Abstract
Traumatic brain injury (TBI) may adversely affect a person's thinking, memory, personality, and behavior. While mild TBI (mTBI) diagnosis is challenging, there is a risk for long-term psychiatric, neurologic, and psychosocial problems in some patients that motivates the search for new and better biomarkers. Recently, diffusion magnetic resonance imaging (dMRI) has shown promise in detecting mTBI, but its validity is still being investigated. Resting state functional network connectivity (rsFNC) is another approach that is emerging as a promising option for the diagnosis of mTBI. The present work investigated the use of rsFNC for mTBI detection compared with dMRI results on the same cohort. Fifty patients with mTBI (25 males) and age-sex matched healthy controls were recruited. Features from dMRI were obtained using all voxels, the enhanced Z-score microstructural assessment for pathology, and the distribution corrected Z-score. Features based on rsFNC were obtained through group independent component analysis and correlation between pairs of resting state networks. A linear support vector machine was used for classification and validated using leave-one-out cross validation. Classification achieved a maximum accuracy of 84.1% for rsFNC and 75.5% for dMRI and 74.5% for both combined. A t test analysis revealed significant increase in rsFNC between cerebellum versus sensorimotor networks and between left angular gyrus versus precuneus in subjects with mTBI. These outcomes suggest that inclusion of both common and unique information is important for classification of mTBI. Results also suggest that rsFNC can yield viable biomarkers that might outperform dMRI and points to connectivity to the cerebellum as an important region for the detection of mTBI.
Keywords: : diffusion tensor imaging, magnetic resonance imaging, traumatic brain injury
Introduction
Traumatic brain injury (TBI) can result in a broad spectrum of symptoms that adversely affect a person's thinking, memory, personality, and behavior. Many complications of TBI manifest shortly after injury even in patients with seemingly mild traumatic brain injury (mTBI) including chronic headaches, dizziness, vertigo, difficulty concentrating, depression, irritability, and impulsiveness.1 In spite of these evident symptoms, the World Health Organization and the National Academy of Neurology recognize that existing mTBI diagnosis methods require a thoughtful and deliberate approach, but provide limited evidence of their validity.2,3 Further, mTBI produces long-term morbidity leading to psychiatric, neurologic, and psychosocial problems for some patients.
The heterogeneity of mTBI symptoms motivates the application of new technologies that might aid in diagnosing mTBI in patients. For example, researchers have turned toward diffusion magnetic resonance imaging (dMRI) as a technique to evaluate injury after mTBI. dMRI has been shown to outperform other imaging methods, such as computed tomography and structural (e.g., T1- and T2-weighted) MRI.4,5 Medical imaging methods for accurate mTBI diagnosis are still being investigated with the hope of better identifying brain injuries and improving clinical outcomes.
Recently, a comparison among several MRI modalities including high resolution structural imaging, diffusion tensor imaging (DTI), magnetization transfer ratio imaging, and magnetic resonance spectroscopic imaging concluded that DTI (a dMRI modality) was the only modality in the analysis sensitive to the acute phase of mTBI.6 In particular, the measure of anisotropic water diffusion known as fractional anisotropy was sensitive to diffuse axonal injuries and correlated with the acute Glasgow Coma Scale.5,7,8
Axonal injuries can be detected using the EZ-MAP method that aims at finding abnormal levels of fractional anisotropy (FA) when compared with a normative set of FA images from healthy persons.4 In discriminating mTBI from healthy brains, the EZ-MAP method (without out-of-sample cross validation) performed somewhere between 65% and 71%.9 The discriminatory power indicates that information extracted from FA is useful as a complementary assessment for brain injuries. A more recent approach, known as DisCo-Z, has been proposed as an alternative to EZ-MAP that is capable of capturing subject-specific abnormalities and further correct for biases on the estimation of z-scores.10
Axonal injuries in white matter seem to concentrate in some important areas of white matter content including genu and splenium of the corpus callosum, the corona radiata, and the internal capsule.5,7,11,12 The integrity of these and other affected areas is important for normal brain function, because white matter structure reflects functional connections between different areas of the brain.13,14 Therefore, brain connectivity across the whole brain of patients with mTBI might be compromised because of disconnections occurring as a consequence of injured white matter tracks.15,16
Using functional MRI (fMRI) to explore functional changes related to gray matter, reduced resting state functional connectivity in the default mode network (DMN) along with increased connectivity within the ventromedial prefrontal cortex has been found in patients with mTBI.17–19 Weaker functional connectivity between important networks has been found in relation to mTBI including DMN-basal ganglia, attention-sensorimotor, attention-frontal, and within the sensorimotor networks.20 Increased resting state functional connectivity in the thalamus has been reported including thalamo-thalamo, thalamo-frontal, and thalamo-temporal circuits.21,22 Other areas where connectivity disruption has been found in subjects with mTBI include the primary visual, hippocampal, and dorsolateral prefrontal cortexes23 as well as increased connectivity in the cerebellum.24
Studying the brain in a more comprehensive way, it has been suggested that stronger connectivity among functional brain networks is a normal response to brain injury.25 Collectively, these results suggest that methods to exploit resting state functional connectivity as a biomarker should be comprehensive rather than specific when including relevant brain areas.
A previous study that used the same sample cohort described here reported no group difference within the DMN or subcortical resting state networks (RSNs) after correcting for false positives using the group independent component analysis (gICA) algorithm, albeit with slightly different pre-processing and parameter selection.26 However, no individual subject classification was performed. The motivation for re-analyzing the data is based on recent studies that suggest there may be important differences depending on the order in which motion and anomalous points are corrected for in resting state analyses.27 Specifically, previous work corrected for motion after the gICA and for anomalous data points (“spikes”) in the pre-processing pipeline.26 We followed the suggestion that resting state functional network connectivity (rsFNC) bias could be minimized by processing head motion variance before using gICA27 rather than after gICA as has been typically done previously in the ICA literature.20,26,28–30
After investigating the effects of several pre-processing pipelines, Vergara and associates31 found that pre-processing head movement before gICA leads to more accurate detection of group differences and higher classification accuracy after testing using both simulated and real data. Simulations in the same study also suggest that pre-processing head movement after gICA results in higher residual correlation with head movement and higher spatial variance between subjects. By following the guidelines presented by Vergara and associates,31 we expected to achieve a more accurate classification because of a smaller residual head movement confounds.
After this data re-processing, we consider two different analyses based on group differences and classification performance. We hypothesized that subjects with mTBI would exhibit functional connectivity changes within the DMN18,32 and abnormalities between: DMN and task positive networks,17,18 cerebellum and sensorimotor areas,24,33 cortical and subcortical regions (thalamus),21,22 DMN and basal ganglia, attentional and sensorimotor areas, attentional and frontal regions, within the sensorimotor areas.20
Methods
Subjects
The cohort included 100 persons (50 patients with mTBI and 50 controls). The DTI and resting state data from this cohort have been used before in which four subjects were excluded because of increased head motion.7,26 The current work represents a re-analysis of the published data. In the current instance, an additional pair of subjects was excluded based on the quality of the DTI data. The final mTBI cohort consisted of 47 (mean age 27.3 ± 9.0 years) patients with mTBI who had been recruited from local emergency departments.
Inclusion criteria were based on the American Congress of Rehabilitation Medicine.26 A total of 47 sex and age (up to three years) matched healthy controls (HC) were included. Subjects classified as mTBI had a Glasgow Coma Scale between 13 and 15 at first contact with medical staff, a maximum of 30 min loss of consciousness (if present), and a maximum of 24 h post-traumatic amnesia (if present).
Subjects were excluded if there was a history of neurological disease, major psychiatric disturbance, and additional closed head injuries with more than 5 min of lost consciousness, additional closed head injury within the past year, learning disorder, attention deficit hyperactivity disorder, or a history of substance abuse/dependence including alcohol. All participants provided informed consent according to the Declaration of Helsinki and the institutional guidelines at the University of New Mexico.
Imaging protocol
All images were collected on a 3 Tesla Siemens Trio scanner. Two DTI scans (b = 800 sec/mm2) were acquired using a twice-re-focused spin echo sequence with 30 diffusion gradients and the b = 0 experiment repeated five times (72 interleaved slices; echo time (TE) = 8.4 sec; repetition time (TR) = 9.0 sec; 90○ flip angle; NEX = 1; slice thickness = 2.0 mm; field of view (FOV) = 256 × 256 mm; matrix size = 128 × 128; voxel resolution = 2 mm3).
Each participant completed a 5-min resting state run using a single-shot, gradient-echo echo planar pulse sequence (TR = 2000 msec; TE = 29 msec; flip angle = 75°; FOV = 240 mm; matrix size = 64 × 64). Foam padding and paper tape were used to restrict motion within the scanner. Thirty-three contiguous, axial 4.55-mm thick slices were selected to provide whole-brain coverage (voxel size: 3.75 × 3.75 × 4.55 mm) during the resting state scan. The first five images were eliminated to account for T1 equilibrium effects; 145 images were selected for further analysis. Presentation software (Neurobehavioral Systems) was used for stimulus presentation. Subjects were instructed to passively stare at a foveally presented fixation cross (visual angle = 1.02°) for approximately 5 min and to keep head movement to a minimum.
dMRI pre-processing
FA images were generated and normalized to a 1 × 1 × 1 mm3 FA template in the Montreal Neurological Institute space using FSL (www.fmrib.ox.ac.uk/fsl/). Age and sex were regressed for each voxel and for all FA images across both cohorts based on a model calculated using HC images only. Subject-specific analyses were conducted on FA data using the Enhanced Z-score Microstructural Assessment for Pathology (EZ-MAP) method4 and the Distribution Corrected Z-score (DisCo-Z) method.10
In the EZ-MAP method, a robust estimation of FA variance from a normative population of HCs is calculated via bootstrap. Bootstrapped variance is then used to produce an enhanced Z score for a particular testing subject based on data from the normative population. A threshold is applied to pinpoint abnormalities, and clustering is used to identify areas with contiguously abnormal voxels.
Following the EZ-MAP method with 1000 bootstrap iterations, voxel-wise mean and enhanced standard deviation images were obtained from HCs and used to calculate a voxel-wise EZ-score image for each subject with mTBI. Abnormal voxels were detected from abnormal clusters of a minimum size of 100 voxels (100 mL) with an EZ-score larger than two. These thresholds including cluster size, number of bootstrap iteration, and minimum EZ-score, were selected following recommendations presented in the EZ-MAP literature for a maximal discrimination of patients and controls.4,9 In the case of HC subjects, the nth HC subject was excluded from mean and enhanced standard deviation calculation. The EZ-score of each HC is a deviation against the other HCs.
For the DisCo-Z calculations, mean and variance were estimated from the HC group and used to transform voxels from both samples into Z-score images. Given the known bias that occurs after this conversion, extreme voxels were calculated using a distributionally corrected threshold of either|Z|> 1.98 for HC and |Z| > 2.04 for mTBI based on the 94 subjects. Similar to the EZ-MAP method, clusters were obtained using a minimum volume of 100 mL. Clusters of smaller size were discarded to reduce possible false positives. The Z-score, characterized by the number of samples, and cluster size thresholds were selected following the method described in Mayer and colleagues.10
fMRI pre-processing
Data were pre-processed using statistical parametric mapping34 (http://www.fil.ion.ucl.ac.uk/spm) including slice-timing correction, realignment, co-registration, spatial normalization, and then transformed to the Montreal Neurological Institute standard space at a resolution of a 3 × 3 × 3 mm3. Despiking consisted of the orthogonalization with respect to spike regressors. Each spike is represented by an independent regressor valued one at the spike time point and zero everywhere else. The DVARS method35 was used to find spike regressors where the root mean square exceeded three standard deviations. Time courses were also orthogonalized with respect to the following: (1) linear, quadratic, and cubic trends; (2) the six realignment parameters; (3) realignment parameters derivatives; and (4) spike regressors.
We did not consider including global, white matter, and CSF regressors because gICA is able to separate these signals in a set of identifiable artifact components.36 In addition, these regressors are not part of the standard pre-processing procedure followed in gICA.26,27,31 A full width half maximum Gaussian kernel of 6 mm was then used for spatial smoothing.
Data from all subjects were subject to a gICA37,38 using the GIFT software (http://mialab.mrn.org/software/gift/) to obtain a set of functionally independent RSNs. The number of components was determined to be 70 using a modified version of ICASSO39,40 such that the overall R-index is close to the minimum and the quality index of any given RSN is above 0.7. This setup was a good consistency trade-off between RSN quality and number of components considered. A fifth-order Butterworth band-pass filter (0.01 0.15) Hz was applied to the time courses of each RSN as has been recommended in the literature.36 Because of the spike orthogonalization, spike time points are zero valued. To apply filtering, spike time points were interpolated using a spline, then set again to zero after filtering.31
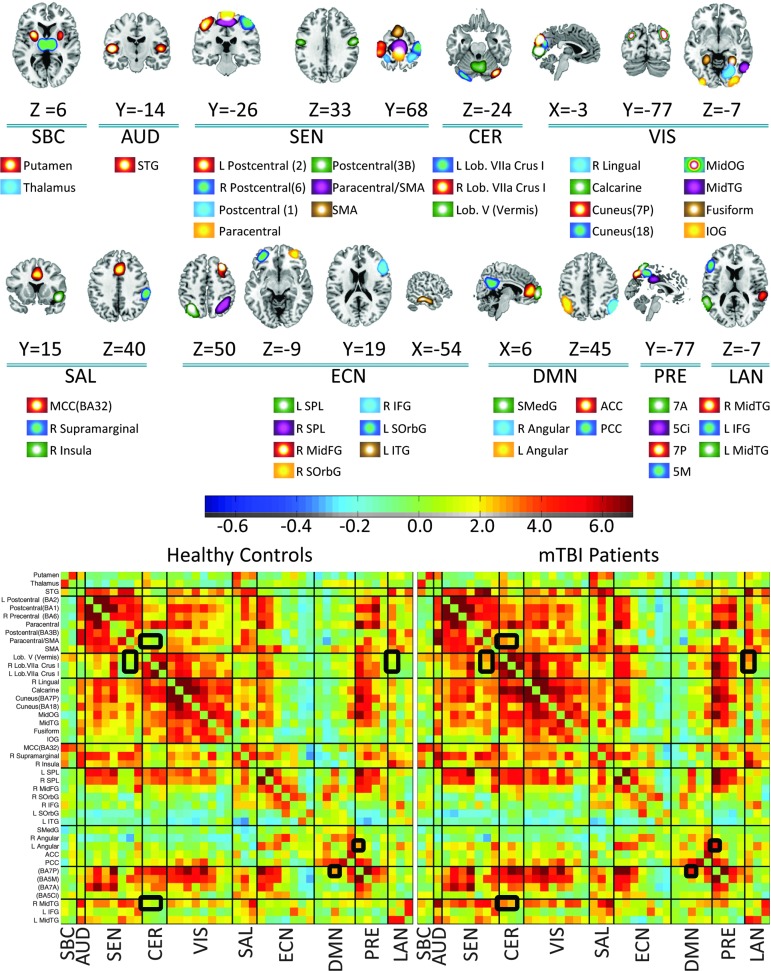
Artifactual RSNs were detected and discarded based on their frequency content following a published method.28 RSNs were also manually inspected and classified into broader categories or discarded if their main activation occurs in areas of white matter or cerebrospinal fluid. Only the subset of 43 non-artifactual RSNs shown in Figure 1 was selected after agreement between three subject matter experts who individually rated the RSNs. More information of each RSN is provided in Supplementary Table 1 (see online supplementary material at ftp.liebertpub.com).
FIG. 1.
Resting state networks (RSNs) and resting state functional network connectivity (rsFNC) matrices used in this study. Ten groups of RSNs were established: SBC (subcortical), AUD (auditory), SEN (sensorimotor), CER (cerebellum), VIS (visual), SAL (salience), ECN (executive control), DMN (default mode), PRE (precuneus), and LAN (language). Repeated RSN names include or use the main Brodmann Area to avoid ambiguity. The rsFNCs with group differences have been highlighted in the rsFNC matrices. Color image is available online at www.liebertpub.com/neu
These RSNs were organized in 10 groups: subcortical, auditory, sensorimotor, cerebellum, visual, salience, executive control, default mode network, precuneus and language. The rsFNC matrix was determined by measuring the correlation coefficient between the time courses of each of the 903 possible RSN pairs. For all RSNs and all subjects, the time points at which spikes were detected were excluded from the calculation of correlation.
Diagnosis performance
Data from the two modalities were used as features to feed a classification algorithm. In FA based features, age and sex effects were removed during dMRI pre-processing in a voxel-wise manner. The voxel-wise regression model can be described as FAx,y,z = FAVx,y,z − (b0 + b1AGE + b2SEX) where (x,y,z) are the coordinates of voxel position, FAV is the voxel's fractional anisotropy value and FA is the regression residual.
After age and sex correction, three different sets of features were used in the dMRI data. In the first set, FA values from all white matter voxels were used as features. The second set used features from the EZ-MAP method consisting of positive EZ-score cluster count, positive EZ-score total volume (based on the number of voxels with abnormally high FA), negative EZ-score cluster count, and negative EZ-score total volume (based on the number of voxels with abnormally low FA). The last dMRI set were DisCo-Z features consisting of the positive Z-score cluster count, positive Z-score total volume, negative Z-score cluster count, and negative Z-score total volume. In the case of fMRI data, the features used consisted of the 903 time course correlations of the rsFNC matrices. The fMRI features were orthogonalized with respect to age and sex before classification.
A linear support vector machine (SVM) was used to classify subjects into mTBIs and HCs. Classification accuracy was assessed using the area under the curve (AUC) measure. Similar to the feature selection process, classification accuracy can change depending on the specifics of the SVM configuration indicating the need for parameter tuning. The process of tuning, however, can lead to over-fitting problems if not properly handled. Over-fitting was avoided by using a nested leave-one-out cross validation (LOOCV) to select the appropriate SVM configuration as it has been done before in the literature.41,42
The sequential minimal optimization (SMO) and least square (LS) methods for solving SVMs were considered. Six different values [0.05, 0.1, 0.5, 1.0, 5.0, 10.0] of SVM's soft margin parameter were also considered. Feature selection for the EZ-MAP and Disco-Z methods was accomplished by extensive search given the small number of features. The numbers of features for the other datasets were too large for extensive search. In the case of FA voxels and rsFNC features, feature selection was performed based on t values obtained from HC versus mTBI group differences. Six thresholds (2.0 1.0 0.75 0.5 0.25 0.0) were considered to discard features based on their t value.
The different SVM implementations were subject to a nested LOOCV loop that delivered a different configuration model for each sample left out. The nested LOOCV optimization loop was implemented in the following way: After omitting one sample, the remaining 93 samples were subject to additional LOOCVs. A total of 72 inner LOOCV loops (2 × 6 × 6) were used in the case of t thresholds corresponding to the possible combinations of two methods (SMO or LS), six soft margin values and six t value thresholds. In the case of extensive search, EZ-MAP and DisCo-Z had 180 (2 × 6 × 15 different combination of four features) inner LOOCV loops. The configuration of the inner LOOCV with the highest AUC value was then used to classify the left out sample in the outer LOOCV loop.
This procedure may change the configuration used to classify each sample unless the inner LOOCV loop chooses the same configuration every time. In practice, we look for a stable SVM model with few changes from sample to sample. Stability is one important piece of information that nested cross-validation provides, which in our case was measured using selection frequency.
Statistical analysis
In addition to classification accuracy, we analyzed group differences for the fMRI data. Group differences of rsFNC were evaluated using two-tailed t tests with 92 degrees of freedom for each RSN pair. A false discovery rate multi-comparison correction was applied to the p values of the 903 rsFNCs t tests. The alpha level for significance was taken as 0.05.
Results
Classification results
Classification accuracies are displayed in Table 1. The highest accuracy of 84.1% was obtained using the rsFNC features. FA features follow in second place with 75.5%. The stability of each SVM parameter is displayed in Figure 2. The most stable model was observed for rsFNC features with a 0.1 soft margin parameter, 0.25 t threshold and LS solving method. Although Figure 2 shows that FA always selected a 0.05 soft-margin parameter, this frequency is an artifact. Closer examination of the data indicates that AUC was the same for all considered soft-margin parameters. The algorithm implementation was instructed to select the first soft-margin parameter in the list, thus creating the artifact selection.
Table 1.
Classification Results for Fractional Anisotropy and Functional Magnetic Resonance Imaging
| Modality | Feature Extraction | Feature Selection | Number of Features (mean ± std) | Feature Type | Sensitivity (%) | Specificity (%) | AUC (%) |
|---|---|---|---|---|---|---|---|
| rsFNC | gICA | t value | 682 ± 150 | rsFNC | 89.4 | 78.8 | 84.1 |
| FA | None | t value | 413k ± 239k | Voxels | 76.6 | 74.5 | 75.5 |
| FA+rsFNC | None+gICA | t value | 388k ± 251k | Voxels+rsFNC | 76.6 | 72.3 | 74.5 |
| FA | EZ-MAP | Extensive | 1.0 ± 0.2 | PN-CV | 70.2 | 61.7 | 66.0 |
| FA | DisCo-Z | Extensive | 2.0 ± 0.2 | PN-CV | 61.7 | 66.0 | 64.8 |
rsFNC, resting state functional network connectivity; FA, fractional anisotropy;
PN-CV, positive and negative clusters and volumes.
FIG. 2.
Results for model selection are presented in the form of frequency histograms. Support vector machine (SVM) model selection was achieved using nested leave-one-out cross-validation. The highest model stabilities were achieved with resting state functional network connectivity (rsFNC) and Distribution Corrected Z-score (DisCo-Z) features. The soft-margin parameter in fractional anisotropy (FA) features did not changed accuracy. Indicated high frequency in FA is an artifact because the algorithm just chooses the first soft-margin from a list where all areas under the curves were the same. Color image is available online at www.liebertpub.com/neu
The t threshold for FA features was less stable than in the rsFNC case. The algorithm consistently selected SMO for FA features with a selection frequency of 100%. In EZ-MAP, SMO was always selected, and the positive EZ-score total volume was selected 97% of the time, but very small stability was observed for the soft-margin parameter. Results for DisCo-Z were fairly stable and always used LS. The most frequent soft-margin parameter in DisCo-Z was 0.05, and the most frequent set of selected features was positive and negative Z-score cluster counts.
The rsFNC features were analyzed further because they delivered the highest accuracy and configuration stability than the other considered methods. We calculated the number of times each rsFNC was selected and plotted the selection percentage in Figure 3, hoping to find a pattern of feature exclusion, but no clearly identifiable pattern could be determined.
FIG. 3.

Matrix showing the number of times (in %) that each resting state functional network connectivity (rsFNC) was selected as a feature for the optimal selection with t values <0.25. Significant t values (false discovery rate corrected p < 0.05) are indicated by white squares. Dark blue rsFNCs were always excluded by feature selection. Dark red rsFNCs were always included. There is not an identifiable pattern of excluded features. SBC, subcortical, AUD, auditory, SEN, sensorimotor, CER, cerebellum, VIS, visual, SAL, salience, ECN, executive control, DMN, default mode, PRE, precuneus, and LAN, language. Color image is available online at www.liebertpub.com/neu
Group differences in rsFNC
Group differences between HC and mTBI rsFNCs are displayed in Figure 4, where significant differences of rsFNC have been highlighted. The actual rsFNC matrices for HC and mTBI are displayed in Figure 1. Large group differences were found between the RSNs in the cerebellum group and RSNs in the other groups. Similarly, high t values can be observed between the left angular gyrus and other RSNs. Five t tests were significant after false discovery rate correction (p < 0.05) considering the 903 RSN pairs. The significant group differences were found among cerebellar RSNs with the supplementary motor area (SMA)/paracentral and right MidTG RSNs. In addition, a significant group difference was observed between left angular gyrus and precuneus. Table 2 shows the group difference information. The spatial content of relevant RSNs can be found in Figure 1.
FIG. 4.

Matrix showing t values for group differences (mild traumatic brain injury [mTBI]–healthy controls [HC]). Significant t values (false discovery rate corrected p < 0.05) are indicated by black squares with mTBI subjects having stronger connectivity than HC subjects. Large rsFNC differences can be observed in the left angular gyrus and cerebellar areas. SBC, subcortical, AUD, auditory, SEN, sensorimotor, CER, cerebellum, VIS, visual, SAL, salience, ECN, executive control, DMN, default mode, PRE, precuneus, and LAN, language. Color image is available online at www.liebertpub.com/neu
Table 2.
Significant Group Differences of Resting State Functional Network Connectivity
| RSN | X | Y | Z | RSN | X | Y | Z | t value | p value* |
|---|---|---|---|---|---|---|---|---|---|
| Lob. V (Vermis) | 0 | −61 | −19 | SMA/Paracentral (BA4A) | 0 | −21 | 62 | 4.4 | 0.023 |
| R Lob.VIIa Crus I | 36 | −83 | −31 | SMA/Paracentral (BA4A) | 0 | −21 | 62 | 4.3 | 0.038 |
| L Angular Gyrus | −44 | 62 | 45 | Precuneus(BA7P) | 0 | −77 | 50 | 4.3 | 0.038 |
| Lob. V (Vermis) | 0 | −61 | −19 | R MidTG | 61 | −35 | 5 | 3.9 | 0.033 |
| R Lob.VIIa Crus I | 36 | −83 | −31 | R MidTG | 61 | −35 | 5 | 3.8 | 0.037 |
RSN, resting state networks.
All p values are false discovery rate corrected.
Discussion
Classification performance
Previous studies have focused on finding abnormalities in white matter5,7 or functional connectivity group differences18,20,26,33 between mTBI and HC subjects. The present work took a different approach where the classification of mTBIs and HCs (i.e., individual subjects via cross-validation) is the central point of the analysis. This methodology directly tests the use of dMRI and fMRI data as biomarkers for mTBI. Although both data modalities are related to the integrity of white matter,13 rsFNC and FA capture complementary information that has been suggested to be different for each modality.43 The present work intended to compare HC–mTBI classification power from proposed techniques to detect abnormalities in white matter with methods to assess abnormalities in functional connectivity.
Looking side by side to results from dMRI versus fMRI (Table 1), we can see that rsFNC features deliver higher classification performance (84.1%) compared with FA (75.5%) after determining optimal SVM parameters. As Figure 3 illustrates, most of the fMRI features were selected in all LOOCV iterations. An average of 24.4% of the rsFNCs features with very small group differences (t values <0.25) was not used for classification. Our results are consonant with the proposal of Stevens and coworkers33 in which rsFNC effects in patients with mTBI can be spread across many parts of the brain.
Part of the success of our procedure is because of the characteristic of the data-driven analysis performed, which is comprehensive rather than narrow. In FA data, the nested LOOCV choose a zero t threshold with the highest frequency suggesting that all of the features were useful for classification. This result is not stable, however, and some iterations rejected a large number of FA features based on the t thresholds of 1.0 and 2.0 (see Fig. 2).
Known methods for feature extraction in dMRI have normally produced few features4,10,44,45 with moderate classification accuracy9 not verified by any cross validation method except for the results presented here. The two feature extraction methods used for the dMRI data, EZ-MAP and DisCo-Z, achieved low classification performances when compared with the use of all FA voxels. DisCo-Z features achieved a slightly lower classification performance than EZ-MAP features. The advantage of DisCo-Z over EZ-MAP in our data, however, strives in a higher stability, as illustrated in Figure 2. The stability in DisCo-Z indicates a higher reliability than EZ-MAP in case these techniques are applied to a different cohort. These outcomes indicate the need for more adequate methods of feature extraction for this modality. The use of all FA voxels did not exclude any of the important brain areas mentioned by other studies,7,11,12,45 supporting the possibility of high classification performance.
The combination of features from dMRI and fMRI did not result in a better performance than FA or rsFNC features alone. SVM configuration and feature selection for the combined modalities case exhibited a similar set of parameter selection than FA, indicating a strong influence of FA features. The characteristics of FA features prevailed over the rsFNC features likely because of their large difference in number. Further research with a focus in multimodal feature extraction should address differences in feature space and other questions associated with combining fMRI and dMRI. There are several data-driven techniques capable of extracting information from latent variables among which we can mention joint-ICA,46 parallel ICA.47 Feature extraction and selection is one important topic that is intrinsically associated with particular latent variables in each modality.
Alterations of rsFNC
Although performed rsFNC analysis included information from many parts of the brain, only five RSN pairs differed between HCs and mTBIs. This fact seems antagonistic with respect to results reported earlier by Vakhtin and associates,20 where several rsFNC group differences were found through the brain, but in that study the number of test comparisons was much lower. In contrast to the results from Vakhtin and associates,20 the study performed by Mayer and colleagues26 did not achieve significance after multi-comparison correction probably because of the larger number of RSNs considered. This difference could be because of the larger number of RSNs considered. In spite of early findings of altered deactivations in the cerebellum and sensorimotor areas,48 little attention has been put into these regions in functional connectivity.
Our analysis found increased connectivity between DMN (left angular gyrus) and posterior precuneus. Connectivity changes associated with the DMN and the precuneus has been observed before.32,49 Dysfunctions associated to the DMN are found in a variety of conditions50 and are likely to be present in the cohort of this study. Four of the significant changes in rsFNC observed in our analysis involved connectivity between cerebellum and sensorimotor RSNs.
A similar result has been observed before by Nathan and coworkers24 in which the functional connectivity between cerebellum and SMA showed an increase in patients with TBI. Another study found functional connectivity of the cerebellum among those correlated with post-concussive complaints.33 Because the cerebellum is useful in movement coordination and it has been linked recently to cognitive functions,51,52 the relationship between cerebellum functional alterations with other cortical areas may be an important topic for future research.
As somewhat expected, the RSN pairs with a significant group differences were also important for classification and were included among the selected features in each LOOCV iteration. That being said, classification results also suggest that better differentiation between HCs and mTBIs requires the inclusion of many parts of the brain at once rather than focusing on isolated regions.
It is important to consider that all patients with mTBI were scanned within 21 days post-injury; thus, observed correlation was observed within this time lapse. This work has limitations often found in studies classifying using fMRI and dMRI including high data dimensionality, small number of samples, and noise.53 Based on these limitations, we selected LOOCV to make a more efficient use of the available cohort size. Nevertheless, nested LOOCV allowed us to provide information about the considered modalities and their difference in SVM stabilities. Another limitation in this study is the relatively simple method used for feature selection. In this work, performance was not improved when using both fMRI and dMRI features; however, advanced data fusion approaches may show benefits when combining both modalities.54
Supplementary Material
Acknowledgments
The authors thank Joseph Ling, Jill Fries, Vikram Rao, Prashanth Nyalakanti, and Sandeep Panta for help with preprocessing the data. This work was funded by the following NIH grants: R24HD050836/R21NS064464/3R21 NS064464 to A.M. and P20GM103472/1R01EB006841 to V.C.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.DeKosky S.T., Ikonomovic M.D., and Gandy S. (2010). Traumatic brain injury: football, warfare, and long-term effects. N. Engl. J. Med. 363, 1293–1296 [DOI] [PubMed] [Google Scholar]
- 2.Ruff R.M., Iverson G.L., Barth J.T., Bush S.S., and Broshek D.K. (2009). Recommendations for diagnosing a mild traumatic brain injury: a National Academy of Neuropsychology education paper. Arch. Clin. Neuropsychol. 24, 3–10 [DOI] [PubMed] [Google Scholar]
- 3.Borg J., Holm L., Cassidy J.D., Peloso P., Carroll L., Von Holst H., and Ericson K. (2004). Diagnostic procedures in mild traumatic brain injury: results of the WHO Collaborating Centre Task Force on Mild Traumatic Brain Injury. J. Rehabil. Med. 36, Suppl 43, 61–75 [DOI] [PubMed] [Google Scholar]
- 4.Lipton M.L., Kim N., Park Y.K., Hulkower M.B., Gardin T.M., Shifteh K., Kim M., Zimmerman M.E., Lipton R.B., and Branch C.A. (2012). Robust detection of traumatic axonal injury in individual mild traumatic brain injury patients: intersubject variation, change over time and bidirectional changes in anisotropy. Brain Imaging Behav. 6, 329–342 [DOI] [PubMed] [Google Scholar]
- 5.Huisman T.A., Schwamm L.H., Schaefer P.W., Koroshetz W.J., Shetty-Alva N., Ozsunar Y., Wu O., and Sorensen A.G. (2004). Diffusion tensor imaging as potential biomarker of white matter injury in diffuse axonal injury. AJNR Am. J. Neuroradiol. 25, 370–376 [PMC free article] [PubMed] [Google Scholar]
- 6.Narayana P.A., Yu X., Hasan K.M., Wilde E.A., Levin H.S., Hunter J.V., Miller E.R., Patel V.K., Robertson C.S., and McCarthy J.J. (2015). Multi-modal MRI of mild traumatic brain injury. NeuroImage Clin. 7, 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ling J.M., Peña A., Yeo R.A., Merideth F.L., Klimaj S., Gasparovic C., and Mayer A.R. (2012). Biomarkers of increased diffusion anisotropy in semi-acute mild traumatic brain injury: a longitudinal perspective. Brain 135, 1281–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huston J.M., and Field A.S. (2013). Clinical applications of diffusion tensor imaging. Magn. Reson. Imaging Clin. North Am. 21, 279–298 [DOI] [PubMed] [Google Scholar]
- 9.Kim N., Branch C.A., Kim M., and Lipton M.L. (2013). Whole brain approaches for identification of microstructural abnormalities in individual patients: comparison of techniques applied to mild traumatic brain injury. PloS One 8, e59382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mayer A.R., Bedrick E.J., Ling J.M., Toulouse T., and Dodd A. (2014). Methods for identifying subject‐specific abnormalities in neuroimaging data. Hum. Brain Mapp. 35, 5457–5470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holli K.K., Wäljas M., Harrison L., Liimatainen S., Luukkaala T., Ryymin P., Eskola H., Soimakallio S., Öhman J., and Dastidar P. (2010). Mild traumatic brain injury: tissue texture analysis correlated to neuropsychological and DTI findings. Acad. Radiol. 17, 1096–1102 [DOI] [PubMed] [Google Scholar]
- 12.Arenth P.M., Russell K.C., Scanlon J.M., Kessler L.J., and Ricker J.H. (2014). Corpus callosum integrity and neuropsychological performance after traumatic brain injury: a diffusion tensor imaging study. J. Head Trauma Rehabil. 29, E1–E10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van den Heuvel M.P., Mandl R.C., Kahn R.S., Hulshoff Pol H.E. (2009). Functionally linked resting‐state networks reflect the underlying structural connectivity architecture of the human brain. Hum. Brain Mapp. 30, 3127–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinnunen K.M., Greenwood R., Powell J.H., Leech R., Hawkins P.C., Bonnelle V., Patel M.C., Counsell S.J., and Sharp D.J. (2010). White matter damage and cognitive impairment after traumatic brain injury. Brain 134, 449–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharp D.J., Scott G., and Leech R. (2014). Network dysfunction after traumatic brain injury. Nat. Rev. Neurol. 10, 156–166 [DOI] [PubMed] [Google Scholar]
- 16.Mayer A.R., Bellgowan P.S., and Hanlon F.M. (2015). Functional magnetic resonance imaging of mild traumatic brain injury. Neurosci. Biobehav. Rev. 49, 8–18 [DOI] [PubMed] [Google Scholar]
- 17.Sours C., Zhuo J., Janowich J., Aarabi B., Shanmuganathan K., and Gullapalli R.P. (2013). Default mode network interference in mild traumatic brain injury–a pilot resting state study. Brain Res. 1537, 201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayer A.R., Mannell M.V., Ling J., Gasparovic C., and Yeo R.A. (2011). Functional connectivity in mild traumatic brain injury. Hum. Brain Mapp. 32, 1825–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Y., Milham M.P., Lui Y.W., Miles L., Reaume J., Sodickson D.K., Grossman R.I., and Ge Y. (2012). Default-mode network disruption in mild traumatic brain injury. Radiology 265, 882–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vakhtin A.A., Calhoun V.D., Jung R.E., Prestopnik J.L., Taylor P.A., and Ford C.C. (2013). Changes in intrinsic functional brain networks following blast-induced mild traumatic brain injury. Brain Inj. 27, 1304–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang L., Ge Y., Sodickson D.K., Miles L., Zhou Y., Reaume J., and Grossman R.I. (2011). Thalamic resting-state functional networks: disruption in patients with mild traumatic brain injury. Radiology 260, 831–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y., Lui Y.W., Zuo X.N., Milham M.P., Reaume J., Grossman R.I., and Ge Y. (2014). Characterization of thalamo‐cortical association using amplitude and connectivity of functional MRI in mild traumatic brain injury. J. Magn. Reson. Imaging 39, 1558–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slobounov S., Gay M., Zhang K., Johnson B., Pennell D., Sebastianelli W., Horovitz S., and Hallett M. (2011). Alteration of brain functional network at rest and in response to YMCA physical stress test in concussed athletes: rsFMRI study. Neuroimage 55, 1716–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nathan D.E., Oakes T.R., Yeh P.H., French L.M., Harper J.F., Liu W., Wolfowitz R.D., Wang B.Q., Graner J.L., and Riedy G. (2014). Exploring variations in functional connectivity of the resting state default mode network in mild traumatic brain injury. Brain Connect. 5, 102–114 [DOI] [PubMed] [Google Scholar]
- 25.Hillary F.G., Rajtmajer S.M., Roman C.A., Medaglia J.D., Slocomb-Dluzen J.E., Calhoun V.D., Good D.C., and Wylie G.R. (2014). The rich get richer: brain injury elicits hyperconnectivity in core subnetworks. PloS One 9, e104021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayer A., Ling J., Allen E.A., Klimaj S., Yeo R. and Hanlon F.M. (2014). Static and dynamic intrinsic connectivity following mild traumatic brain injury. J. Neurotrauma. Epub ahead of print [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Damaraju E., Allen E.A. and Calhoun V.D. (2014). Impact of head motion on ICA-derived functional connectivity measures, in: Abstracts Fourth Biennial Conference on Resting State Brain Connectivity. Brain Connectivity: Boston/Cambridge, Massachusetts [Google Scholar]
- 28.Allen E.A., Erhardt E.B., Damaraju E., Gruner W., Segall J.M., Silva R.F., Havlicek M., Rachakonda S., Fries J., Kalyanam R., Michael A.M., Caprihan A., Turner J.A., Eichele T., Adelsheim S., Bryan A.D., Bustillo J., Clark V.P., Feldstein Ewing S.W., Filbey F., Ford C.C., Hutchison K., Jung R.E., Kiehl K.A., Kodituwakku P., Komesu Y.M., Mayer A.R., Pearlson G.D., Phillips J.P., Sadek J.R., Stevens M., Teuscher U., Thoma R.J., and Calhoun V.D. (2011). A baseline for the multivariate comparison of resting-state networks. Front. Syst. Neurosci. 5, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan K., Qin W., Yu D., Bi Y., Xing L., Jin C. and Tian J. (2015). Core brain networks interactions and cognitive control in internet gaming disorder individuals in late adolescence/early adulthood. Brain Struct. Funct. 221, 1427–1442 [DOI] [PubMed] [Google Scholar]
- 30.Rocca M.A., Turconi A.C., Strazzer S., Absinta M., Valsasina P., Beretta E., Copetti M., Cazzagon M., Falini A., and Filippi M. (2013). MRI predicts efficacy of constraint-induced movement therapy in children with brain injury. Neurotherapeutics 10, 511–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vergara V.M., Mayer A.R., Damaraju E., Hutchison K., and Calhoun V.D. (2016). The effect of preprocessing pipelines in subject classification and detection of abnormal resting state functional network connectivity using group ICA. Neuroimage. Epub ahead of print [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharp D.J., Beckmann C.F., Greenwood R., Kinnunen K.M., Bonnelle V., De Boissezon X., Powell J.H., Counsell S.J., Patel M.C., and Leech R. (2011). Default mode network functional and structural connectivity after traumatic brain injury. Brain 134, 2233–2247 [DOI] [PubMed] [Google Scholar]
- 33.Stevens M.C., Lovejoy D., Kim J., Oakes H., Kureshi I., and Witt S.T. (2012). Multiple resting state network functional connectivity abnormalities in mild traumatic brain injury. Brain Imaging Behav. 6, 293–318 [DOI] [PubMed] [Google Scholar]
- 34.Friston K.J. (2003). Statistical parametric mapping. In: Neuroscience Databases. Springer: New York, pps. 237–250 [Google Scholar]
- 35.Power J.D., Barnes K.A., Snyder A.Z., Schlaggar B.L., and Petersen S.E. (2012). Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage 59, 2142–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen E.A., Erhardt E.B., Damaraju E., Gruner W., Segall J.M., Silva R.F., Havlicek M., Rachakonda S., Fries J., Kalyanam R., Michael AM, Caprihan A, Turner JA, Eichele T, Adelsheim S, Bryan AD, Bustillo J, Clark VP, Feldstein Ewing SW, Filbey F, Ford CC, Hutchison K, Jung RE, Kiehl KA, Kodituwakku P, Komesu YM, Mayer AR, Pearlson GD, Phillips JP, Sadek JR, Stevens M, Teuscher U, Thoma RJ, Calhoun VD. (2011). A baseline for the multivariate comparison of resting-state networks. Front. Syst. Neurosci. 5, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calhoun V., Adali T., Pearlson G., and Pekar J. (2001). A method for making group inferences from functional MRI data using independent component analysis. Hum. Brain Mapp. 14, 140–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calhoun V.D., and Adali T. (2012). Multisubject independent component analysis of fMRI: a decade of intrinsic networks, default mode, and neurodiagnostic discovery. IEEE Rev. Biomed. Eng. 5, 60–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Himberg J., Hyvärinen A., and Esposito F. (2004). Validating the independent components of neuroimaging time series via clustering and visualization. Neuroimage 22, 1214–1222 [DOI] [PubMed] [Google Scholar]
- 40.Ma S., Correa N.M., Li X.L., Eichele T., Calhoun V.D., and Adali T. (2011). Automatic identification of functional clusters in FMRI data using spatial dependence. IEEE Trans. Biomed. Eng. 58, 3406–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whelan R., Watts R., Orr C.A., Althoff R.R., Artiges E., Banaschewski T., Barker G.J., Bokde A.L., Büchel C., Carvalho F.M., Conrod P.J., Flor H., Fauth-Bühler M., Frouin V, Gallinat J., Gan G., Gowland P., Heinz A., Ittermann B., Lawrence C., Mann K., Martinot J.L., Nees F., Ortiz N., Paillère-Martinot M.L., Paus T., Pausova Z., Rietschel M., Robbins T.W., Smolka M.N., Ströhle A., Schumann G, Garavan H.; IMAGEN Consortium. (2014). Neuropsychosocial profiles of current and future adolescent alcohol misusers. Nature 512, 185–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hahn T., Kircher T., Straube B., Wittchen H.U., Konrad C., Ströhle A., Wittmann A., Pfleiderer B., Reif A., Arolt V., and Lueken U. (2015). Predicting treatment response to cognitive behavioral therapy in panic disorder with agoraphobia by integrating local neural information. JAMA Psychiatry 72, 68–74 [DOI] [PubMed] [Google Scholar]
- 43.Koch M.A., Norris D.G., and Hund-Georgiadis M. (2002). An investigation of functional and anatomical connectivity using magnetic resonance imaging. Neuroimage 16, 241–250 [DOI] [PubMed] [Google Scholar]
- 44.Yuh E.L., Cooper S.R., Mukherjee P., Yue J.K., Lingsma H.F., Gordon W.A., Valadka A.B., Okonkwo D.O., Schnyer D.M., Vassar M.J., Maas AI, Manley GT; TRACK-TBI Investigators. (2014). Diffusion tensor imaging for outcome prediction in mild traumatic brain injury: a TRACK-TBI study. J. Neurotrauma 31, 1457–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shenton M., Hamoda H., Schneiderman J., Bouix S., Pasternak O., Rathi Y., Vu M.A., Purohit M., Helmer K., Koerte I., Lin AP, Westin CF, Kikinis R, Kubicki M, Stern RA, and Zafonte R. (2012). A review of magnetic resonance imaging and diffusion tensor imaging findings in mild traumatic brain injury. Brain Imaging Behav. 6, 137–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sui J., Pearlson G., Caprihan A., Adali T., Kiehl K.A., Liu J., Yamamoto J., and Calhoun V.D. (2011). Discriminating schizophrenia and bipolar disorder by fusing fMRI and DTI in a multimodal CCA+ joint ICA model. Neuroimage 57, 839–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J.Y., Pearlson G., Windemuth A., Ruano G., Perrone-Bizzozero N.I., and Calhoun V. (2009). Combining fMRI and SNP data to investigate connections between brain function and genetics using parallel ICA. Hum. Brain Mapp. 30, 241–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kasahara M., Menon D., Salmond C., Outtrim J., Tavares J.T., Carpenter T., Pickard J., Sahakian B., and Stamatakis E. (2010). Altered functional connectivity in the motor network after traumatic brain injury. Neurology 75, 168–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bharath R.D., Munivenkatappa A., Gohel S., Panda R., Saini J., Rajeswaran J., Shukla D., Bhagavatula I.D., and Biswal B.B. (2015). Recovery of resting brain connectivity ensuing mild traumatic brain injury. Front. Hum. Neurosci. 9, 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leech R., and Sharp D.J. (2014). The role of the posterior cingulate cortex in cognition and disease. Brain 137, 12–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Timmann D., and Daum I. (2007). Cerebellar contributions to cognitive functions: a progress report after two decades of research. Cerebellum 6, 159–162 [DOI] [PubMed] [Google Scholar]
- 52.Schmahmann J.D., and Caplan D. (2006). Cognition, emotion and the cerebellum. Brain 129, 290–292 [DOI] [PubMed] [Google Scholar]
- 53.Shen H., Wang L., Liu Y., and Hu D. (2010). Discriminative analysis of resting-state functional connectivity patterns of schizophrenia using low dimensional embedding of fMRI. Neuroimage 49, 3110–3121 [DOI] [PubMed] [Google Scholar]
- 54.Calhoun V.D., and Sui J. (2016). Multimodal fusion of brain imaging data: a key to finding the missing link (s) in complex mental illness. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 1, 230–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.