sterol-derived electrophile(s) that serve as reactive adduct-forming agents. The alkynyl tag was used in these experiments to pull down sterol-protein adducts, and the adducted proteins were identified by proteomic analysis of the entire peptide set generated after trypsin digestion. Identification of the reactive electrophile(s) will likely require a more targeted approach, with a proteome or single protein exposure to specific electrophiles such as those shown in Fig. 8. Targeted experiments of this type are more likely to lead to the identification of relevant electrophile-adducted tryptic peptides than the more biologically relevant exposures reported here. The current studies do, however, provide a biosynthetic-based rationale for further experiments and identify likely protein targets of sterol-derived electrophiles. The fact that fibroblasts from SLOS patients give results with a-Lan that parallel the metabolic transformations found in Dhcr7-deficient Neuro2a cells (see Table 1), suggests that exploration of adduction in these primary cells will also be worthwhile.
The identification of sterol-protein covalent adducts along with the derived tryptic peptides bound to the sterol electrophile could provide biomarkers for pathologies associated with elevated 7-DHC such as SLOS. SLOS therapies that reduce 7-DHC levels or 7-DHC oxysterol formation, for example, could be monitored by targeted adduct-peptide analysis. By the same token, exposures that increase oxidative stress and/or endogenous levels of 7-DHC could result in the formation of 7-DHC-derived protein adducts that could be monitored by targeted analysis of derivative peptides. In this regard, it is of some interest that recent studies have found a link between exposure to a number of pharmaceuticals and plasma levels of 7-DHC [35], [36]. It seems likely that the result of such exposures will result in the formation of sterol adducts of the proteins identified in the current study, thus providing an additional stimulus for further investigation.
4. Material and methods
4.1. Synthetic procedures
1H and 13C NMR spectra were collected on a 300 or 400 MHz NMR. HRMS analyses were carried out at the University of Notre Dame. Purification by column chromatography was carried out on silica gel and TLC plates were visualized with UV and stained with phosphomolybdic acid. Lanosterol was purchased from Pfaltz & Bauer as a mixture of sterols. It contains approximately 55% lanosterol, 40% dihydrolanosterol, and 5% dihydroagnosterol. Ergosterol was purchased from TCI America. Full synthetic procedures for the alkynyl lipids are described in Supporting Information. All cell culture reagents were from Mediatech (Manassas, VA), Life Technologies (Grand Island, NY), and Greiner Bio-One GmBH (Monroe, NC).
4.2. Cell cultures: Neuro2a and human fibroblasts
The neuroblastoma cell line Neuro2a was purchased from American Type Culture Collection (Rockville, MD). Dhcr7-deficient Neuro2a cells were generated as previously described [37]. Control (GM05758) SLOS (GM05788) and NPC1 (GM18436) human fibroblasts were purchased from the Coriell Institute. All cultured control, SLOS, and NPC1 (Niemann-Pick disease, type C1) human fibroblasts used were passages of 8–20. All cells were subcultured once a week, and the culture medium was changed every two days. All cell lines were maintained in DMEM supplemented with L-glutamine, 10% fetal bovine serum (FBS; Thermo Scientific HyClone, Logan, UT), and penicillin/streptomycin at 37 °C and 5% CO2.
4.3. Alkynyl lipid treatment, extraction and HPLC-MS analysis of sterols
Neuro2a cells and Dhcr7-deficient Neuro2a cells were treated with alkynyl sterols for 24 h in a N2 supplemented medium and human fibroblasts were treated for 6 days in cholesterol-deficient medium. At the end of experiment the cells were pelleted as previously described [24]. To the cell pellets was added 5β-cholestan-3α-ol (36 nmol) as an internal standard, a Folch solution [600 μL, CHCl3:MeOH (2:1) containing 1 mM BHT and PPh3], and saline (300 μL, 0.9% aqueous NaCl). The resulting mixture was vortexed for 1 min and centrifuged at 3200 rpm for 1 min to separate the layers. The lower organic phase was recovered and divided into equal aliquots for LC-MS and GC analysis. One aliquot was concentrated on a SpeedVac concentrator and resuspended in MeOH (100 μL) for LC-MS analysis. Samples were analyzed on a Thermofinnigan LTQ or LCQ ion trap mass spectrometer using a Supelco Discovery C18 column (150×2.1 mm, 5 µM). The sterols were eluted with 100% MeOH (0.1% acetic acid) at a flow rate of 200 μL/min. The MS was operated in the positive-ion mode using atmospheric pressure chemical ionization (APCI). Response factors for each sterol relative to 5β-cholestan-3α-ol were determined: a-Lan (0.032), a-Lath (0.84), a−7-DHC (0.041), a-Chol (0.071), Lan (0.032), Lath (0.84), 7-DHC (0.030), and Chol (0.061). Since a-Chol and a-Lath coelute on HPLC, quantitation was carried out using the following equation with the Lath/Chol ratio determined by GC: (AreaLath×rfLath)/(AreaChol×rfChol)=Lath/Chol. This correction allows the contribution by a-Chol and a-Lath to the total area to be determined.
4.4. GC analysis of sterols
The remaining aliquot from above was concentrated on a SpeedVac concentrator and resuspended in BSTFA (100 μL) for GC analysis. Analysis was carried out on a Supelco DB-5 column with FID detection using the following temperature program: 220 °C to 275 °C (15 °C/min) to 280 °C (1 °C/min) maintained for 2 min to 290 °C (5 °C/min) and remain for 10 min. Response factors for each sterol relative to 5β-cholestan-3α-ol were determined: a-Lan (1.2), aLath (1.2), a-7-DHC (1.5), a-Chol (1.1), Lan (0.86), Lath (1.1), 7-DHC (0.98), and Chol (0.97).
4.5. Cell lysis and click reaction
The cells were lysed in 1 mL of lysis buffer (50 mM HEPES, 150 mM NaCl, 0.1% TritonX100, pH=7.0 containing protease inhibitors). The cells were sonicated and left on ice for 20 min then centrifuged at 15,000g for 10 min at 4 °C. The supernatant was collected and protein concentration determined (Thermo Scientific BCA kit, Rgt A 23221, Rgt B 23224). All protein concentrations were adjusted to 2 mg/mL and reduced with sodium borohydride (final concentration 5 mM) for 1 h on ice to stabilize adducts followed by quenching the reaction with 1.0 M HCl (adjust pH~6). The following click reagents were added to each of the samples: azido-biotin reagent (0.2 mM), tris(3-hydroxypropyltriazolylmethyl)amine (THPTA) ligand (0.2 mM), copper sulfate (1 mM), and sodium ascorbate (1 mM), and the samples were vortexed and allowed to rotate for 2 h at room temperature in the dark. Streptavidin visualization of modified proteins, protein capture, and photorelease were carried out as previously described [20].
4.6. Proteomics analysis
Filtrates of photoreleased proteins were collected and dried under vacuum. The dried photoreleased proteins were dissolved in a sample loading buffer (10 μL LDS+2 μL of 1 M DTT+28 μL PBS) and loaded onto SDS-Page and run for 4 min at 200 V. The gel was then stained, bands were excised and cut into cubes. The cubes were washed three times with 25 mM AMBIC and then added to 100 μL of 25 mM AMBIC containing DTT (final conc. 10 mM) and incubated at 50 °C for 10 min followed by iodoacetamide treatment (final conc. 20 mM) at RT for 10 min in the dark. The supernatant was removed and 100 μL of MeCN/25 mM AMBIC (1/1, v/v) was added. After 20 min, the liquid was removed and this procedure was repeated twice. After removal of the final supernatant, 100 μL 25 mM AMBIC containing 0.5 μg trypsin/sample was added and the mixture left at 37 °C for 20 h. After this time, an additional 0.2 μg of trypsin was added and the mixture was incubated at 37 °C for 4 h, after which the supernatant was collected. The gels were dehydrated twice with 50 μL of MeCN: 1% H2O (6:4) and all supernatants were combined and then dried on a SpeedVac concentrator. Peptides were re-suspended in 50 μL of 0.1% formic acid and 3 μL was injected on to an Eksigent NanoLC Ultra HPLC with a capillary reverse-phase analytical column. The analytical column was packed with C18 resin (Jupiter, 3 µm, 300 Å, Phenomenex) in a capillary column (20 cm length, 360 µm O.D.×100 µm I.D.). Peptides were eluted at a flow rate of 350 nL/min, and the mobile phase solvents consisted of water containing 0.1% formic acid (solvent A) and acetonitrile containing 0.1% formic acid (solvent B) with a 95 min gradient consisting of the following: 0–6 min, 2% B; 6–74 min, 2–99% B; 74–84 min, 99% B; 84–84.5 min, to 2% B; and 84.5–95 min, 2% B. Eluting peptides were mass analyzed on an Q Exactive hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Scientific), equipped with a nano-electrospray ionization source. The instrument was operated using a data dependent method with dynamic exclusion enabled. Full-scan (m/z 375–1500) spectra were acquired with the Orbitrap mass analyzer (resolution 70,000) and an automatic gain control (AGC) target is 3×106. The 15 most intense precursor ions in each MS scan were selected for data dependent mode for higher-energy collision dissociation (HCD) MS/MS analysis.
4.7. MS/MS data analysis and informatics tools
Mouse proteomes database (mouseRefSeq_150512.fasta; 25885 protein entry) was downloaded from the NCBI website (ftp://ftp.ncbi.nlm.nih.gov/refseq/release). Peptide Identification: An open source ProteoWizard msConvert tool [38] was used to convert RAW data to mzML format, followed by the use of MSGF [39], [40], generating mzid. MSGF was configured to have fixed modifications of +57.0215 for carbamidomethyl to Cys and dynamic modifications of +15.9949 to Met, and −17.0265 to N-term Gln. The sequence database was reversed so that each protein sequence appeared in both normal and reversed orientations, enabling false discovery rate estimation. The identified proteins and peptides from 3 biological replica were assembled and visualized using IDPicker 3.1.9288 64-bit [41]. Protein assembly criteria are as follows: 2% maximum FDR for peptide-spectrum match, 1 minimum spectra/peptide, 1 minimum spectra/match, 2 minimum distinct peptides/protein, 1 minimum additional peptide and 2 minimum spectra per protein. IDPicker reports were exported to QuasiTel [42] to calculate enrichment factors between DMSO control and alkynyl-sterol samples. The cut-off for QuasiTel analysis used was greater than 2 of log2 (count1*/count2*), which count1*=total spectral count (count1)+1 for alkynyl-sterol treated cells and count2*=total spectral count (count2)+1 for DMSO control cells. The majority of count2 values are zeros because there are no proteins adducted by alkynyl-sterols identified. An artificial number 1 is added for both counts to calculate enrichment factors in this case.
Notes
The authors declare no competing financial interest.
Acknowledgements
The National Institutes of Health (NICHD R01 HD064727 to NAP, NIEHS R01 ES024133 to NAP and ZK, R21 ES024666 to NAP).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2017.02.013.
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- 1.Nes W.D. Biosynthesis of cholesterol and other sterols. Chem. Rev. 2011;111(10):6423–6451. doi: 10.1021/cr200021m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.G.E. Herman, Disorders of cholesterol biosynthesis: prototypic metabolic malformation syndromes, Hum. Mol. Genet. 12 Spec No 1, 2003, R75-88. [DOI] [PubMed]
- 3.Porter F.D., Herman G.E. Malformation syndromes caused by disorders of cholesterol synthesis. J. Lipid Res. 2011;52(1):6–34. doi: 10.1194/jlr.R009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Platt F.M., Wassif C., Colaco A., Dardis A., Lloyd-Evans E., Bembi B., Porter F.D. Disorders of cholesterol metabolism and their unanticipated convergent mechanisms of disease. Ann. Rev. Genom. Hum. Gen. 2014;15:173–194. doi: 10.1146/annurev-genom-091212-153412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dias C., Rupps R., Millar B., Choi K., Marra M., Demos M., Kratz L.E., Boerkoel C.F. Desmosterolosis: an illustration of diagnostic ambiguity of cholesterol synthesis disorders. Orphanet J. Rare Dis. 2014;9(1):94. doi: 10.1186/1750-1172-9-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cross J.L., Iben J., Simpson C.L., Thurm A., Swedo S., Tierney E., Bailey-Wilson J.E., Biesecker L.G., Porter F.D., Wassif C.A. Determination of the allelic frequency in Smith-Lemli-Opitz syndrome by analysis of massively parallel sequencing data sets. Clin. Gen. 2015;87(6):570–575. doi: 10.1111/cge.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Witsch-Baumgartner M., Schwentner I., Gruber M., Benlian P., Bertranpetit J., Bieth E., Chevy F., Clusellas N., Estivill X., Gasparini G., Giros M., Kelley R.I., Krajewska-Walasek M., Menzel J., Miettinen T., Ogorelkova M., Rossi M., Scala I., Schinzel A., Schmidt K., Schonitzer D., Seemanova E., Sperling K., Syrrou M., Talmud P.J., Wollnik B., Krawczak M., Labuda D., Utermann G. Age and origin of major Smith-Lemli-Opitz syndrome (SLOS) mutations in European populations. J. Med. Gen. 2008;45(4):200–209. doi: 10.1136/jmg.2007.053520. [DOI] [PubMed] [Google Scholar]
- 8.Porter F. Smith-Lemli-Opitz syndrome: pathogenesis, diagnosis and management. Eur. J. Hum. Genet. 2008;16(5):535–541. doi: 10.1038/ejhg.2008.10. [DOI] [PubMed] [Google Scholar]
- 9.Wassif C.A., Zhu P., Kratz L., Krakowiak P.A., Battaile K.P., Weight F.F., Grinberg A., Steiner R.D., Nwokoro N.A., Kelley R.I., Stewart R.R., Porter F. Biochemical, phenotypic and neurophysiological characterization of a genetic mouse model of RSH/Smith–Lemli–Opitz syndrome. Hum. Mol. Genet. 2001;10(6):555–564. doi: 10.1093/hmg/10.6.555. [DOI] [PubMed] [Google Scholar]
- 10.Waterham H.R., Hennekam R.C.M. Mutational spectrum of Smith-Lemli-Opitz syndrome. Am. J. Med. Gen. 2012;160C(4):263–284. doi: 10.1002/ajmg.c.31346. [DOI] [PubMed] [Google Scholar]
- 11.Kanungo S., Soares N., He M., Steiner R.D. Sterol metabolism disorders and neurodevelopment—an update. Dev. Dis. Res. Rev. 2013;17:197–210. doi: 10.1002/ddrr.1114. [DOI] [PubMed] [Google Scholar]
- 12.Vaughan D.K., Peachey N.S., Richards M.J., Buchan B., Fliesler S.J. Light-induced exacerbation of retinal degeneration in a rat model of Smith-Lemli-Opitz syndrome. Exp. Eye Res. 2006;82(3):496–504. doi: 10.1016/j.exer.2005.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richards M.J., Nagel B.A., Fliesler S.J. Lipid hydroperoxide formation in the retina: correlation with retinal degeneration and light damage in a rat model of Smith-Lemli-Opitz syndrome. Exp. Eye Res. 2006;82(3):538–541. doi: 10.1016/j.exer.2005.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu L., Korade Z., Porter N.A. Oxysterols from free radical chain oxidation of 7-dehydrocholesterol: product and mechanistic studies. J. Am. Chem. Soc. 2010;132(7):2222–2232. doi: 10.1021/ja9080265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu L., Davis T.A., Porter N.A. Rate constants for peroxidation of polyunsaturated fatty acids and sterols in solution and in liposomes. J. Am. Chem. Soc. 2009;131(36):13037–13044. doi: 10.1021/ja9029076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korade Z., Xu L., Mirnics K., Porter N.A. Lipid biomarkers of oxidative stress in a genetic mouse model of Smith-Lemli-Opitz syndrome. J. Inherit. Metab. Dis. 2013;36:113–122. doi: 10.1007/s10545-012-9504-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korade Z., Xu L., Shelton R., Porter N.A. Biological activities of 7-dehydrocholesterol-derived oxysterols: implications for Smith-Lemli-Opitz syndrome. J. Lipid Res. 2010;51(11):3259–3269. doi: 10.1194/jlr.M009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Codreanu S.G., Ullery J.C., Zhu J., Tallman K.A., Beavers W.N., Porter N.A., Marnett L.J., Zhang B., Liebler D.C. Alkylation damage by lipid electrophiles targets functional protein systems. Mol. Cell. Proteom. 2014;13(3):849–859. doi: 10.1074/mcp.M113.032953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Codreanu S.G., Kim H.Y., Porter N.A., Liebler D.C. Biotinylated probes for the analysis of protein modification by electrophiles. Methods Mol. Biol. 2012;803:77–95. doi: 10.1007/978-1-61779-364-6_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim H.-Y.H., Tallman K.A., Liebler D.C., Porter N.A. An azido-biotin reagent for use in the isolation of protein adducts of lipid-derived electrophiles by streptavidin catch and photorelease. Mol. Cell. Proteom. 2009;8(9):2080–2089. doi: 10.1074/mcp.M900121-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofmann K., Thiele C., Schott H.F., Gaebler A., Schoene M., Kiver Y., Friedrichs S., Lutjohann D., Kuerschner L. A novel alkyne cholesterol to trace cellular cholesterol metabolism and localization. J. Lipid Res. 2014;55(3):583–591. doi: 10.1194/jlr.D044727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ciepla P., Konitsiotis A.D., Serwa R.A., Masumoto N., Leong W.P., Dallman M.J., Magee A.I., Tate E.W. New chemical probes targeting cholesterylation of Sonic Hedgehog in human cells and zebrafish. Chem. Sci. 2014;5(11):4249–4259. doi: 10.1039/c4sc01600a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hulce J.J., Cognetta A.B., Niphakis M.J., Tully S.E., Cravatt B.F. Proteome-wide mapping of cholesterol-interacting proteins in mammalian cells. Nat. Methods. 2013;10(3):259–264. doi: 10.1038/nmeth.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Windsor K., Genaro-Mattos T.C., Kim H.Y., Liu W., Tallman K.A., Miyamoto S., Korade Z., Porter N.A. Probing lipid-protein adduction with alkynyl surrogates: application to Smith-Lemli-Opitz syndrome. J. Lipid Res. 2013;54:2842–2850. doi: 10.1194/jlr.M041061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodríguez I.R., Fliesler S.J. Photodamage generates 7‐keto‐ and 7‐hydroxycholesterol in the rat retina via a free radical‐mediated mechanism. Photochem. Photobiol. 2009;85(5):1116–1125. doi: 10.1111/j.1751-1097.2009.00568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu L., Porter N.A. Reactivities and products of free radical oxidation of cholestadienols. J. Am. Chem. Soc. 2014;136(14):5443–5450. doi: 10.1021/ja5011674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin H., Xu L., Porter N.A. Free Radical Lipid Peroxidation: Mechanisms and Analysis. Chem. Rev. 2011;111(10):5944–5972. doi: 10.1021/cr200084z. [DOI] [PubMed] [Google Scholar]
- 28.Rantakari P., Lagerbohm H., Kaimainen M., Suomela J.P., Strauss L., Sainio K., Pakarinen P., Poutanen M. Hydroxysteroid (17{beta}) dehydrogenase 12 is essential for mouse organogenesis and embryonic survival. Endocrinology. 2010;151(4):1893–1901. doi: 10.1210/en.2009-0929. [DOI] [PubMed] [Google Scholar]
- 29.Takashima K., Saitoh A., Funabashi T., Hirose S., Yagi C., Nozaki S., Sato R., Shin H.W., Nakayama K. COPI-mediated retrieval of SCAP is crucial for regulating lipogenesis under basal and sterol-deficient conditions. J. Cell Sci. 2015;128(15):2805–2815. doi: 10.1242/jcs.164137. [DOI] [PubMed] [Google Scholar]
- 30.Ruiz J.L., Fernandes L.R., Levy D., Bydlowski S.P. Interrelationship between ATP-binding cassette transporters and oxysterols. Biochem. Pharmacol. 2013;86(1):80–88. doi: 10.1016/j.bcp.2013.02.033. [DOI] [PubMed] [Google Scholar]
- 31.Chang S., Ren G., Steiner R.D., Merkens L., Roullet J.B., Korade Z., DiMuzio P.J., Tulenko T.N. Elevated autophagy and mitochondrial dysfunction in the Smith-Lemli-Opitz syndrome. Mol. Genet. Metab. Rep. 2014;1:431–442. doi: 10.1016/j.ymgmr.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neve E.P., Nordling A., Andersson T.B., Hellman U., Diczfalusy U., Johansson I., Ingelman-Sundberg M. Amidoxime reductase system containing cytochrome b5 type B (CYB5B) and MOSC2 is of importance for lipid synthesis in adipocyte mitochondria. J. Biol. Chem. 2012;287(9):6307–6317. doi: 10.1074/jbc.M111.328237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benderdour M., Charron G., DeBlois D., Comte B., Des Rosiers C. Cardiac mitochondrial NADP+-isocitrate dehydrogenase is inactivated through 4-hydroxynonenal adduct formation: an event that precedes hypertrophy development. J. Biol. Chem. 2003;278(46):45154–45159. doi: 10.1074/jbc.M306285200. [DOI] [PubMed] [Google Scholar]
- 34.Jiang X.-S., Backlund P.S., Wassif C.A., Yergey A.L., Porter F.D. Quantitative proteomics analysis of inborn errors of cholesterol synthesis: identification of altered metabolic pathways in DHCR7 and SC5D deficiency. Mol. Cell. Proteom. 2010;9(7):1461–1475. doi: 10.1074/mcp.M900548-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall P., Michels V., Gavrilov D., Matern D., Oglesbee D., Raymond K., Rinaldo P., Tortorelli S. Aripiprazole and trazodone cause elevations of 7-dehydrocholesterol in the absence of Smith-Lemli-Opitz Syndrome. Mol. Genet. Metab. 2013;110(1–2):176–178. doi: 10.1016/j.ymgme.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 36.Kim H.-Y.H., Korade Z., Tallman K.A., Liu W., Weaver C.D., Mirnics K., Porter N.A. Inhibitors of 7-dehydrocholesterol reductase: screening of a collection of pharmacologically active compounds in Neuro2a cells. Chem. Res. Toxicol. Just Accept. Manuscr. 2016 doi: 10.1021/acs.chemrestox.6b00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korade Z., Kenworthy A.K., Mirnics K. Molecular consequences of altered neuronal cholesterol biosynthesis. J. Neurosci. Res. 2009;87(4):866–875. doi: 10.1002/jnr.21917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kessner D., Chambers M., Burke R., Agus D., Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24(21):2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim S., Pevzner P. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 2014;5:5277–5287. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim S., Gupta N., Pevzner P. Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J. Proteome Res. 2008;7:3354–3363. doi: 10.1021/pr8001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma Z., Dasari S., Chambers M., Litton M., Sobecki S., Zimmerman L., Halvey P., Schilling B., Drake P., Gibson B., Tabb D. IDPicker 2.0: Improved protein assembly with high discrimination peptide identification filtering. J. Proteome Res. 2009;8:3872–3881. doi: 10.1021/pr900360j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li M., Gray W., Zhang H., Chung C., Billheimer D., Yarbrough W., Liebler D., Shyr Y., Slebos R. Comparative shotgun proteomics using spectral count data and quasi-likelihood modeling. J. Proteome Res. 2010;9:4295–4305. doi: 10.1021/pr100527g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material