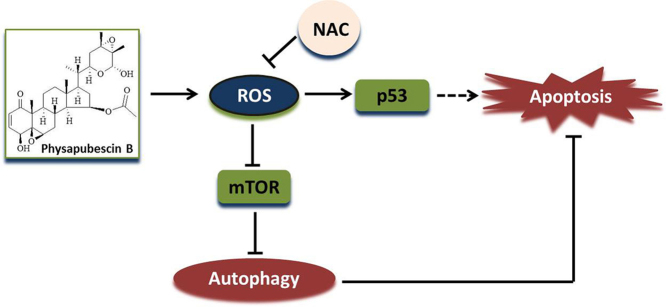
Abstract
Physapubescin B, a steroidal compound extracted from the plant Physalis pubescens L. (Solanaceae), has been reported to possess anti-cancer potential, whereas the molecular mechanism remains elusive. In this study, we first demonstrated that physapubescin B induced autophagy in human cancer cells based on the evidence that physapubescin B increased lipidation of microtubule-associated protein 1 light chain 3 (LC3) as well as number of GFP-LC3 puncta. We further examined the molecular mechanisms and found that physapubescin B enhanced the autophagic flux through promotion of reactive oxygen species (ROS)-mediated suppression of mammalian target of rapamycin complex I (mTORC1), the key negative regulator of autophagy. Additionally, excessive ROS caused by physapubescin B also induced p53-dependent apoptotic cell death. Furthermore, we provided evidence that inhibition of autophagy either by a chemical inhibitor or gene silencing promoted physapubescin B-induced apoptotic cell death, indicating that autophagy serves as a cell survival mechanism to protect cell death. Thus, our data provide a clue that inhibition of autophagy would serve as a novel strategy for enhancing the anti-cancer potential of physapubescin B.
Abbreviations: GβL, G protein beta subunit-like; PRAS40, Proline-rich AKT1 substrate 40; Raptor, regulatory-associated protein of mTOR
Keywords: Physapubescin B, Autophagy, ROS, MTORC1, Apoptotic cell death
Graphical abstract
Highlights
-
•
Physapubescin B dramatically induces autophagy as well as apoptosis in human cancer cell lines HCT116 and HeLa.
-
•
Physapubescin B suppresses mTORC1 and induces autophagy through ROS generation.
-
•
Physapubescin B-induced cell death is partly dependent on ROS-p53 axis.
-
•
Treatment with autophagy inhibitor or knockdown of autophagy-related gene could enhance physapubescin B-induced cell death.
1. Introduction
Physalis pubescens L. (Solanaceae) is an herbal plant distributed abundantly worldwide. Its calyces have been widely used in traditional Chinese medicine due to the high abundance of steroids, among which withanolides are the major steroidal constituents [1], [2]. In the past several decades, more than a dozen withanolides were isolated from Physalis species such as Physalis angulata, Physalis alkekengi, Physalis lancifolia and are shown to have anti-inflammatory [3], antimicrobial [4], [5], antiparasitic [6], immunomodulatory [7] and anti-tumor [8], [9] effects. Physapubescin B (C30H42O8, MW. 530) is one of the withanolides extracted from Physalis pubescens L. (Solanaceae), which possesses quinone reductase induction activity and inhibits the proliferation of mouse hepatoma Hepa1c1c7 cells [10]. It has also been reported to exhibit anti-tumor activity against human prostate cancer involving the G2/M phase cell cycle arrest [11]. Besides, its isomer physapubescin has been shown to inhibit the viability of renal cell carcinoma (RCC) cells through down-regulation of Hypoxia Inducible Factor (HIF)−2α [12]. At present, the exact mechanisms underlying the anti-cancer potential of physapubescin B remain to be further investigated.
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved cellular catabolic process responsible for degrading damaged organelles and long-lived proteins in response to stress conditions such as starvation (nutrient deprivation) so as to maintain cell homeostasis [13], [14]. A set of autophagy-related genes (Atg genes) are involved in the process of autophagy: Initiation, nucleation, maturation and fusion of autophagosome with lysosome for degradation [15], [16]. Up to date, it has been well established that autophagy plays a key role in a variety of cellular processes such as cell stress response, metabolism and cell death/survival [17], [18]. More importantly, autophagy is closely involved in the etiology of many important human diseases such as infectious diseases, neurodegenerative diseases and cancers [19]. At present, the role of autophagy in cancer remains controversial. In the early stage, autophagy is an important anti-cancer mechanism to prevent cancer initiation, while autophagy is believed to support cancer promotion and progression via its pro-survival function in cancer cells [20].
Autophagy is known to be tightly regulated by a network of upstream signaling cascades [21]. Among them, the mammalian target of rapamycin (mTOR) has been identified as a critical negative regulator of autophagy [22], [23]. mTOR is a serine/threonine protein kinase and serves as a key component of two functionally distinct complexes, mTORC1 and mTORC2, depending on their respective binding partners. mTORC1 comprises mTOR, GβL, PRAS40 and Raptor and plays a bigger role in the regulation of autophagy [24]. The Atg1-Atg13-FIP200 complex is essential in autophagosome formation. Activated mTORC1 leads to phosphorylation of Atg13 which prevents its binding with Atg1 so as to disrupt autophagosome formation and consequently inhibit autophagy [25].
Reactive oxygen species (ROS) are produced as natural byproducts during the metabolism of oxygen and play a vital role in cellular homeostasis. In addition to endogenous sources, ROS level can also increase due to stress such as UV, heat exposure and chemical stimulation [26]. ROS are known to play important roles in various physiological and pathological processes such as autophagy and cell death [27], [28], [29]. The regulation of autophagy by ROS can be summarized as transcriptional and post-transcriptional regulation. As to transcriptional regulation, cellular accumulation of ROS activates transcription factors such as p53, HIF-1, Nuclear factor-like 2 (NRF2) and Forkhead box O3 (FOXO3) which up-regulate the transcription of several proteins involved in autophagy [30]. For post-transcriptional regulation, mounting evidence suggests that the down-regulation of mTOR activity is associated with ROS generation. ROS may inhibit mTOR activity through PI3K/Akt pathway [31], AMPK [32] or a BNIP3-dependent manner [33] to induce autophagy. Direct oxidation and inhibition of Atg4 by ROS have also been reported [34]. Autophagy, in turn, contributes to ROS elimination under various stress conditions [35].
In this study, we elucidated the effect of physapubescin B on autophagy and the underlying mechanisms. Our data demonstrate that physapubescin B promotes intracellular ROS generation, leading to mTORC1 inhibition and autophagy induction. Suppression of autophagy is able to enhance physapubescin B-induced apoptotic cell death, indicating the pro-survival function of autophagy. Our study thus identifies a novel function of a natural product physapubescin B and indicates that suppression of autophagy is able to enhance its anti-cancer potential.
2. Materials and methods
2.1. Compounds
Physapubescin B was isolated from the dried fruits of Physalis pubescens L. in our laboratory and was identified in our previous paper [10]. We have determined its purity to be 98% by HPLC. The compound was dissolved in DMSO to make a 100 mM stock solution, aliquoted and stored at -30 ℃.
2.2. Reagents and antibodies
We used the following reagents in our experiments: Z-VAD (OMe)-FMK (Santa Cruz, CAS 187389-52-2), Chloroquine diphosphate (CQ, Sigma, C6628), N-Acetyl Cysteine (NAC, Merck), Propidium Iodide (PI, Sigma P4170), CM-H2DCFDA (Invitrogen, C6827). The antibodies in our experiments include: anti-PARP antibody (CST, 9542), anti-caspase-3 antibody (CST, 9662), anti-β-actin antibody (Sigma, A5441), anti-microtubule-associated protein 1 light chain 3/LC3 antibody (Sigma, L7543), anti-phospho-p70 S6 Kinase (Thr389) antibody (CST, 9205), anti-p70 S6 Kinase antibody (CST, 9202), anti-phospho-4E-BP1 (Ser65) antibody (CST, 2855 P), anti-4E-BP1 antibody (CST, 9644), anti-ATG7 antibody (CST, 2631), anti-p53 antibody (PharMingen, 554170), anti-p21 Wat 1/Cip1 (DCS60) antibody (CST, 2946).
2.3. Cell culture
HCT116 cells and HeLa cells were obtained from American Type Culture Collection (ATCC). HeLa cells stably expressing GFP-LC3 were kindly provided by Dr. N Mizushima (University of Tokyo, Japan). An isogenic pair of HCT116 colon cancer cell lines (tp53+/+ and tp53-/-) was a gift from Dr. Bert Vogelstein (Johns Hopkins University, USA). All cell lines were maintained in DMEM (Sigma, D1152) containing 10% fetal bovine serum (HyClone, SV30160.03) and 1% penicillin/streptomycin (Invitrogen) in a 5% CO2 atmosphere at 37 °C.
2.4. Western blotting
After all the designated treatments, cells were collected and rinsed with chilled PBS, then lysed by the Laemmli buffer (62.5 mM Tris-HCl, pH 6.8, 20% glycerol, 2% SDS, 2 mM DTT, phosphatase inhibitor and proteinase inhibitor cocktail). Cell lysates were boiled for 10 min. An equal amount of protein was resolved by SDS-PAGE gel and transferred onto PVDF membrane (Bio-Rad, 162–0177). Membranes were blocked in blocking buffer (Thermo Scientific, 37538) for 30 min, then followed by incubation with the designated first and second antibodies and finally developed with the enhanced chemiluminescence method (Thermo Scientific, 34076) and visualized by a Kodak Image Station 4000 R (Kodak). The expression of proteins was quantitatively evaluated by ImageJ.
2.5. Detection of cell death
Cell death was estimated qualitatively by morphological changes under a microscope (Leica) and quantitatively by propidium iodide (PI, 5 µg/ml) live cell exclusion assay coupled with flow cytometry (BD Biosciences). Western blot was also used to detect PARP and caspase-3 cleavage.
2.6. Confocal imaging
GFP-LC3 puncta were observed under the Olympus Fluoview FV1000 Confocal Microscope. Cells were seeded in an 8-well Lab-Tek™ Chambered Coverglass (Thermo Scientific, 155411). After the designated treatment, images were taken with 60x objective lens and analyzed by ImageJ.
2.7. Small interfering RNA (siRNA) and transient transfection
The Atg7 siRNA (Dharmacon, SMARTpool, ON-TARGET plus human ATG7, L-020112-00-0005, Target Sequences:
CCAACACACUCGAGUCUUU; GAUCUAAAUCUCAAACUGA; GCCCACAGAUGGAGUAGCA; GCCAGAGGAUUCAACAUGA) and scramble siRNA (Cell Signaling Technology, 6242) were transfected into HCT116 cells by Lipofectamine 3000 (Life Technologies, L3000015) according to the manufacturer's instructions.
2.8. Measurement of ROS production
Cells were cultured in a 12-well plate and treated as indicated. CM-H2DCFDA probe (Life technologies, C6827) was added 30 min before harvest. Images were captured by a Leica microscope to show the intensity of green fluorescence. The cells were then digested by trypsin and subjected to flow cytometry system (BD Biosciences) for analysis.
2.9. Statistical analysis
All the statistical data are presented as means±SD from 3 independent experiments and analyzed using the Student's t-test to determine the differences which are considered statistically significant at P≤0.05.
3. Results
3.1. Physapubescin B induces autophagy
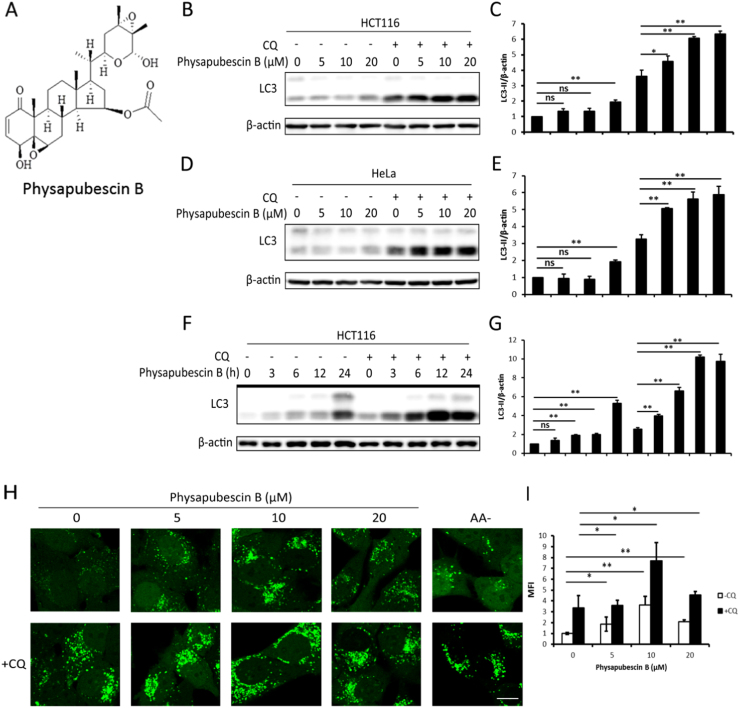
We first evaluated the effect of physapubescin B on autophagy. Fig. 1A shows the structure of physapubescin B. As shown in Fig. 1B, C, physapubescin B markedly increased the lipidation of LC3 (LC3-II) in a dose-dependent manner. Since both autophagy induction and blockage of autophagosomal degradation cause LC3-II accumulation, we inhibited autophagosomal degradation by the lysosome inhibitor chloroquine (CQ) and found a further increase in LC3-II, indicating an increase in autophagic flux [22] (Fig. 1B, C). Similar results were observed in HeLa cells (Fig. 1D, E). In addition, the effect of physapubescin B on autophagy induction was also time-dependent, as shown by the increase in LC3‑II in a time-dependent manner (Fig. 1F, G). Moreover, we used HeLa cells stably expressing GFP-LC3 to further confirm the effect of physapubescin B on autophagy induction. The GFP-LC3 puncta were used to represent autophagosomes [36]. As shown in Fig. 1H, there was a remarkable increase of GFP-LC3 puncta after physapubescin B treatment, which was further increased by the presence of CQ. The number of GFP-LC3 puncta was even comparable to amino acid starvation in the presence of CQ. The data in Fig. 1H were quantified and presented in Fig. 1I. All these data show that physapubescin B is a potent autophagy inducer.
Fig. 1.
Physapubescin B increases autophagic flux. (A) The chemical structure of physapubescin B. (B) HCT116 (D) HeLa cells were treated with the indicated doses of physapubescin B with or without chloroquine (CQ, 50 μM) for 3 h. Cell lysates were collected and subjected to western blot for detecting LC3 conversion. β-actin was used as a loading control. (C) HCT116 (E) HeLa proteins expression from (B) and (D) was evaluated by ImageJ, means±SD were presented (*, P<0.05; **, P<0.01; ns, P>0.05; Student's t-test). (F) HCT116 cells were treated with physapubescin B (20 μM) for the indicated time points with or without CQ (50 μM). Cell lysates were collected and subjected to western blot for detecting LC3 conversion. β-actin was used as a loading control, and (G) the proteins expression was evaluated by ImageJ, means±SD were presented (**, P<0.01; ns, P>0.05; Student's t-test). (H) HeLa cells stably expressing GFP-LC3 were incubated with the indicated concentrations of physapubescin B with or without CQ (50 μM) for 3 h. Autophagosomes were inferred by the presence of GFP-LC3 puncta under confocal microscopy (×600). Scale bar, 5 µm. Amino acid starvation (AA-) was used as a positive control for induction of autophagy. (I) The mean fluorescence intensity (MFI) was analyzed by ImageJ, means±SD were presented (*, P<0.05; **, P<0.01, Student's t-test).
3.2. Physapubescin B inhibits mTORC1 activity
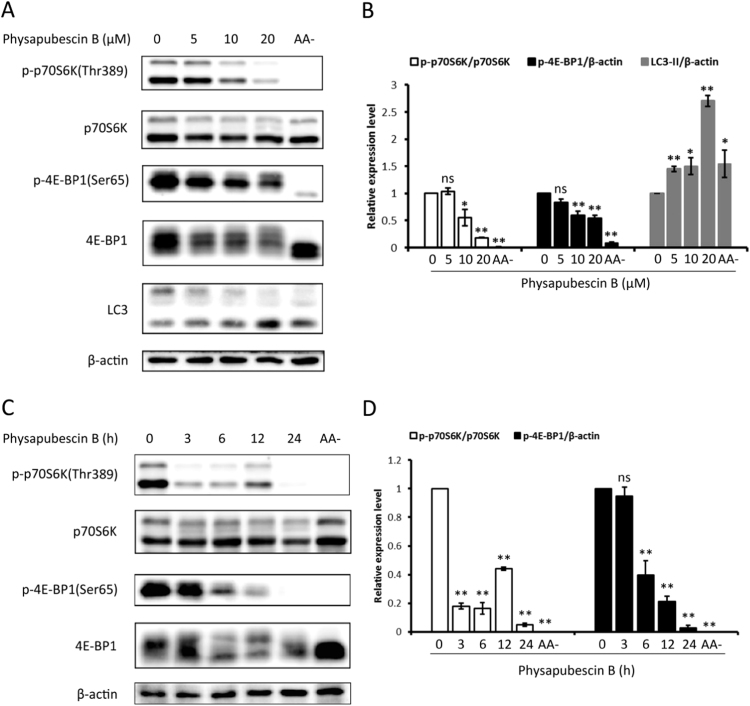
The mTORC1 complex is a negative regulator of autophagy by phosphorylating and inhibiting the function of the ULK1 complex [22], [23], [25]. We next investigated the effect of physapubescin B on the mTORC1 signaling pathway. The p70 S6 kinase (p70S6K) and the translation repressor protein 4E-BP1 are direct substrates of mTORC1. Specifically, the mTORC1 phosphorylates p70S6K at Thr389 and 4E-BP1 at Ser65. Therefore, the phosphorylation status of p70S6K (Thr389) and 4E-BP1 (Ser65) can be used to determine the mTORC1 activity. As shown in Fig. 2A, B and 2 C, D, treatment with physapubescin B led to a significant reduction in p-p70S6K (Thr389) and p-4E-BP1 (Ser65) in a dose-dependent and time-dependent manner. The suppressive effect of physapubescin B on mTORC1 was comparable to amino acid starvation. Collectively, these data indicate the possibility that physapubescin B may induce autophagy by suppression of mTORC1.
Fig. 2.
Physapubescin B inhibits mTORC1 activity. (A) HCT116 cells were treated with the indicated concentrations of physapubescin B for 3 h. Cell lysates were collected and subjected to western blot. Amino acid-free DMEM (AA-, 3 h) was used as a positive control for mTORC1 suppression. And (B) proteins expression was evaluated by ImageJ, means±SD were presented (*, P<0.05; **, P<0.01; ns, P>0.05; Student's t-test). (C) HCT116 cells were treated with physapubescin B (20 μM) for various time points as indicated or amino acid-free DMEM (AA-) for 3 h. Cell lysates were collected for western blot analysis. And (D) proteins expression was evaluated by ImageJ, means±SD were presented (**, P<0.01; ns, P>0.05; Student's t-test).
3.3. Physapubescin B suppresses mTORC1 and induces autophagy through ROS generation
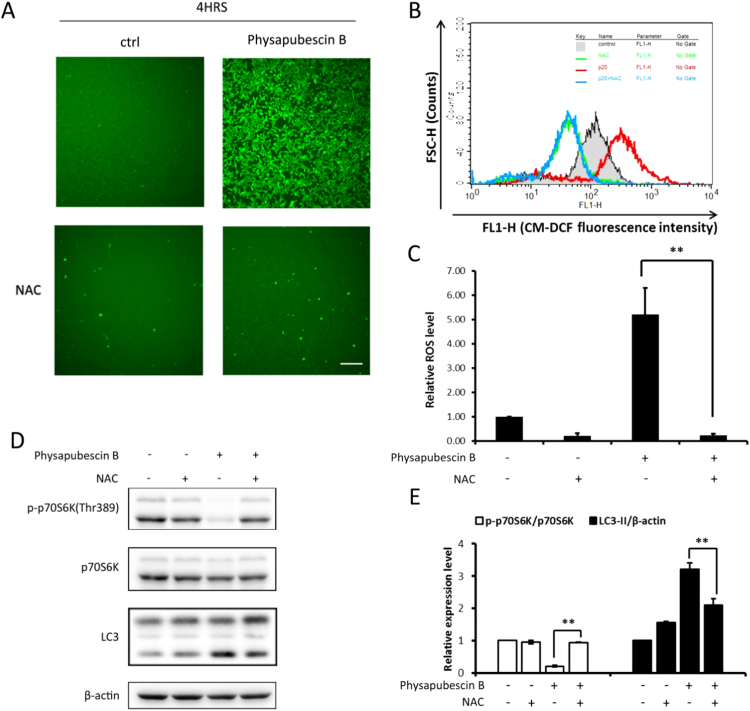
In order to understand the mechanisms underlying the inhibitory effect of physapubescin B on mTORC1, here we measured the change of ROS level in physapubescin B-treated cells using the CM-H2DCFDA probe [37]. As shown in Fig. 3A, a significant increase of green fluorescence representing ROS production was observed after physapubescin B treatment. N-Acetyl Cysteine (NAC) is a cell penetrating antioxidant that replenishes intracellular GSH so as to prevent the cells from oxidative stress [38]. Co-treatment of NAC totally blocked the ROS generation (Fig. 3A). Therefore, physapubescin B can induce ROS generation. Measuring the CM-H2DCFDA fluorescence intensity by flow cytometry further supported this conclusion (Fig. 3B, C). Based on the fact that physapubescin B suppresses mTORC1 activity and consequently activates autophagy, as shown earlier, and that ROS are known to induce autophagy [28], we then tested whether physapubescin B's effect on mTORC1 and autophagy was mediated via increasing ROS production. To do this, we treated HCT116 cells with physapubescin B and NAC. NAC treatment abolished physapubescin B-induced mTORC1 suppression, and partly reduced the LC3-II level (Fig. 3D, E). Therefore, data from this part of our study indicate that physapubescin B suppresses mTORC1 and induces autophagy through ROS generation.
Fig. 3.
Physapubescin B suppresses mTORC1 and induces autophagy through ROS generation. (A) HCT116 cells were treated with physapubescin B (20 μM) with or without NAC (5 mM) for 4 h followed by incubation with CM-H2DCFDA. The cells were then observed under a fluorescence microscope (x100). Scale bar, 200 µm. (B) HCT116 cells were treated as in (A). The fluorescence intensity was analyzed by flow cytometry. (C) Statistical analysis of the fluorescence intensity of HCT116 cells as treated in (A). Data (mean±SD) are representative of three independent experiments. (**, P<0.01, Student's t-test). (D) HCT116 cells were treated as in (A). Cell lysate was harvested and subjected to western blot, and (E) proteins expression was evaluated by ImageJ, means±SD were presented (**, P<0.01, Student's t-test).
3.4. Physapubescin B induces apoptotic cell death
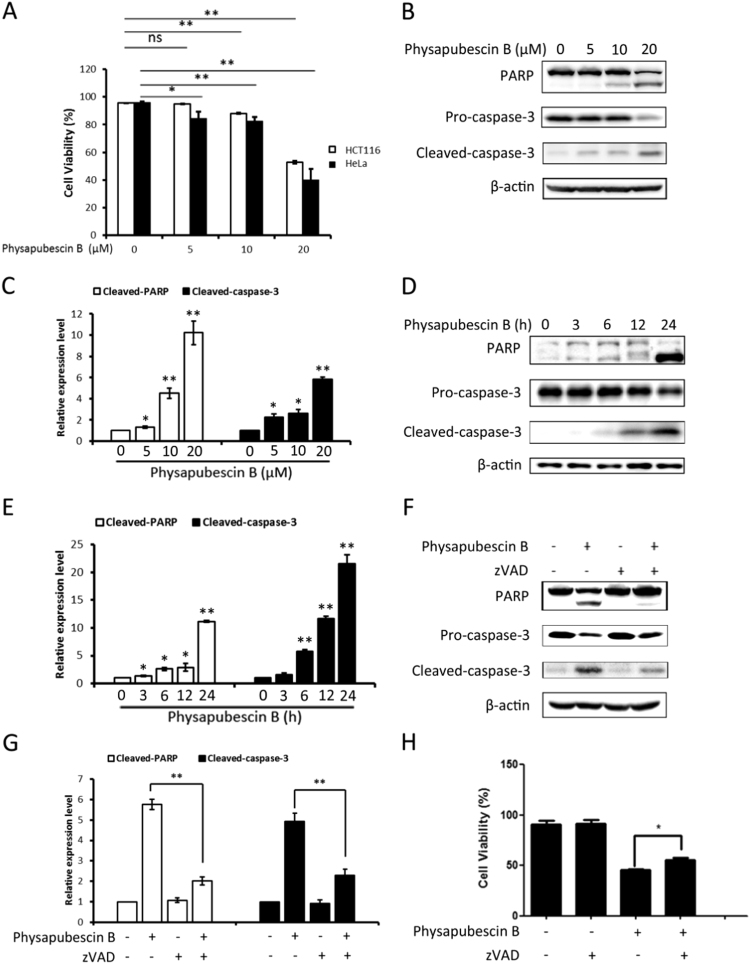
To study the effect of physapubescin B on cell viability, we then evaluated the cytotoxicity of physapubescin B on HCT116 cells and HeLa cells. As shown in Fig. 4A, physapubescin B reduced the cell viability in a dose-dependent manner, indicating the cytotoxic effect of physapubescin B. To investigate the type of cell death induced by physapubescin B, we treated HCT116 cells with physapubescin B and observed an increase in PARP and caspase-3 cleavage in a dose-dependent (Fig. 4B, C) and time-dependent manner (Fig. 4D, E), indicating apoptotic cell death. Z-Val-Ala-Asp(OMe)-fluoromethylketone (zVAD) is an irreversible and cell permeable broad-spectrum caspase inhibitor. To further confirm the pro-apoptotic effect of physapubescin B, we next investigated whether zVAD could abolish the PARP and caspase-3 cleavage caused by physapubescin B. As shown in Fig. 4F, G, zVAD dramatically blocked PARP and caspase-3 cleavage even in the presence of physapubescin B. Further, the cell death caused by physapubescin B could be partially rescued by zVAD treatment (Fig. 4H). Together, our data suggest that physapubescin B can effectively induce apoptotic cell death in human cancer cells.
Fig. 4.
Physapubescin B induces apoptotic cell death. (A) HCT116 and HeLa cells were treated with physapubescin B (0 5 10 20 μM) for 24 h. After treatment, the cells were then followed by propidium iodide (PI, 5 μg/ml) live cell exclusion staining for quantification of cell death. Data (mean±SD) are representative of three independent experiments. (*, P<0.05; **, P<0.01; ns, P>0.05; Student's t-test). (B) HCT116 cells were treated with physapubescin B (0 5 10 20 μM) for 24 h. The cells were then harvested and subjected to western blot analysis, and (C) proteins expression was evaluated by ImageJ, means±SD were presented (*, P<0.05; **, P<0.01, Student's t-test). (D) HCT116 cells were harvested after treatment with physapubescin B (20 μM) for the indicated periods of time, western blot was then performed for the detection of various apoptosis markers. β-actin was used as a loading control. And (E) proteins expression was evaluated by ImageJ, means±SD were presented (*, P<0.05; **, P<0.01, Student's t-test). (F) HCT116 cells were treated with physapubescin B (20 μM) alone or combined with zVAD (20 μM) for 24 h followed by western blot to detect apoptosis markers, and (G) proteins expression was evaluated by ImageJ (**, P<0.01, Student's t-test). (H) HCT116 cells were treated as in (F) and the cell death was quantified by flow cytometry after PI staining (*, P<0.05, Student's t-test).
3.5. Physapubescin B-induced cell death is partly dependent on ROS-p53 axis
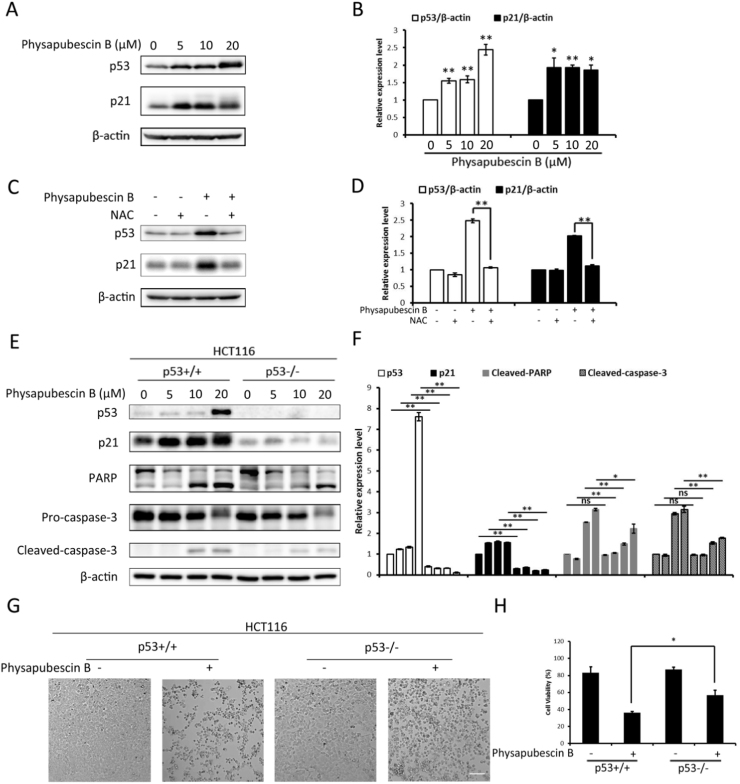
p53-mediated apoptosis is a very well established central mechanism for tumor suppression. We next aimed to study whether the apoptotic cell death induced by physapubescin B was associated with p53 function. We observed increased protein level of p53 as well as its target gene, p21 post physapubescin B treatment in a dose-dependent manner, indicating an activation of the tumor suppressor p53 (Fig. 5A, B). We then treated cells with physapubescin B alone or combined with NAC and p53 activation can be reversed by NAC co-treatment, implying p53 was activated by elevated ROS level (Fig. 5C, D). To directly investigate the relevance of p53 to physapubescin B-induced cell death, we used an isogenic pair of HCT116 colon cancer cell lines (tp53+/+ and tp53-/-). Notably, we observed less PARP and caspase-3 cleavage in HCT116 (tp53-/-), indicating the cell death caused by physapubescin B was at least partly dependent on p53 (Fig. 5E, F). Consistently, the cell viability increased by 20% in HCT116 (tp53-/-) compared to HCT116 (tp53+/+), but there was still some cell death caused by physapubescin B in p53 KO cells (Fig. 5G, H), suggesting p53 is necessary but not sufficient in physapubescin B-induced cell death.
Fig. 5.
Physapubescin B-induced cell death is partly dependent on ROS-p53 axis. (A) HCT116 cells were incubated with physapubescin B (0 5 10 20 μM) for 24 h, western blot analysis was performed to detect p53, p21 levels, and (B) proteins expression was evaluated by ImageJ, means±SD were presented (*, P<0.05; **, P<0.01, Student's t-test). (C) HCT116 cells were treated with physapubescin B (20 μM) with or without NAC (5 mM) for 24 h. Protein levels of p53, p21 and β-actin were evaluated by western blot, and (D) proteins expression was evaluated by ImageJ, means±SD were presented (**, P<0.01, Student's t-test). (E) HCT116 p53+/+(WT) and p53-/- (KO) cells were treated as indicated, cell lysate was collected and subjected to western blot, and (F) proteins expression was evaluated by ImageJ, means±SD were presented (*, P<0.05; **, P<0.01; ns, P>0.05; Student's t-test). (G) HCT116 p53+/+(WT) and p53-/- (KO) cells were treated with physapubescin B (20 μM) for 24 h. Cell morphology was observed by a microscope (x200). Scale bar, 100 µm. (H) Cell viability of (G) was quantified by flow cytometry combined with PI staining. Data (mean±SD) are representative of three independent experiments (*, P<0.05, Student's t-test).
3.6. Physapubescin B-induced autophagy acts as a cell survival mechanism
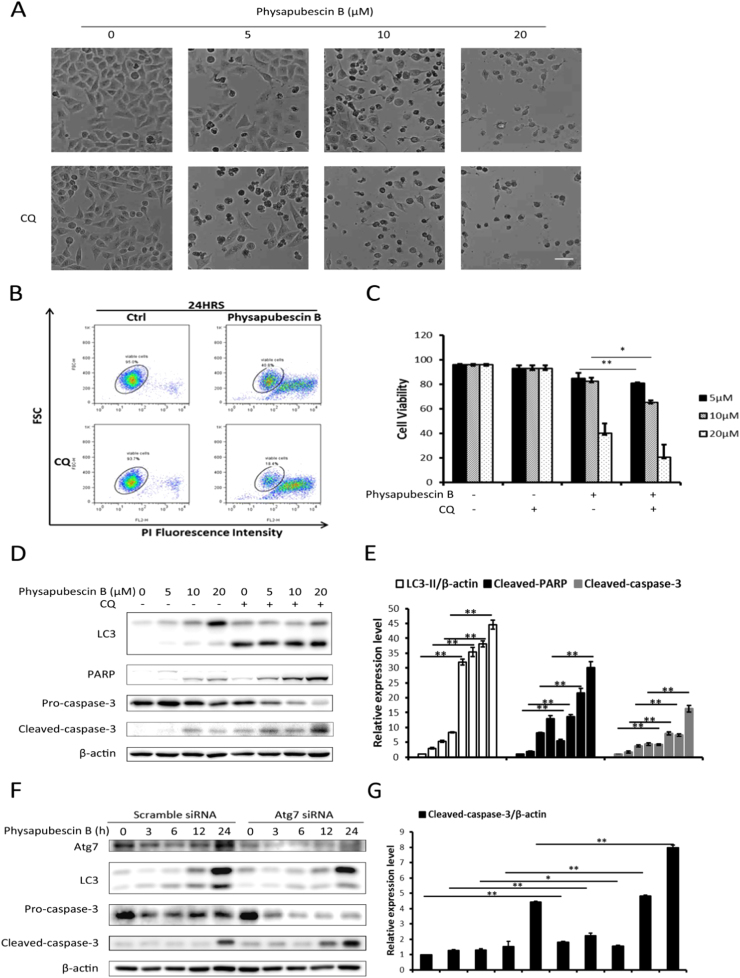
In this part of our study, we aimed to elucidate the role of autophagy in physapubescin B-induced cell death. Firstly, we used a lysosome inhibitor chloroquine (CQ) to inhibit autophagy. As shown in Fig. 6A, CQ treatment significantly increased cell death induced by physapubescin B. Quantitative data of cell death by flow cytometry further confirmed this result (Fig. 6B, C). In addition, the western blot result showed that CQ plus physapubescin B treatment strongly enhanced the cleavage of PARP and caspase-3, indicating an increase in apoptotic cell death post inhibition of autophagy (Fig. 6D, E). Moreover, knock down of Atg7, which strongly inhibits autophagy, resulted in an increase in PARP and caspase-3 cleavage, indicating autophagy inhibition enhanced physapubescin B-induced cell death (Fig. 6F, G). In summary, these data suggest that autophagy acts as a cell survival mechanism in physapubescin B-induced cell death.
Fig. 6.
Physapubescin B-induced autophagy acts as a cell survival mechanism. (A) HeLa cells were treated with the indicated doses of physapubescin B with or without CQ (50 μM) for 24 h. Cell morphology was observed under a microscope. Scale bar, 50 µm. (B) Shown are representative dot-plots of flow cytometry data of the PI exclusion assay. HeLa cells were incubated in physapubescin B (20 μM) with or without CQ (50 μM) for 24 h. (C) Quantification of the cell viability data from panel A is shown. Data were presented as the means±SD of three independent experiments (*, P<0.05; **, P<0.01, Student's t-test). (D) HeLa cells were treated as in (A), cell lysate was collected for western blot analysis, and (E) proteins expression was evaluated by ImageJ, means±SD were presented (**, P<0.01, Student's t-test). (F) HCT116 cells were transiently transfected with a nonspecific siRNA or Atg7-targeted siRNA for 48 h and treated with physapubescin B (20 μM) for the indicated time periods. Cell lysate was harvested and subjected to western blot, and (G) proteins expression was evaluated by ImageJ, means±SD were presented (*, P<0.05; **, P<0.01, Student's t-test).
4. Discussion
Recently, the commonly used clinical therapies for cancer comprise physical treatment such as radiotherapy, surgical therapy as well as chemical treatment [39]. However, the effect of these methods is not always satisfactory. The number of compounds in clinical practice is quite limited. Some patients are even not sensitive to the existing chemotherapeutics [40]. Thus it is urgent to develop more pharmacologically active compounds. Natural products are widely used in traditional Chinese medicine and have been verified to possess a wide variety of biological activities, including anti-cancer effect [41]. They could also be taken as the leading compounds for structural modification in drug development.
Previous studies have revealed that, physapubescin B, a steroidal compound extracted from Physalis pubescens L. (Solanaceae), has an anti-proliferative activity against different cancer cell lines with unclear mechanisms [10], [11]. Here we firstly observed more GFP-LC3 puncta in HeLa cells stably expressing GFP-LC3 when treated with physapubescin B with or without CQ, indicating an increase of autophagic flux (Fig. 1H). Decreased phosphorylation of mTORC1 substrates revealed that mTORC1 was suppressed by physapubescin B (Fig. 2). There are already a number of reports showing that ROS may regulate autophagy via mTOR-dependent mechanisms [31], [32], [33]. Withanolides isolated from Physalis pubescens L. (Solanaceae) such as physalin A [42], [43], physalin F [44] are often shown to induce ROS production. We then used CM-H2DCFDA probe to confirm this speculation. ROS level was notably increased by physapubescin B (Fig. 3A). Furthermore, the suppression of mTORC1 activity and the increase of autophagic flux were caused by elevated ROS level (Fig. 3D). It is the first report to link physapubescin B with autophagy and to establish the critical role of oxidative stress in this process.
Consistently with previous studies [10], [11], we also observed obvious cytotoxicity of physapubescin B in two cancer cell lines, human colorectal cancer cell line HCT116 and human cervical carcinoma cell line HeLa (Fig. 4A). We observed an increase in PARP and caspase-3 cleavage, indicating that apoptotic cell death was induced by physapubescin B (Fig. 4B, D). We then used a pan-caspase inhibitor zVAD to rescue the cell death. Although zVAD dramatically blocked the cleavage of PARP and caspase-3 (Fig. 4F), it failed to block the cell death induced by physapubescin B to a substantial extent (Fig. 4H). Such a finding indicates the possibility that some other death mechanisms may also involve in this process.
The cell morphological change caused by physapubescin B occurred within 6 h. This rapid change promoted us to explore the underlying mechanisms by which physapubescin B induced cell death. p53 activation was detected with physapubescin B treatment (Fig. 5A). ROS were shown to be the upstream of p53 (Fig. 5C). We then introduced an isogenic pair of HCT116 colon cancer cell lines (tp53+/+ and tp53-/-) and found that cell death was largely attenuated in p53 KO cells (Fig. 5H), suggesting that physapubescin B-induced cell death was dependent on ROS-p53 axis. Physapubescin B has been shown to possess quinone reductase induction activities so that it can conjugate with intracellular GSH [10]. This probably accounts for the dramatically elevated ROS caused by physapubescin B. Other withanolides possess an α, β-unsaturated ketone moiety in the A ring may have the similar biological activities. Ding et al. reported that physapubescin B induced ROS-triggered G2/M cell cycle arrest in PC3 cells and Du145 cells at a lower concentration [11]. Given these evidence, we surmise that lower dose of physapubescin B induces G2/M cell cycle arrest mediated by Cdc25C degradation. But when oxidative stress is aggravated by higher dose of physapubescin B, apoptosis pathway would be triggered. ROS-p53 axis is likely to play a pivotal role in both of these two processes. And that's the main mechanism that physapubescin B can be anti-cancer. However, although less, PARP and caspase-3 cleavages were still observed in p53 KO cells (Fig. 5E). p53 can partially account for the apoptotic cell death. The exact mechanisms by which physapubescin B induces apoptosis remain to be further explored. This observation indicates a possibility that physapubescin B may also be effective in other human cancers which are p53 mutant.
In the last part, we provided the evidence showing that physapubescin B-induced autophagy acted as a protective role based on the observations that either autophagy inhibitor CQ treatment or knockdown of Atg7 was able to exacerbate physapubescin B-mediated cell death (Fig. 6). Accumulated ROS are able to trigger autophagy pathway which, in turn, eliminates excessive ROS to maintain cellular homeostasis. This probably indicates a self-protective feedback loop for tumor cells. Recently, autophagy inhibitors such as CQ have been subjected to clinical trials as chemoradiotherapeutic sensitizers [45]. Our convincing evidence also points out that the anti-cancer potential of physapubescin B can be enhanced when combined with autophagy inhibitors.
In summary, we report that physapubescin B, a natural product from fruit, induces cancer cell death mediated by ROS. At the same time, the increased ROS induce autophagy via mTORC1 inhibition, which promotes cancer cell survival. Thus the combination of physapubescin B with autophagy inhibitors would be a promising strategy for cancer therapy.
Conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgements
We would like to express our heartfelt gratitude to Dr. N Mizushima (University of Tokyo, Japan) for providing the HeLa cells stably expressing GFP-LC3 and Dr. Bert Vogelstein (Johns Hopkins University, USA) for providing an isogenic pair of HCT116 colon cancer cell lines (tp53+/+ and tp53-/-). This work was supported by the National Natural Science Foundation of China [grant numbers 31471297, 81302455].
Contributor Information
Han-Ming Shen, Email: han-ming_shen@nuhs.edu.sg.
Dajing Xia, Email: dxia@zju.edu.cn.
References
- 1.Huang Y., Cui J., Chen S., Gan C., Zhou A. Synthesis and antiproliferative activity of some steroidal lactams. Steroids. 2011;76:1346–1350. doi: 10.1016/j.steroids.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Ji L., Yuan Y., Luo L., Chen Z., Ma X. Physalins with anti-inflammatory activity are present in Physalis alkekengi var. franchetii and can function as Michael reaction acceptors. Steroids. 2012;77:441–447. doi: 10.1016/j.steroids.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 3.Yang B.-Y., Guo R., Li T., Wu J.-J., Zhang J. New anti-inflammatory withanolides from the leaves of Datura metel L. Steroids. 2014;87:26–34. doi: 10.1016/j.steroids.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Silva M.T., Simas S.M., Batista T.G., Cardarelli P., Tomassini T.C. Studies on antimicrobial activity, in vitro, of Physalis angulata L.(Solanaceae) fraction and physalin B bringing out the importance of assay determination. Mem. Inst. Oswaldo Cruz. 2005;100(7):779–782. doi: 10.1590/s0074-02762005000700018. [DOI] [PubMed] [Google Scholar]
- 5.Yang Y.K., Xie S.D., Xu W.X., Nian Y., Liu X.L. Six new physalins from Physalis alkekengi var. franchetii and their cytotoxicity and antibacterial activity. Fitoterapia. 2016;112:144–152. doi: 10.1016/j.fitote.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 6.Choudhary M.I., Yousaf S., Ahmed S., Yasmeen K. Antileishmanial Physalins from Physalis minima. Chem. Biodivers. 2005;2(9):1164–1173. doi: 10.1002/cbdv.200590086. [DOI] [PubMed] [Google Scholar]
- 7.Barthel A., Vogel H., Pauchet Y., Pauls G., Kunert G. Immune modulation enables a specialist insect to benefit from antibacterial withanolides in its host plant. Nat. Commun. 2016;7:12530. doi: 10.1038/ncomms12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henrich C.J., Brooks A.D., Erickson K.L., Thomas C.L., Bokesch H.R. Withanolide E sensitizes renal carcinoma cells to TRAIL-induced apoptosis by increasing cFLIP degradation. Cell Death Dis. 2015;6:e1666. doi: 10.1038/cddis.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma T., Fan B.Y., Zhang C., Zhao H.J., Han C. Metabonomics applied in exploring the antitumour mechanism of physapubenolide on hepatocellular carcinoma cells by targeting glycolysis through the Akt-p53 pathway. Sci. Rep. 2016;6:29926. doi: 10.1038/srep29926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji L., Yuan Y., Ma Z., Chen Z., Gan L. Induction of quinone reductase (QR) by withanolides isolated from Physalis pubescens L. (Solanaceae) Steroids. 2013;78:860–865. doi: 10.1016/j.steroids.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 11.Ding W., Hu Z., Zhang Z., Ma Q., Tang H. Physapubescin B Exhibits Potent Activity against Human Prostate Cancer In Vitro and In Vivo. J. Agric Food Chem. 2015;63:9504–9512. doi: 10.1021/acs.jafc.5b03045. [DOI] [PubMed] [Google Scholar]
- 12.Chen L., Xia G., Qiu F., Wu C., Denmon A.P. Physapubescin selectively induces apoptosis in VHL-null renal cell carcinoma cells through down-regulation of HIF-2α and inhibits tumor growth. Sci. Rep. 2016;6:32582. doi: 10.1038/srep32582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 14.Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He C., Klionsky D.J. Regulation mechanisms and signaling pathways of autophagy. Annu Rev. Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan S.H., Shui G., Zhou J., Li J.J., Bay B.H. Induction of autophagy by palmitic acid via protein kinase C-mediated signaling pathway independent of mTOR (mammalian target of rapamycin) J. Biol. Chem. 2012;287:14364–14376. doi: 10.1074/jbc.M111.294157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galluzzi L., Pietrocola F., Levine B., Kroemer G. Metabolic Control of Autophagy. Cell. 2014;159:1263–1276. doi: 10.1016/j.cell.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong Z., Sanchez-Lopez E., Karin M. Autophagy, Inflammation, and Immunity: a troika governing cancer and its treatment. Cell. 2016;166:288–298. doi: 10.1016/j.cell.2016.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang P., Mizushima N. Autophagy and human diseases. Cell Res. 2014;24:69–79. doi: 10.1038/cr.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo J.Y., Xia B., White E. Autophagy-mediated tumor promotion. Cell. 2013;155:1216–1219. doi: 10.1016/j.cell.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abada A., Elazar Z. Getting ready for building: signaling and autophagosome biogenesis. EMBO Rep. 2014;15:839–852. doi: 10.15252/embr.201439076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizushima N., Yoshimori T., Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sivendran S., Agarwal N., Gartrell B., Ying J., Boucher K.M. Metabolic complications with the use of mTOR inhibitors for cancer therapy. Cancer Treat. Rev. 2014;40:190–196. doi: 10.1016/j.ctrv.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wullschleger S., Loewith R., Hall M.N. TOR Signaling in Growth and Metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 25.Alers S., Loffler A.S., Wesselborg S., Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol. Cell Biol. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Devasagayam T.P.A.T.J.C., Boloor K.K. Free radicals and antioxidants in human health: current status and future prospects. Japi. 2004;52:794–804. [PubMed] [Google Scholar]
- 27.Gibson S.B. A matter of balance between life and death: targeting reactive oxygen species (ROS)-induced autophagy for cancer therapy. Autophagy. 2010;6:835–837. doi: 10.4161/auto.6.7.13335. [DOI] [PubMed] [Google Scholar]
- 28.Scherz-Shouval R., Elazar Z. Regulation of autophagy by ROS: physiology and pathology. Trends Biochem Sci. 2011;36:30–38. doi: 10.1016/j.tibs.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Shen H.-M., Liu Z-g JNK signaling pathway is a key modulator in cell death mediated by reactive oxygen and nitrogen species. Free Radic. Biol. Med. 2006;40:928–939. doi: 10.1016/j.freeradbiomed.2005.10.056. [DOI] [PubMed] [Google Scholar]
- 30.Li L., Tan J., Miao Y., Lei P., Zhang Q. ROS and Autophagy: Interactions and Molecular Regulatory Mechanisms. Cell Mol. Neurobiol. 2015;35:615–621. doi: 10.1007/s10571-015-0166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L., Wang H., Xu J., Zhu J., Ding K. Inhibition of cathepsin S induces autophagy and apoptosis in human glioblastoma cell lines through ROS-mediated PI3K/AKT/mTOR/p70S6K and JNK signaling pathways. Toxicol. Lett. 2014;228:248–259. doi: 10.1016/j.toxlet.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 32.Marin J.J., Lozano E., Perez M.J. Lack of mitochondrial DNA impairs chemical hypoxia-induced autophagy in liver tumor cells through ROS-AMPK-ULK1 signaling dysregulation independently of HIF-1alpha. Free Radic. Biol. Med. 2016;101:71–84. doi: 10.1016/j.freeradbiomed.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 33.Byun Y.J., Kim S.K., Kim Y.M., Chae G.T., Jeong S.W. Hydrogen peroxide induces autophagic cell death in C6 glioma cells via BNIP3-mediated suppression of the mTOR pathway. Neurosci. Lett. 2009;461:131–135. doi: 10.1016/j.neulet.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 34.Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. Embo J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L., Ishdorj G., Gibson S.B. Reactive oxygen species regulation of autophagy in cancer: implications for cancer treatment. Free Radic. Biol. Med. 2012;53:1399–1410. doi: 10.1016/j.freeradbiomed.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 36.Klionsky D.J., Abdelmohsen K., Abe A., Abedin M.J., Abeliovich H. Vol. 12. 2016. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) pp. 1–222. (Autophagy). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalyanaraman B., Darley-Usmar V., Davies K.J., Dennery P.A., Forman H.J. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic. Biol. Med. 2012;52:1–6. doi: 10.1016/j.freeradbiomed.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalyanaraman B. Teaching the basics of redox biology to medical and graduate students: oxidants, antioxidants and disease mechanisms. Redox Biol. 2013;1:244–257. doi: 10.1016/j.redox.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He H., Zang L.H., Feng Y.S., Wang J., Liu W.W. Physalin A induces apoptotic cell death and protective autophagy in HT1080 human fibrosarcoma cells. J. Nat. Prod. 2013;76:880–888. doi: 10.1021/np400017k. [DOI] [PubMed] [Google Scholar]
- 40.AlQathama A., Prieto J.M. Natural products with therapeutic potential in melanoma metastasis. Nat. Prod. Rep. 2015;32:1170–1182. doi: 10.1039/c4np00130c. [DOI] [PubMed] [Google Scholar]
- 41.Newman D.J., Cragg G.M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang N., Jian J.F., Cao S.J., Zhang Q., Mao Y.W. Physalin A induces G2/M phase cell cycle arrest in human non-small cell lung cancer cells: involvement of the p38 MAPK/ROS pathway. Mol. Cell Biochem. 2016;415:145–155. doi: 10.1007/s11010-016-2686-1. [DOI] [PubMed] [Google Scholar]
- 43.He H., Zang L.H., Feng Y.S., Chen L.X., Kang N. Physalin A induces apoptosis via p53-Noxa-mediated ROS generation, and autophagy plays a protective role against apoptosis through p38-NF-κB survival pathway in A375-S2 cells. J. Ethnopharmacol. 2013;148:544–555. doi: 10.1016/j.jep.2013.04.051. [DOI] [PubMed] [Google Scholar]
- 44.Wu S.Y., Leu Y.L., Chang Y.L., Wu T.S., Kuo P.C. Physalin F induces cell apoptosis in human renal carcinoma cells by targeting NF-kappaB and generating reactive oxygen species. PLoS One. 2012;7:e40727. doi: 10.1371/journal.pone.0040727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eldredge H.B., DeNittis A., DuHadaway J.B., Chernick M., Metz R. Concurrent whole brain radiotherapy and short-course chloroquine IN patients with brain metastases: a pilot trial. J. Radiat. Oncol. 2013;2 doi: 10.1007/s13566-013-0111-x. [DOI] [PMC free article] [PubMed] [Google Scholar]