Abstract
The gain of eccrine sweat glands in hairy body skin has empowered humans to run marathons and tolerate temperature extremes. Epithelial-mesenchymal cross-talk is integral to the diverse patterning of skin appendages, but the molecular events underlying their specification remain largely unknown. Using genome-wide analyses and functional studies, we show that sweat glands are specified by mesenchymal-derived bone morphogenetic proteins (BMPs) and fibroblast growth factors that signal to epithelial buds and suppress epithelial-derived sonic hedgehog (SHH) production. Conversely, hair follicles are specified when mesenchymal BMP signaling is blocked, permitting SHH production. Fate determination is confined to a critical developmental window and is regionally specified in mice. In contrast, a shift from hair to gland fates is achieved in humans when a spike in BMP silences SHH during the final embryonic wave(s) of bud morphogenesis.
Epithelial appendages—including hair follicles (HFs) and teeth as well as mammary, sweat, and salivary glands—begin to form during embryogenesis when Wnt signaling triggers progenitors within the epithelial sheet to organize spatially into morphologically similar placodes. Whereas most mammals restrict the specification of epidermal appendages regionally, HFs and sweat glands (SwGs) coexist throughout much of the skin of primates. The acquisition of SwGs and their importance in thermoregulation are underscored by humans who suffer from a life-threatening condition when SwGs are eliminated, either from loss in severe burns or from the genetic disorder hypohidrotic ectodermal dysplasia (HED).
Patients with HED display mutations in genes encoding proteins such as ectodermal dysplasia antigen (EDA), the EDA receptor (EDAR), and WNT10a, a ligand for Wnt signaling. These findings have illuminated a critical role for these pathways in controlling the development of a number of epidermal appendages, including SwGs and coarse hairs (1, 2). Conversely, an EDAR gain-of-function variant has been expanding among the Southeast Asian population, where excessive SwGs are desirable because of the hot and humid climate of that region (3). In this regard, both Wnt and EDA/EDAR pathways appear to function in promoting placode formation, increasing the density of SwGs and several other appendage types (4).
Classical tissue recombination experiments have revealed that mesenchyme plays a critical role in dictating the divergent downstream events that determine appendage selection (5, 6). For example, salivary mesenchyme combined with mammary epithelium generates an epithelial morphology resembling salivary glands (7), and regardless of its regional origin, embryonic chick epidermis develops feathers if combined with feather-forming dermis but develops scales if the epidermis is exposed to scale-forming dermis (8). Similarly, rabbit corneal epithelium generates either HFs or SwGs depending on whether it is exposed to the mesenchyme of dorsal back skin or ventral foot skin, respectively (9). Despite the existence of many such examples, relatively little is known about the specific spatiotemporal cross-talk and molecular mechanisms that underlie epithelial fate specification in response to mesenchymal exposure.
Regional differences in BMP pathway genes in skin mesenchymes
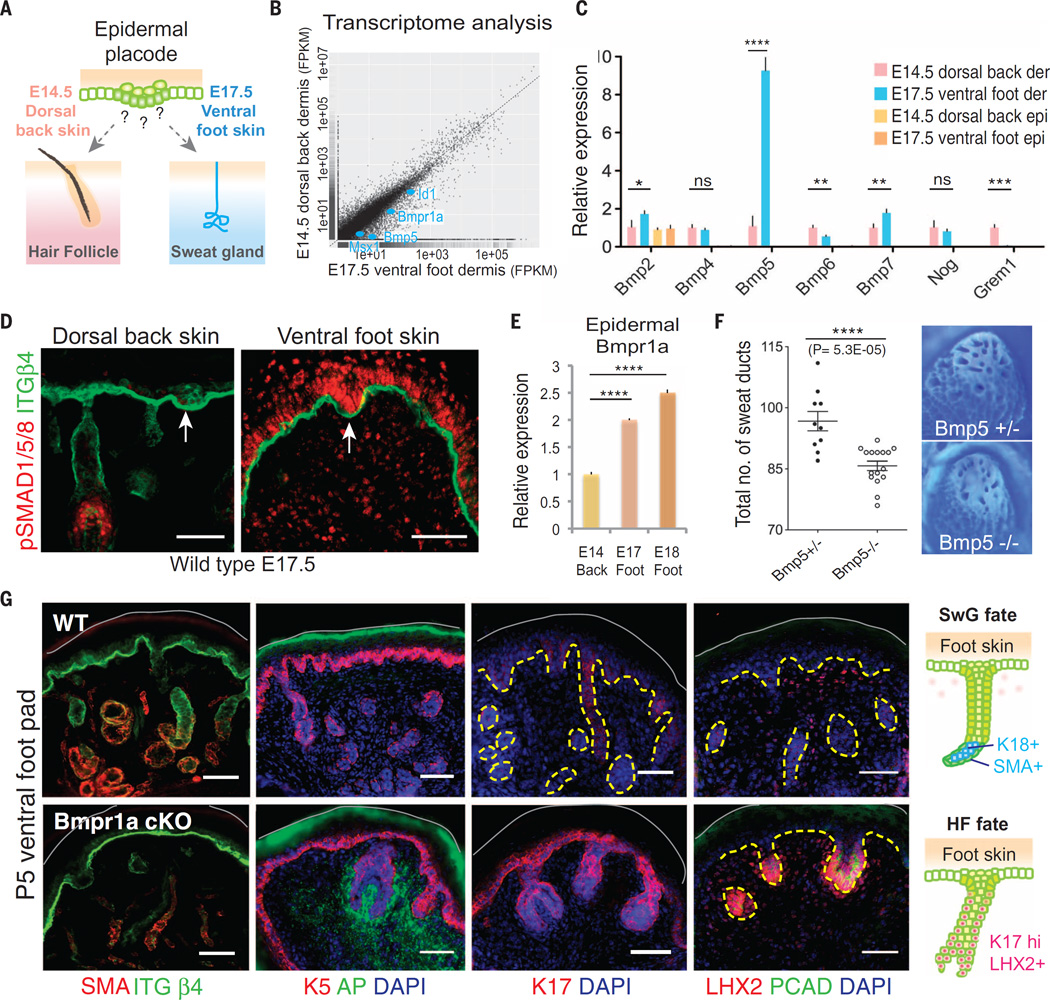
To understand how epithelial fate is specified by mesenchymal signals, we began by exploiting the fact that in mice, dorsal back skin supports only HF morphogenesis, whereas eccrine SwG morphogenesis is restricted to ventral foot skin. We first pinpointed the appearance of WNT-induced epidermal placodes at these body sites, reasoning that the underlying mesenchymes become competent to specify the differential fates of these placodes. We then carried out genomewide, high-throughput RNA sequencing (RNAseq) on body site–specific dermal mesenchymes at the embryonic ages when their respective placodes emerge [embryonic day 14.5 (E14.5), dorsal back skin; E17.5, ventral foot skin] (Fig. 1A).
Fig. 1. Mesenchymal-epithelial BMP signaling is required for sweat gland fate.
(A) Schematics of spatiotemporally distinct development of mouse sweat glands and hair follicles. (B) Scatter plot of RNA-seq analysis of E14.5 dorsal back skin and E17.5 ventral foot skin dermis. Bmpr1a, Bmp5, Id1, and Msx1 (targets of BMP signaling) are enriched in ventral foot skin dermis (blue dots). (C) Quantitative PCR for various BMP ligands and inhibitors in E14.5 dorsal back skin (HF-permissive) and E17.5 ventral foot skin (SwG-permissive) epidermis and dermis. Shown is a strong up-regulation of Bmp5 in mouse foot skin dermis. (D) Immunofluorescence of phospho-SMAD1/5/8 in dorsal back and ventral foot skins. Arrows denote hair and sweat placodes, respectively. Scale bars, 50 µm. (E) Quantitative PCR for epidermal Bmpr1a mRNAs at indicated embryonic stages and regional skins. (F) (Left) Quantification of sweat ducts in adult Bmp5 heterozygous and null mice. n = 10 and 16 foot skin samples, respectively. (Right) Representative images of adult foot pad epidermis stained with Nile blue A in order to visualize sweat ducts. (G) Immunofluorescence images of foot pads of wild type (WT) and Krt14-Cre; Bmpr1a conditional knockout (cKO) at P5. SMA, smooth muscle actin, a myoepithelial glandular marker; K5, keratin 5, which marks the basal layer of epidermal progenitors; ITGβ4, integrinβ4, which marks the basal surface of the epidermis; AP, alkaline phosphatase, a marker for HF dermal papillae; K17, LHX2 and PCAD, markers of the HF. Gray solid lines denote skin surface. Dashed lines indicate dermo-epidermal border. Scale bars, 50 µm. (Right) Schematics of SwG fate and HF fate in the foot skin.
Comparative bioinformatics revealed that bone morphogenetic protein (BMP) signaling pathway genes were significantly enriched [≥2×, P < 0.05, false-discovery rate (q value) < 0.05] among the transcripts [with fragments per kilobase per million mapped reads (FPKM) > 1] in ventral foot skin versus dorsal back skin dermis (Fig. 1B). Of these, the BMP5 ligand stood out for its regional-specific expression, confirmed with quantitative polymerase chain reaction (PCR) (Fig. 1C). Also surfacing in our differential transcriptome analysis were Bmpr1a, encoding a key component of the BMP receptor kinase, and Id1 and Msx1, two downstream targets of its activated transcriptional effectors, phospho-SMAD1/5/8 (Fig. 1B).
Anti-phospho-SMAD1/5/8 immunofluorescence confirmed the marked differential elevation in BMP signaling within ventral foot skin versus dorsal back skin during the fate-permissive state of their epithelial buds (Fig. 1D). However, there was also strong nuclear immunolabeling in the overlying SwG buds, not seen in HF buds. Bmpr1a transcripts were also markedly elevated in SwG-permissive epidermis, relative to HF-permissive epidermis (Fig. 1E). We therefore examined BMPs and BMP inhibitors in both mesenchyme and epidermis. Despite expression of other BMPs and BMP inhibitors in both mesenchyme and epidermis, only mesenchymal BMP5 exhibited a regional-specific expression that might account for the striking difference in BMP signaling that we observed between SwG and HF buds (Fig. 1C).
Mesenchymal-epithelial BMP signaling in specifying SwGs
If paracrine mesenchymal-epithelial BMP signaling in foot skin occurs, as our data suggest, and if this cross-talk is physiologically important, then loss of mesenchymal Bmp5 should affect overlying SwG development. Ablating Bmp5 significantly reduced SwG numbers in ventral foot skin and resulted in decreased epithelial BMP signaling (pSMAD1) (Fig. 1F and fig. S1A). Although the effect was robust, SwGs still developed and BMP signaling was still seen in both mesenchymal and epithelial compartments of ventral foot skin (fig. S1A), suggesting that other mesenchymal BMPs may partially compensate for the loss of BMP5. To test this possibility and assess the full functional relevance of the elevated BMP signaling in the footpads, we used K14-Cre mice to conditionally ablate Bmpr1a in embryonic skin epidermis. Immuno-pSMAD1/5/8 labeling confirmed that BMP signaling was completely abrogated in the epithelium, whereas it was still sustained as expected in the underlying mesenchyme (fig. S1B). Under these conditions, no SwGs were generated (fig. S1C).
Instead of forming a coiled gland, the Bmpr1a-null epithelial down-growths in the ventral foot pads enveloped the associated dermal mesenchyme and assumed a morphology, biochemistry, and proliferative status more typical of HF bulbs (Fig. 1G and fig. S1D). By postnatal day 5 (P5), K17, LHX2, and P-cadherin—hallmarks of HFs (10) and not expressed by SwGs—were all ectopically expressed in Bmpr1a-null foot skin epithelial down-growths. In contrast, smooth muscle actin, featured by the myoepithelial cells of the gland, was absent (Fig. 1G). Further reflective that an epithelial fate switch had occurred, we observed condensation of alkaline phosphatase–positive mesenchymal cells into a structure resembling a dermal papilla (DP), the cluster of mesenchymal cells associated with the HF that are essential for its growth and maintenance (11). These data not only document the physiological relevance of elevating epithelial BMP signaling in developing SwGs but also suggest that paracrine cross-talk between mesenchymal BMPs and BMP receptorhigh epithelial buds is critical for SwG development.
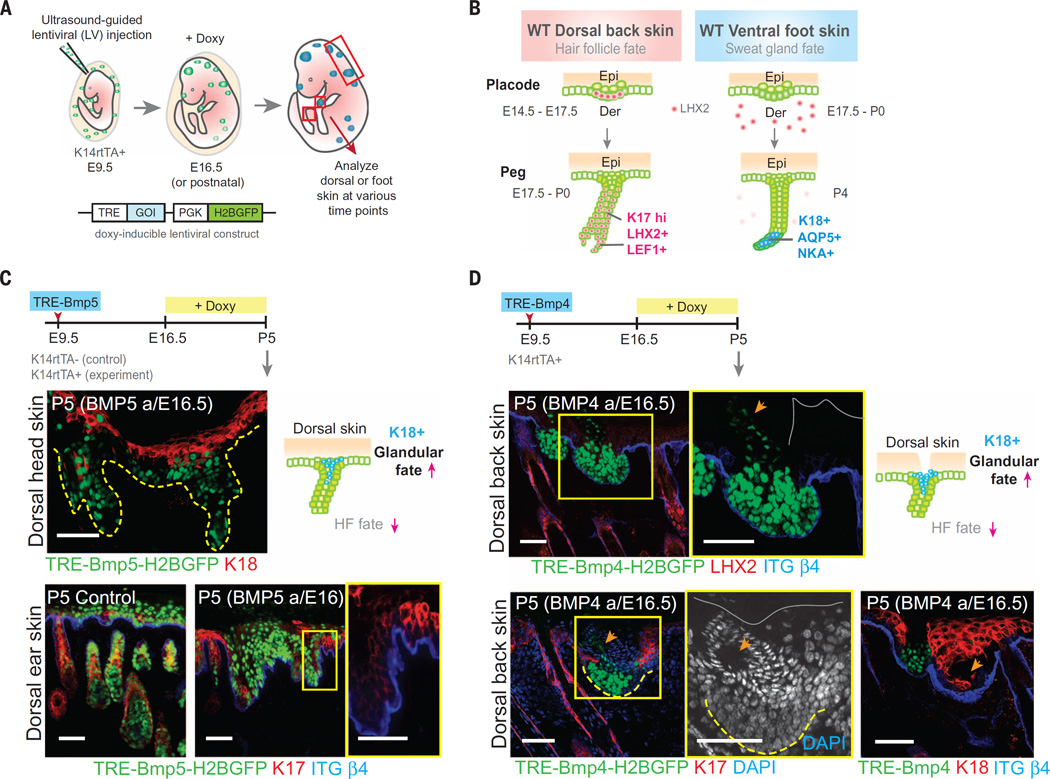
BMP signaling can cause a hair follicle to glandular fate switch
To further test whether these regional differences in BMP signaling are key to SwG versus HF appendage fate, we generated lentiviruses harboring aPGK-H2BGFP marker and doxycycline-inducible transgenes encoding BMPs and BMP inhibitor. We then used an ultrasound-guided in utero injection system(12) to introduce lentivirus into the amniotic sacs of living E9.5 K14rtTA mouse embryos (Fig. 2A). Within 24 hours, the transgene and green fluorescent protein (GFP) marker selectively and stably integrate into the host genome of epidermal progenitor cells and their progeny (12). Transgene expression can then be controlled by the timing of doxycycline administration.
Fig. 2. Ectopic elevation of BMP signaling in dorsal skin elicits a HF: glandular fate shift.
(A) Schematic of ultrasound-guided in utero lentiviral injections and doxycycline (Doxy)–inducible ectopic expression system. K14-rtTA embryos express the Doxy-inducible transactivator beginning at ~E13.5. (B) Schematic of wild-type hair and sweat placode and peg stages. HF and SwG markers are shown in pink and blue, respectively. (C and D) Timeline of lentiviral injection, BMP5 (C) and BMP4 (D) induction and analyses. Immunofluorescence images of dorsal skin show loss of HF markers and ectopic expression of glandular luminal cell marker K18. Lentiviral injections were performed at least two times, and two to four animals were collected and analyzed at each time point. Boxed areas are shown in higher magnification at right. Gray solid lines mark the skin surface. Dashed lines indicate dermoepidermal border and arrows indicate ductal structures associated with BMP4-expressing epidermal cells. Scale bars, 50 µm; except (C) lower right, 25 µm.
We first tested the consequences of elevating BMPs in dorsal hairy skin. After transducing with TRE-Bmp5, we administered doxycycline at E16.5 of gestation. Transgene expression was robust by E17.5 during permissive phases of sweat buds and final waves of HF buds (Fig. 2, B and C). By analyzing at P5, we learned that BMP5 dramatically perturbed back skin HF morphogenesis, and HF markers were lost (Fig. 2C). A similar block in HF morphogenesis was seen upon activation of TRE-Bmp4 in embryonic back skin epidermis (Fig. 2D). Elevating BMP levels not only blocked HF morphogenesis in dorsal back skin but also ectopically induced glandular and ductal-like structures, accompanied by expression of glandular keratin, K18 (Fig. 2D). Together with our loss-of-function data, these results suggest that collective BMP levels, rather than specific ligand, affect fate choice.
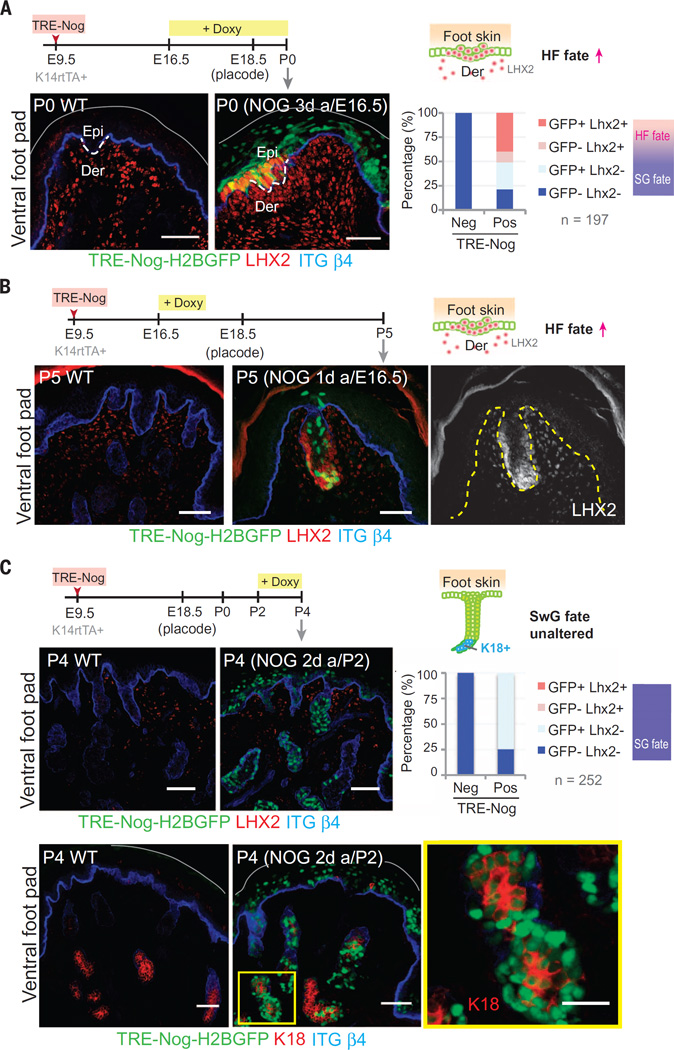
BMP inhibition acts within a critical window to cause SwG to HF fate switch
We next examined the consequences of inhibiting BMP signaling throughout the ventral foot skin by ectopically expressing the secreted BMP inhibitor NOGGIN. Prior studies on back skin revealed a role for NOGGIN in promoting HF fate (13), and it was previously postulated that ectopic NOGGIN might trigger “transdifferentiation” of SwGs into hairs (14). Our findings raised the additional possibility that by intercepting mesenchymal BMPs, NOGGIN might act by blocking signaling directly to the epithelium, redirecting foot skin buds to a HF fate. To test this possibility, it was necessary to control NOGGIN expression.
We first induced a TRE-Noggin transgene at E16.5 of embryogenesis just before sweat bud formation. Soon thereafter, the epithelial foot skin buds showed signs of HF morphogenesis, including LIM/homeobox protein (LHX2) expression (Fig. 3A). Quantification revealed that even at these early times after Noggin activation, ~50% of transduced (GFP+) cells were already LHX2+, an early HF fate indicator. These changes occurred before signs of dermal condensation.
Fig. 3. Ectopic inhibition of BMP acts during a critical window to suppress SwG fate in the foot skin.
(A to C) Timeline of LV injection, NOGGIN induction, and analyses. (A) Immunofluorescence images and quantification of LHX2+ and LHX2– cells in the LV-infected and uninfected epidermis from three different foot pads (cell number as indicated). When BMP is inhibited during early stages of epithelial bud formation, a switch from SwG to HF fate occurs, reflected by the appearance of LHX2 in the epithelium. (B) Immunofluorescence images of LHX2. A brief pulse of NOGGIN expression at the placode stage is sufficient to cause a fate change that lasts postnatally. (C) Immunofluorescence images and quantification of LHX2+ and LHX2– cells in the LV-infected and uninfected epidermis from three different foot pads (cell number as indicated). Inhibiting BMP after the placode stage fails to elicit a fate change, as reflected by the sustained expression of SwG luminal cell marker K18 in the NOGGIN+ ventral foot skin. Lentiviral injections were performed at least two times, and two to four animals were collected and analyzed at each time point. Gray solid lines mark the skin surface. Dashed lines indicate dermoepidermal border. Boxed area is shown in higher magnification at right. Scale bars, 50 µm and (bottom far right) 25 µm.
A short pulse of NOGGIN expression at this critical placode stage was sufficient to trigger these events, which then lasted postnatally, as evidenced by the LHX2-positive hair pegs within P5 foot skin (Fig. 3B). In contrast, when we waited until P2 to initiate NOGGIN expression, no adverse effects were seen in the foot skin (Fig. 3C). Glands were still formed and expressed only SwG and not HF markers, without any trace of early HF marker LHX2 in NOGGIN-expressing cells. Together, these results extend earlier findings of Plikus et al. (14) but add three key new points: First, BMP activation in back skin or BMP inhibition in foot skin potently affects SwG versus HF fate determination. Second, the impact of BMP signaling on SwG fate choice occurs exclusively within a narrow temporal window when ventral foot skin buds appear. Third, the BMP-mediated changes in the epithelium precede those in mesenchyme, underscoring the importance of mesenchymal-epithelial cross-talk and offering molecular insights into the classical tissue recombination experiments.
BMP signaling enhances mesenchymal but dampens epithelial Wnt signaling
Canonical Wnt signaling, mediated through its effector β-catenin and its transcriptional coactivator lymphoid enhancer–binding factor 1 (LEF1), is essential for placode formation irrespective of appendage type (15–18). Indeed, epidermal-specific loss of β-catenin abrogated sweat bud formation (fig. S2A) (18). Cross-talk at the epidermal-dermal border involving Wnt signaling is also well established (19). However, we were curious to know how the subsequent elevation in BMP signaling throughout the ventral foot skin would affect this cross-talk; although BMP inhibition is known to elevate Wnt signaling and stabilize LEF1 in hair buds (20, 21), back skin explant studies have suggested a positive role for BMP signaling in mesenchymal Lef1 expression (22). If these prior studies are functionally relevant, the heightened BMP signaling throughout the ventral foot skin should skew the Wnt cross-talk at the epidermal-dermal border, elevating mesenchymal Wnt signaling and dampening epithelial Wnt signaling downstream from placode formation.
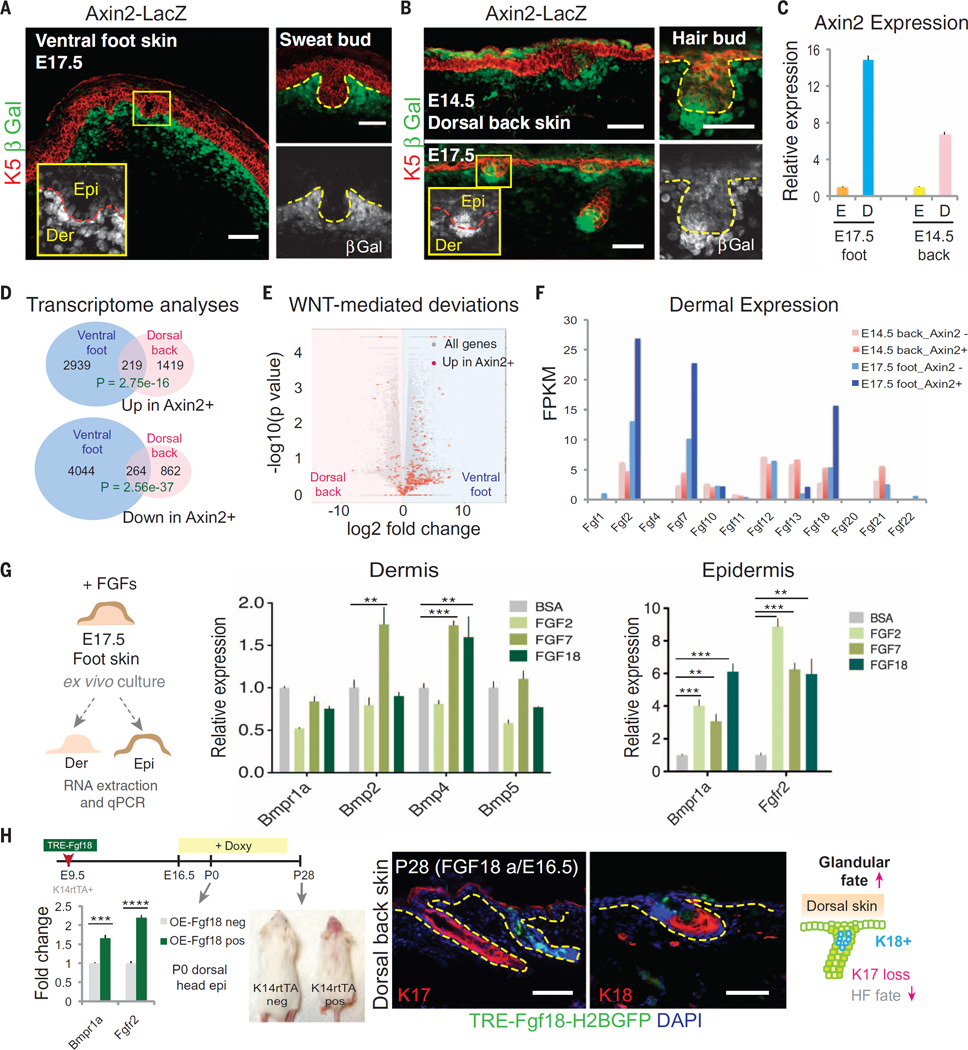
Indeed, during the bud-permissive phase of ventral foot skin, WNT reporter (Axin2-LacZ) activity was markedly stronger in the mesenchyme than in epithelium (Fig. 4A). Closer inspection revealed its presence in the epithelial placodes, but as buds developed, it became even weaker (Fig. 4A) and only reappeared in sweat epithelium when sweat ducts were formed (fig. S2B). In contrast, during the bud-permissive phase of dorsal back skin, WNT reporter activity was present in both mesenchyme and epithelium (Fig. 4B) (23). Similarly, nuclear LEF1, a key canonical Wnt effector, showed nuclear LEF1 transiently in SwG buds and diminished LEF1 in down-growths of ventral foot skin, whereas in dorsal back skin, LEF1 was high in the epithelial buds and sustained at the leading front of the down-growths, where it was seen in both the epithelium and the DP (fig. S3A). Quantification by means of quantitative PCR confirmed that the Wnt gradient was sharper in the ventral foot skin than in the dorsal back skin during the bud-permissive phase (Fig. 4C).
Fig. 4. WNT responsiveness is elevated in mesenchyme and dampened in epithelium of BMP-high ventral foot skin, leading to dermal and epidermal changes in fate determinants of SwG-permissive skin.
(A and B) Axin2-LacZ (WNT) reporter expression reveals a sharpened epithelial-mesenchymal WNT differential in ventral foot skin (E17.5) (A), compared with dorsal back skin (E14.5 and E17.5) (B). Boxed areas highlight marked differences in Wnt signaling between sweat and hair buds (from E17.5 foot skin and back skin, respectively). Epi, epidermis; Der, dermis. Dashed lines indicate dermo-epidermal border. Scale bars, 50 µm. (C) Quantitative PCR for Axin2 expression in the epidermis (E) and dermis (D) of E17.5 foot skin and E14.5 back skin, confirming the sharpened epithelial-mesenchymal WNT differential in ventral foot skin (E17.5), compared with dorsal back skin (E14.5). Dermal values are normalized to the corresponding epidermis. (D) Venn diagrams summarizing transcriptome analyses of genes upregulated or down-regulated in Axin2-LacZ–positive ventral compared with dorsal dermis. (E) Volcano plot showing all transcripts that are log2-fold changed in dorsal versus ventral dermis. Transcripts greater than fivefold changed between Axin2-positive and Axin2-negative cells of each dermal population are labeled in red. WNT-responsive genes are generally more highly expressed in ventral foot skin relative to dorsal back skin dermis. (F) Expression of FGF ligand family member transcripts in WNT-responding and -nonresponding fibroblasts from dorsal and ventral dermis at their respective embryonic stages when epidermal placodes emerge. Shown are strong upregulation of Fgf2, Fgf7, and Fgf18 in Axin2-LacZ–positive versus –negative foot dermis. (G) Ex vivo effects of FGFs on E17.5 foot skin. (Left) After 16 to 20 hours of treatment, dermis and epidermis were manually separated and analyzed by means of quantitative PCR (middle and right, respectively) for transcripts indicated. Shown is a FGF-mediated Bmp elevation in dermis; Bmpr and Fgfr2 in epidermis. n = 6 to 8 ventral foot skin explants from three embryos. (H) Timeline of lentiviral injection,TRE-FGF18 induction, and analyses of dorsal skin. Quantitative PCR shows FGF18-induced up-regulation of Bmpr1a and Fgfr2 in vivo. Hair loss is visible in the highly transduced head skin as a result of ectopic FGF18 expression. (Right) Immunofluorescence images and diagram showing loss of HF marker K17 and gain of glandular marker K18 upon FGF18 induction. Dashed lines indicate dermo-epidermal border. Scale bars, 50 µm.
During the time when ventral foot skin was permissive for sweat bud development, the elevation in BMP and Wnt signaling was accompanied by other features that distinguished this mesenchyme from its dorsal counterpart. Reflecting the elevation in BMP signaling at this time, alkaline phosphatase exhibited a broad mesenchymal pattern similar to LEF1 (fig. S3B). LHX2, which in back skin is restricted to the WNT-high cells of the hair bud (10), was also mesenchymally expressed in permissive ventral foot skin dermis (fig. S3C). Postnatally, as the window of sweat bud fate specification waned, so too did all of these features (Fig. 1G and fig. S3).
Elevated Wnt signaling through BMP signaling contributes to divergent mesenchymes
Probing further into how differences in mesenchymal Wnt signaling at the epidermal-dermal border regionally influence the transcriptional landscape of body-site mesenchymes that support distinct bud fates, we transcriptionally profiled the Axin2-positive and Axin2-negative dermal cells after their fluorescence-activated cell sorting (FACS) purification from E17.5 ventral foot skin and E14.5 dorsal back skin (fig. S4). RNA-seq analyses revealed a small percentage (≤10%) of WNT-responsive genes that are shared between glandular and follicular-competent mesenchyme (Fig. 4D). Volcano plot analysis further revealed that transcripts greater than fivefold up-regulated in the Axin2-positive versus Axin2-negative dermal subfractions were distributed away from the midline (Fig. 4E), suggesting that elevated WNT activation through BMP signaling contributes to divergence between glandular and follicular-competent mesenchymes.
Axin2-reporter and Gene Ontology (GO)–term analyses revealed that although Wnt pathway genes were enriched in WNT-receiving cells of both foot and back skin dermis, a number of differences emerged (fig. S5A). Wnt5a and Wnt10a transcripts more highly expressed in foot skin than back skin dermal cells (fig. S5B):WNT5A is known to induce Krt9 (24), encoding a signature keratin of palmoplantar epidermis, and WNT10A is the only known Wnt ligand whose mutations have been implicated in human HED (2, 25). However, among the family of BMP ligands, none showed changes between the Axin2+ and Axin2− population (fig. S5C), which is consistent with the notion that BMP signaling is upstream of Wnt signaling in skin mesenchyme (22).
Wnt signaling elicits elevated FGF signaling in foot skin mesenchyme
There was a difference in mRNAs encoding fibroblast growth factor (FGF) ligands and other Ras–mitogen-activated protein kinase (MAPK) pathway members. In particular, Fgf2, Fgf7, and Fgf18 were more highly transcribed in Wnt-responsive ventral foot skin dermis than in any other dermal fraction examined (Fig. 4F and fig. S5A). We therefore turned to addressing whether FGFs might contribute to sweat fate specification.
To assess FGF functional relevance, we exposed E17.5 foot skin explants for 16 to 20 hours with each of these FGFs and then separated dermis and epithelium and analyzed the transcriptional consequences. FGFs showed little or minor effect on the dermal expression of BMP ligands or BMP receptor (Fig. 4G, middle). In contrast, their effects on epidermal expression of Bmpr1a and Fgfr2 were appreciable (Fig. 4G, right). Although these FGFs are known to activate different receptors and elicit distinct molecular effects in different contexts, their action on these epidermal receptors was similar. Together, these findings suggest that FGFs are selectively up-regulated in BMP-hi and Wnt-receiving foot skin mesenchyme at the dermoepidermal border and act directly on the overlying epithelium to sensitize it to paracrine-generated BMPs and FGFs. Indeed, when we transduced embryos with TRE-Fgf18 and activated FGF18 expression during the HF-permissive phase of dorsal skin, Bmpr1a and Fgfr2 were up-regulated (Fig. 4H). In addition, like BMP5 and BMP4, ectopic FGF18 caused hair loss in dorsal head skin, suppressed expression of HF marker K17, and induced expression of glandular marker K18, altogether acting toward glandular fate.
Expression of Engrailed during SwG fate specification: Parallels to fly wing development
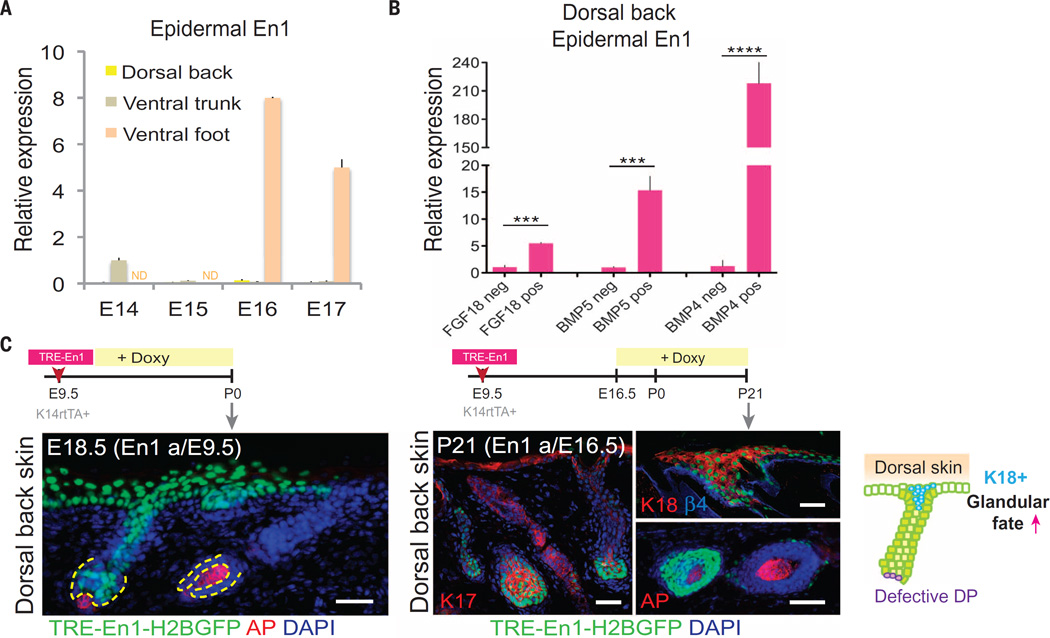
The BMP-induced sharpened differential that we observed in Wnt signaling at the dermoepidermal border of permissive ventral foot skin was reminiscent of Drosophila wing development, in which strong Wnt signaling cells are juxtapositioned with low-Wnt signaling cells, a condition that in turn was necessary to sustain expression of the transcription repressor Engrailed specifically in the low-Wnt-signaling cells (26). We found that En1 was highly expressed in mouse ventral epithelium during the SwG fate–permissive stage of foot skin but not in the dorsal epithelium during the HF fate–permissive stage of back skin (Fig. 5A). En1 was also expressed in early ventral trunk (belly) skin, and its expression waned after E14.5 (Fig. 5A). This timing coincided with the completion of mammary gland fate specification (E13.5) and was just before hair follicle fate specification (E15.5) in the belly skin.
Fig. 5. Engrailed-1 is expressed during SwG fate specification in ventral foot skin and interferes with DP formation when ectopically expressed in dorsal back skin.
(A) En1 quantitative PCR of dorsal back, ventral trunk, and ventral foot epidermal mRNAs during placode-permissive stages. (B) En1 is elevated in dorsal back skin epidermis of ectopic Fgf18-, Bmp5-, and Bmp4-expressing embryos after 3 days of doxy-induction. (C) Timeline of lentiviral injection, TRE-En1 induction, and analyses of dorsal back skin after ectopic expression. Immunofluorescence images for AP (alkaline phosphatase, a marker for DP) and K18 (a marker for glandular fate). There was diminished DP and ectopic induction of K18 in En1-expressing epidermal down-growths of dorsal back skin. Dashed lines indicate dermo-epidermal border. Scale bars, 50 µm.
These data point to a complex circuitry triggered by BMP elevation in ventral foot skin mesenchyme that enhances differential Wnt signaling to reach a permissive threshold to result in En1 expression. In agreement with this hypothesis, TRE-Bmp5 and TRE-Bmp4 activation resulted in a dramatic up-regulation of En1 in dorsal skin epidermis, as did TRE-Fgf18 (Fig. 5B). Moreover, when En1 was ectopically induced, it resulted in transformations consistent with a more glandular fate, including diminished DP and K18 induction (Fig. 5C). Together, these data suggest a role for epidermal ENGRAILED-1 in promoting the SwG fate; we found this noteworthy because mice lacking Engrailed-1 dorsalize the ventral skin (27), and Engrailed-1 heterozygote mutant mice display fewer SwGs in their ventral foot skin (28).
All roads lead to suppression of SHH signaling in SwG-permissive skin
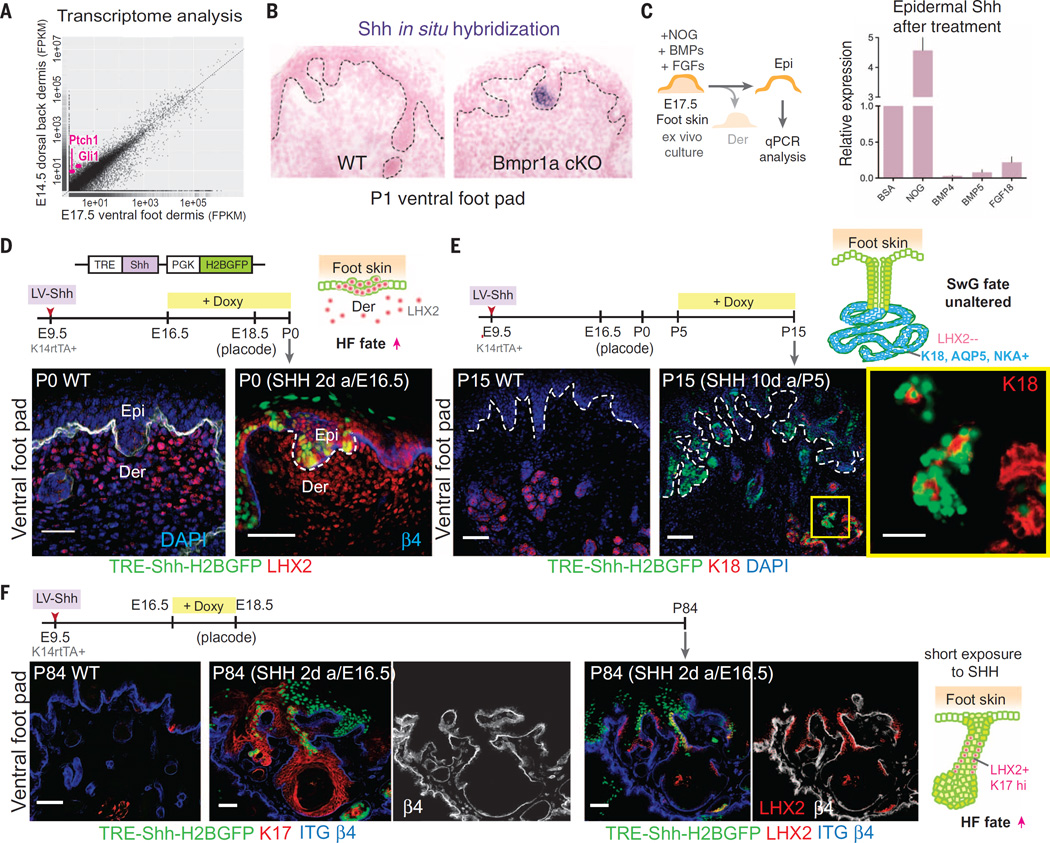
Studies on the HF have emphasized the role of epithelial sonic hedgehog (SHH), downstream of Wnt signaling, in stimulating mesenchymal clustering to form the Noggin-expressing DP (29, 30). Because sweat buds display reduced epithelial Wnt signaling and also lack this mesenchymal cluster, we posited that in sweat buds, Shh is silenced, which in turn should be reflected in the failure of foot skin dermis to activate downstream SHH-signaling target genes. Our RNA-seq analyses showed that indeed, Gli1 and Ptch1, the classic downstream target genes of SHH signaling, are highly enriched in dorsal back skin (HF-competent) versus ventral foot skin (SwG-competent) dermis (Fig. 6A). Quantitative PCR confirmed the strong expression of Shh in E14.5 dorsal epidermis relative to ventral foot skin epidermis (fig. S6A).
Fig. 6. SHH signaling is suppressed in SwG buds and upon ectopic induction and is only effective within a critical window in switching sweat to hair fate.
(A) Scatter plot of RNA-seq transcriptome analyses from E14.5 dorsal back skin and E17.5 ventral foot skin dermis. Targets of SHH signaling, Gli1 and Ptch1, are enriched in dorsal versus ventral dermis (pink dots). (B) Shh in situ hybridizations of ventral foot pads of WT and Bmpr1a cKO P1 pups, revealing the role of BMP in suppressing Shh in SwG-buds. (C) Ex vivo culture of E17.5 foot skin with addition of BMP-inhibitor, NOGGIN, BMPs, or FGF18. Epidermal Shh expression was analyzed by means of quantitative PCR. n = 6 to 8 foot skin samples analyzed from three embryos per experiment. (D) Timeline of lentiviral injection, Shh induction, and analyses. Immunofluorescence images showing that when induction occurs at the epithelial bud stage, hair bud marker LHX2 is ectopically induced in the foot pad epidermis, which is reflective of a switch from SwG to HF fate. Dashed line indicates dermo-epidermal border. Scale bar, 50 µm. (E) Timeline of lentiviral injection, Shh induction, and analyses. Immunofluorescence images showing that when induction occurs after the epithelial bud stage, appendage fate is unaltered in the ventral foot skin. Boxed area is shown in higher magnification at right. Scale bar, 50 µm and (far right) 25 µm. (F) Timeline of lentiviral injection, Shh induction, and analyses. Immunofluorescence images showing that short exposure to SHH during foot skin epithelial bud formation is sufficient to cause long-lasting effects on fate change and expression of HF markers K17 and LHX2. Dashed lines indicate dermo-epidermal border. Scale bars, 50 µm.
Because Bmpr1a-null foot skin buds displayed signs of mesenchymal cell clustering (Fig. 1G), we hypothesized that Shh expression must rely not only on epithelial Wnt signaling (21, 30) but also on inhibition of mesenchymal BMP signaling to the epithelial buds. In other words, when mesenchymal BMPs actively signal to the epithelial buds, as in the case of ventral foot skin, Shh expression must be suppressed. In situ hybridizations and quantitative PCR showed that Shh, although not detected in wild-type ventral foot skin, was indeed activated in Bmpr1a-null ventral foot skin buds in vivo, as well as in E17.5 foot skin explants exposed to NOGGIN (Fig. 6, B and C). In contrast, exposure of foot skin explants to additional BMPs further suppressed already low expression of Shh (Fig. 6C). Furthermore, elevating NOGGIN in dorsal back skin resulted in higher activation of Shh, as well as HF markers Lhx2 and K17 (fig. S6B).
Shh repression in ventral foot skin buds seemed also to be influenced by FGF-MAPK signaling, perhaps through its ability to sensitize buds to BMP signaling (Figs. 4G and 6C). In particular, FGF18 seemed to have the strongest effect on Shh repression as shown in explants and in vivo when activated in dorsal back skin (Fig. 6C and fig. S6, C and D). The combinatorial effect of FGFs inShh suppression could be partially rescued by MAPK inhibitor SB203580 (fig. S6D). These results correlated with GO-term analyses of WNT-responsive changes, revealing that the MAPK pathway is positively enriched in ventral foot skin but negatively enriched in dorsal back skin (fig. S5A).
Our data on FGFs further bolstered the importance of mesenchymal-to-epithelial signaling circuits for Shh suppression in foot skin. That said, Wnt signaling is required and upstream of Shh expression in hair buds (20, 21) and is repressed in SwG buds (Fig. 4A and fig. S3A). Additionally, ENGRAILED has been previously reported to act directly to transcriptionally repress Shh and/or the genes encoding its downstream transcriptional effectors (31–34). All of these pathways appeared to converge on repression of Shh and SHH signaling in SwG-permissive skin.
Regional BMP:SHH antagonism dictates SwG versus HF fates
To evaluate whether Shh up-regulation in ventral foot skin epithelium can prevent SwG fate, we again turned to the inducible lentiviral system. Analogous to our experimental design for Noggin, we induced Shh for several days at E16.5. By P0, foot skin epithelial buds ectopically induced LHX2 and other HF markers and failed to progress to SwGs (Fig. 6D and fig. S6E, left), whereas in the underlying mesenchyme, signs of SHH signaling were seen, including a rise in BMP inhibitors (fig. S6E, right). Similar to our findings with Noggin induction, when Shh was induced after sweat buds were already established, gland formation progressed, as evidenced by morphology, luminal marker K18, and SwG markers NKA and AQUAPORIN-5 (Fig. 6E and fig. S6F). This was true even after 10 days of a high level of Shh expression (Fig. 6E), suggesting that once the narrow window of permissiveness is passed, glandular fate is no longer sensitive to SHH.
Last, we tested whether ectopic SHH administered during the permissive window has long-term consequences on appendage choice. We activated epithelial Shh in embryos for only 2 days at the sweat bud–formation stage and then stopped Doxy treatment and examined the foot pads of adult mice ~3 months later. Although morphology was clearly abnormal, the activation of hair-specific markers and absence of gland-specific markers provided compelling evidence that brief exposure to SHH during the critical window of gland bud morphogenesis was sufficient to cause irreversible changes in developing buds, preventing SwG fate and promoting features typically associated with HF fate (Fig. 6F). Additionally, when we generated mouse embryos whose skin contained mosaic patches of a constitutively active SHH receptor (SmoM2) (35) in the epithelium, only wild-type foot skin patches displayed K18+ SwGs, whereas SmoM2 patches expressed LHX2+ budlike structures (fig. S7). This suggested that selective activation of SHH signaling within the ventral foot skin epithelium is sufficient to trigger the fate switch, regardless of dermal signaling feedback.
Temporal regulation of BMP:SHH antagonism endows human skin with both SwGs and HFs
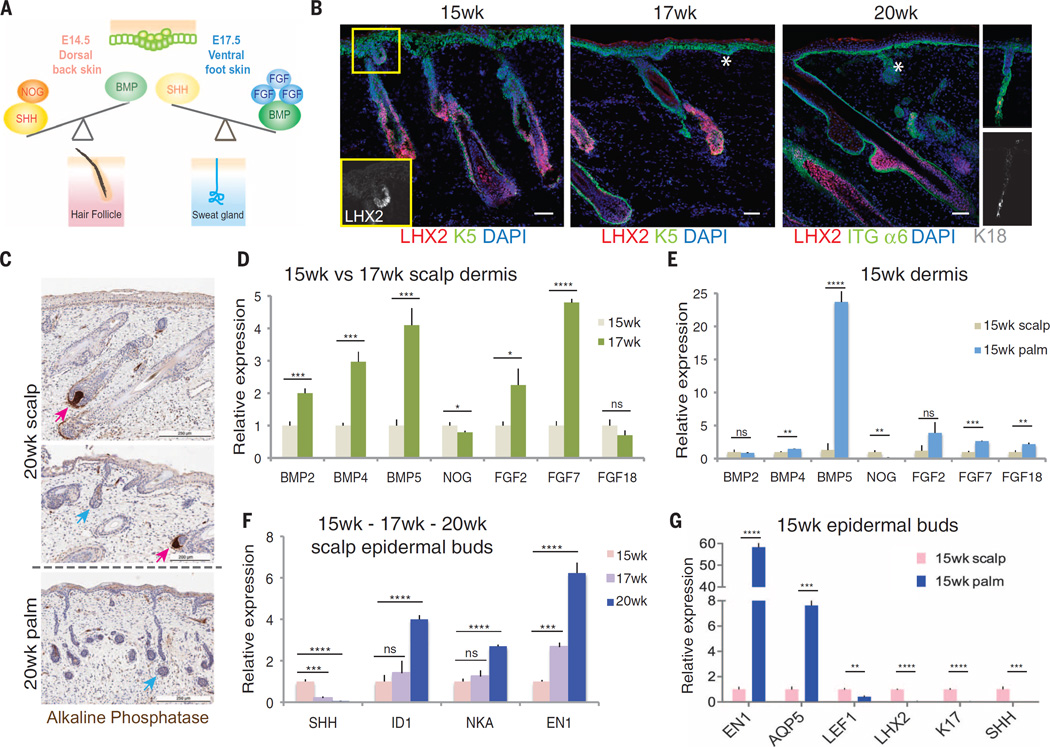
Our data are summarized in the model shown in Fig. 7A. We tested this model by addressing whether it can be used to explain how human skin is able to accommodate both SwGs and HFs within a common dermis. The organized patterning of glands and HFs within human skin argues against the notion that its dermis simply maintains intermediate levels of FGFs and BMPs, permissive for both types of epidermal appendages. Rather, our mouse data predict that within embryonic human dermis, there must be a temporal switch in dermal permissiveness, which allows for the sequential emergence of each appendage type. Indeed, upon analyzing human fetal scalp skin at gestational ages 15, 17, and 20 weeks with our markers established from mouse studies, we observed temporal segregation of waves of HF and SwG formation, with hair bud morphogenesis beginning to phase out between 15 and 17 weeks and sweat buds emerging at 17 weeks (Fig. 7, B and C). By 20 weeks, sweat buds were still forming, in the absence of further signs of de novo HF morphogenesis.
Fig. 7. Human dorsal skin temporally shifts from BMPloFGFloSHHhi to BMPhiFGFhiSHHlo during embryogenesis, yielding HF and SwG coexistence.
(A) Model showing differences in the balance between BMP/FGF and SHH signaling specify the fate of HFs or SwGs. (B) Immunofluorescence images of human fetal dorsal scalp skin at 15, 17, and 20 weeks of gestation. Boxed area shows emerging LHX2+ HF buds. Stars indicate LHX2neg placode and bud. HFs and SwG buds are distinguished by hair bud–specific marker LHX2. (Far right) Appearance of K18-positivematuring sweat gland by 20weeks. Scale bars, 100 µm. (C) Histology of dorsal scalp skin and ventral palm skin, costained for alkaline phosphatase activity to identify dermal papillae (pink arrows), absent in sweat buds and glands (blue arrows).Visible is coexistence of glands and HFs in dorsal scalp skin but only SwGs in palm skin. Scale bars, 200 µm. (D to G) Quantitative PCR of transcripts isolated from dermis and epithelial buds of scalp skin and palm skin, at ages indicated. (D) Increase in BMP and FGF transcripts in scalp dermis between 15 and 17 weeks. (E) High level of BMP5 expression in 15-week ventral palm (SwG only) versus 15-week dorsal scalp (HF only) dermis. (F) Early decline of SHH production and later increase in BMP signaling (ID1) and SwG markers (NKA) in emerging epithelial buds between 15 and 20 weeks. Also shown is the emergence of EN1 in dorsal scalp skin, which contrasts with mice, where EN1 is confined to ventral foot skin. (G) At 15 weeks, SwG markers are already high in palm but not in scalp epidermis.
To test whether a temporal change in BMP, FGF, and SHH signaling drives the intriguing developmental switch from hair to sweat fate within human scalp skin, we FACS-purified epithelial bud and mesenchymal cells (fig. S8, A and B) and analyzed their relative gene expression at these key windows of development. Within scalp dermis, expression of BMP2, BMP4, BMP5, FGF2, and FGF7 were all significantly up-regulated at week 17 relative to week 15, which is coincident with the shift from hair to sweat bud formation (Fig. 7D). Thus, in contrast to mice, in which these dermal BMPs and FGFs were regionally restricted to the ventral foot skin, in humans the same genes were now temporally restricted to establish discrete hair versus sweat bud windows of permissiveness during embryogenesis of dorsal scalp skin.
To determine whether humans maintained evolutionary vestiges of regional skin differences, we also compared human scalp and palm skins at these gestational stages. As shown in Fig. 7E, the sweat bud–promoting mesenchymal signals that were elevated temporally by week 17 in human scalp skin were already present by week 15 in human palm skin, which was exclusive for SwG appendages (fig. S8C). Like in mouse foot skin, BMP5 was the most prominent BMP expressed in palmar dermis (Fig. 7E), and LHX2 was exclusively expressed in palmar dermis and scalp HF epithelium (fig. S8C).
Consistent with the antagonistic regulatory circuitry that we unveiled in mouse, SHH expression decreased dramatically within the scalp epithelial buds as mesenchymal BMPs and FGFs, and epithelial ENGRAILED-1, were elevated (Fig. 7F). Additional signs of the developmental switch in appendage specification were revealed by temporal up-regulation of BMP target gene ID1 and SwG marker NKA (Na/K-ATPase) (Fig. 7F). The sequential appearance of the appendages was similar but temporally delayed in dorsal limb skin, which like scalp is typified by both hair and SwGs (fig. S8D). In contrast in human palm skin, which (like its mouse counterpart) develops exclusively SwGs, SHH was already markedly suppressed regionally by week 15, which is coincident with the low level of WNT-(LEF1)–signaling and high level of ENGRAILED-1 at this early time (Fig. 7G). Overall, our findings underscore the distinct temporal flip in BMP:SHH signaling that occurs in human development and endows human skin with the ability to permit sweat bud fate commitment as the final wave(s) of bud development in otherwise hairy skin.
Discussion
In the evolutionary developmental biology view of BMP signaling and fate specification of integument, both chicken scales (36) and mouse and human SwGs (our study) require BMP signaling to specify their fate, whereas feathers (37) and HFs (13) must suppress it. Thus, the differential impact of BMP signaling on appendage fate specification has ancient roots and occurs repeatedly throughout vertebrate evolution in making epidermal bud fate decisions. In addition to its role in ectodermal fate specification, BMP-SHH antagonism is also critical for the patterning of chicken endodermal digestive epithelia (38, 39), especially in the glandular region of the stomach. Together with other findings on BMP requirement for various ectodermal glands (mammary and meibomian) (40, 41), a particular role begins to emerge for BMP signaling in promoting glandular fate, typically occurring on the ventral side of the body. Our studies provide insights into how elevated mesenchymal BMP signaling drives a thereafter self-perpetuating molecular cascade that culminates in silencing SHH signaling to suppress one appendage fate and specify another. By spatially regulating BMP:SHH antagonism, mammals can specify HF and glandular fates in distinct body sites. With additional temporal regulation of BMP:SHH antagonism, human skin has the special capacity first to generate waves of HF fates and then to switch midstream to specify glandular fates. Our findings pave the way for future therapeutic advances in skin regeneration with dual epithelial appendages.
Materials and methods
Mice and in utero lentiviral injection
All animals used for the experiments in this manuscript were generated previously: K14Cre (Fuchs lab) (42), Bmpr1afl/fl (kindly from Y. Mishina) (43), SEA/GnJ (The Jackson Laboratory) (44), K14-rtTA (Fuchs lab) (45), Smofl/fl (The Jackson Laboratory) (46), R26SmoM2 (The Jackson Laboratory) (47), R26Flox-Stop-Flox-YFP (The Jackson Laboratory) (48), Ctnnblfl/fl (kindly from R. Kemler) (49), Axin2-LacZ (The Jackson Laboratory) (50). In utero ultrasound-guided lentiviral injection procedures and preparation of high-titer lentivirus have been described (12). K14rtTA transgenic male (CD1 strain) were mated with CD1 wild type female, and their embryos were injected with high-titer lentivirus at E9.5. Each lentiviral injection experiments were performed at least two times and all foot skins from 2–4 animals were collected and analyzed at each time points. Animals were maintained in an Association for Assessment and Accreditation of Laboratory Animal Care (AALAC)-accredited animal facility, and procedures were performed with Institutional Animal Care and Use Committee (IACUC)-approved protocols. rtTA transcription factor was activated by feeding mice with doxycycline (2 mg/kg) chow at times specified. Lentiviral doxycycline-inducible Shh expression construct (pLKO-TRE-Shh-PGK-H2BGFP) has been previously described (30). Construct for the lentiviral Cre transductions has been previously described (12). Mouse Noggin was cloned by PCR from pMgB950 (kindly from R. Harland) to replace Shh in the pLKO-TRE-Shh-PGK-H2BGFP to generate pLKO-TRE-Nog-PGK-H2BGFP. Mouse Bmp5 and Fgf18 cDNAs were purchased from Sino Biological Inc. and were cloned into pLKO-TRE-PGK-H2BGFP vector.
Human tissue procurement
Fetal skin samples (scalp and palm) of 15, 17, 20 wks (2–3 samples for each stage) were procured by Novogenix Laboratories (Los Angeles, CA) and Advanced Bioscience Resources (Alameda, CA) in compliance with federal and state laws, and National Institute of Health guidelines. Tissues were processed and paraffin-embedded by HistoWiz, Inc. Standard histology and Alkaline Phosphatase immunohistochemistry staining were also performed at HistoWiz (Brooklyn, NY).
Quantification of sweat ducts
Experimental details have been described previously (3). Briefly, mouse hind foot skins were removed by dissection and incubated in EDTA (50 mM) 37°C for 30 min to separate epidermis and dermis. Under these conditions, sweat ducts remain associated with the epidermal cell sheet. Whole mount histochemical staining was with 0/1% Nile Blue A (Sigma). After washing, sweat ducts showed intense blue staining and were scored and imaged using a Leica M165 FC stereomicroscope.
In situ hybridization
In situ hybridization for Shh was performed as described previously (19).
Immunofluorescence and imaging
Skin (dorsal back and ventral foot) samples from 3–4 animals for each experiment were dissected and embedded in OCT (Tissue Tek) and cryo-sectioned at a thickness of 10–12 µm. Tissue sections were fixed in 4% PFA and then blocked in gelatin block (1% gelatin, 2.5% normal donkey serum, 1% BSA, 0.3%Triton in PBS) 1 hour at RT. Primary antibodies were incubated at 4°C overnight. After washing with PBS, samples were incubated for 2 hours at room temperature with secondary antibodies conjugated with Alexa488, RRX, or Alexa647 (1:800, 1:400, and 1:400, respectively, Life Technologies) and 4′6-diamidino-2-phenylindole (DAPI). Samples were washed again with PBS and then mounted in Prolong Gold (Invitrogen). EdU incorporation was detected by Click-It EdU AlexaFluor594 Imaging Kit (Invitrogen) per manufacturer’s instructions. The following primary antibodies were used: P-Cadherin (Rat, 1:50, Fuchs lab), E-Cadherin (Rat, 1:100, Fuchs lab), phospho-Smad1/5/8 (rabbit, 1:800, Cell signaling), LEF1 (rabbit, 1:300, Fuchs Lab), K5 (rabbit, 1:600, Fuchs Lab), K5 (guinea pig, 1:500, Fuchs Lab), K17 (rabbit, 1:600, Fuchs Lab), K18 (rabbit, 1:400, Fuchs Lab), LHX2 (rabbit, 1:2000, Fuchs Lab), anti-GFP/YFP (chicken, 1:2000, Abcam), ITGα6 (Rat, 1:100, BD), ITGβ4 (Rat, 1:100, BD), Alkaline phosphatase (Goat, 1:100, R&D), β-Galactosidase (chicken, 1:2000, Abcam), NKA (Rabbit, 1:8000, Abcam), AQP5 (rabbit, 1:2000, Abcam), EpCam-Alexa488 (mouse, 1:100, Cell signaling). Epifluorescence images were acquired with an AxioOberver.Z1 microscope equipped with a Hamamatsu ORCA-ER camera (Hamamatsu Photonics), and with an ApoTome.2 (Carl Zeiss) slider that reduces the light scatter in the fluorescent samples. 20× and 40× objectives were used. Data collection was controlled by Zen software (Carl Zeiss).
Tissue preparation and fluorescence-activated cell sorting
Mouse E14.5 dorsal back skin and E17.5 ventral foot skin of Axin2-LacZ reporter mice were dissected and incubated in EDTA (50mM), 37°C for 30 min to allow separation of epidermis and dermis. Note that for collecting E17.5 foot skin, only front feet were used, to avoid potential contamination of HF-competent dermal cells in the inter-pad region of the hind feet. Since these are skin tissues harvested at the placode formation stage, epidermal placodes were mostly removed with epidermal fraction and minimum epidermal cells remained with dermal fraction. The dermal tissues were then digested in Hanks Balanced Salt Solution (Lonza) with 1000 U/ml collagenase and 300 U/ml hyaluronadase (Sigma) and washed and re-suspended in PBS with 4% FBS to make single cell suspension. The following antibodies were used for fluorescence-activated cell sorting (FACS): α6-PE (CD49f, 1:1000, eBioscience), CD140a-APC (1:100, eBioscience). Staining of intracellular β-galactosidase was performed using The FluoReporter lacZ Flow Cytometry Kit (Molecular Probes) as per manufacturer’s instructions. DAPI was used to exclude dead cells. Cell isolations were performed on FACSAria sorters equipped with DIVA software (BD Biosciences).
Human skin tissues were digested in LiberaseTL (Roche), 37°C for 1 hour 20min. Digested tissues were resuspended and washed in PBS with 4%FBS to make single cell suspensions. The following antibodies were used for FACS: CD140a-PE (1:50, BioLegnad), EpCAM-Alexa488 (1:100, Cell signaling), CD49f-PECy7(1:1000, eBioscience), CD133- APC (1:100, Miltenyi). DAPI was used to exclude dead cells. Cell isolations were performed on FACSAria sorters equipped with DIVA software (BD Biosciences).
Explant culture and RNA extraction and RT-qPCR
Six to eight pieces of E17.5 foot skin from >3 different mouse embryos per condition were placed as explants into culture medium. The culture conditions were modified from the hanging-drop method (51), and 100–120 µl of culture medium were used for each drop. The final concentrations of recombinant proteins and inhibitors used were: bFGF (50 ng/ml), FGF7 (50 ng/ml), FGF18 (50 ng/ml), BMP4 (100 ng/ml), BMP5 (100 ng/ml), Noggin (4 µg/ml), MEKi (U0126, 10 µM), MAPKi (SB203580, 1 µM). After overnight culture (16–18 hour), skin tissues were incubated in EDTA (50 mM), 37°C for 20 min, and epidermis and dermis were separated and immediately frozen in liquid nitrogen. For embryonic foot skin tissues from lentiviral injection experiments to be analyzed by RT-qPCR, note that only front foot skins were collected because of higher infection efficiency compared to hind foot skin, and at least 6–8 pieces from 3–4 embryos were collected for each experiment. For embryonic dorsal skin from lentiviral injection experiments, samples from head region (highest infection efficiency) were collected from at least two different embryos. All these samples were similarly incubated in EDTA (50 mM), 37°C for 20 min, to separate epidermis/dermis and were immediately frozen. The frozen tissues were homogenized using Bessman Tissue Pulverizer (Spectrum™) and collected in Trizol (Invitrogen). RNA was extracted with a Direct-zol RNA MiniPrep kit (Zymo Research) as per manufacturer’s instructions. Equivalent amounts of RNA were reverse-transcribed by SuperScript VILO cDNA Synthesis Kit (Invitrogen). cDNAs were mixed with indicated primers and Power SYBR Green PCR Master Mix (AppliedBiosystems), and quantitative PCR (qPCR) was performed on a Applied Biosystems 7900HT Fast Real-Time PCR system. cDNAs were normalized to equal amounts using primers against Hprt or Ppib. Primer sequences for RT-PCR were obtained using NCBI Primer-BLAST webtool.
RNA-seq analyses and statistics
For RNA-seq, RNA quality was determined using Agilent 2100 Bioanalyzer. Samples used for sequencing had RNA integrity numbers (RIN) > 8. Library preparation using Illumina TrueSeq mRNA sample preparation kit was performed at the Weill Cornell Medical College Genomic Core facility, and RNAs were sequenced on Illumina HiSeq 2000 machines. RNA reads were aligned to the mm9 build of the mouse genome using Tophat. Transcript assembly and differential expression was done using Cufflinks with Refseq mRNAs to guide assembly (52). Analysis of RNA-seq data was performed using the cummeRbund package in R (53). DAVID bioinformatic database was used to find enriched functional GO-term annotations in RNA-seq data (https://david.ncifcrf.gov/content.jsp?file=citation.htm). Box-and-whisker plots were generated using Prism5 software (GraphPad) to determine the significance between two groups. Error bars plotted on graphs denote SD. Unpaired student’s t test was used to determine the significance between two groups: *P<0.05; **P<0.01; ***P < 0.001; ****P<0.0001; ns, not significant.
Supplementary Material
Acknowledgments
We thank Fuchs laboratory members, particularly M. Genander, T. Ouspenskaia, H.A. Pasolli, H. Yang, V. Fiore, S. Liu, and Y. Ge for reagents and helpful discussions, and J. Levorse, J. Racelis, and S. Chai for technical assistance. We thank Rockefeller Comparative Bioscience Center (Association for Assessment and Accreditation of Laboratory Animal Care–accredited) for care of mice in accordance with National Institutes of Health (NIH) guidelines, Bioimaging Resource Center (A. North, director), and Flow Cytometry Resource Center (S. Mazel, director, and S. Semova, S. Tadesse, and S. Han). E.F. is an investigator of the Howard Hughes Medical Institute. C.P.L. is a recipient of fellowships from the National Institutes of Health (National Institute of Arthritis and Musculoskeletal and Skin Diseases grant K01-AR066073) and Robertson Therapeutic Development Fund. This work was supported by NIH grant R01-AR050452 (E.F.). All reagents engineered in the course of this study are available from jdelacruz@rockefeller.edu under a materials transfer agreement with the Rockefeller University. C.P.L. and E.F. conceived the project, designed the experiments, and wrote the manuscript. C.P.L. performed the experiments. C.P.L. and L.P. performed mouse surgical procedures. B.E.K. and C.P.L. performed RNA-seq data analyses.
Footnotes
RNAseq data are deposited in the Gene Expression Omnibus under accession number GSE85249 (www.ncbi.nlm.nih.gov/geo).
The authors declare no competing financial interests.
REFERENCES AND NOTES
- 1.Kere J, et al. X-linked anhidrotic (hypohidrotic) ectodermal dysplasia is caused by mutation in a novel transmembrane protein. Nat. Genet. 1996;13:409–416. doi: 10.1038/ng0895-409. pmid: 8696334. [DOI] [PubMed] [Google Scholar]
- 2.Cluzeau C, et al. Only four genes (EDA1, EDAR, EDARADD, and WNT10A) account for 90% of hypohidrotic/anhidrotic ectodermal dysplasia cases. Hum. Mutat. 2011;32:70–72. doi: 10.1002/humu.21384. pmid: 20979233. [DOI] [PubMed] [Google Scholar]
- 3.Kamberov YG, et al. Modeling recent human evolution in mice by expression of a selected EDAR variant. Cell. 2013;152:691–702. doi: 10.1016/j.cell.2013.01.016. pmid: 23415220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mustonen T, et al. Stimulation of ectodermal organ development by Ectodysplasin-A1. Dev. Biol. 2003;259:123–136. doi: 10.1016/s0012-1606(03)00157-x. pmid: 12812793. [DOI] [PubMed] [Google Scholar]
- 5.Sengel P. Pattern formation in skin development. Int. J. Dev. Biol. 1990;34:33–50. pmid: 2203463. [PubMed] [Google Scholar]
- 6.Chuong CM, Chodankar R, Widelitz RB, Jiang TX. Evo-devo of feathers and scales: Building complex epithelial appendages. Curr. Opin. Genet. Dev. 2000;10:449–456. doi: 10.1016/s0959-437x(00)00111-8. pmid: 11023302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kratochwil K. Organ specificity in mesenchymal induction demonstrated in the embryonic development of the mammary gland of the mouse. Dev. Biol. 1969;20:46–71. doi: 10.1016/0012-1606(69)90004-9. pmid: 5795848. [DOI] [PubMed] [Google Scholar]
- 8.Dhouailly D, Rogers GE, Sengel P. The specification of feather and scale protein synthesis in epidermal-dermal recombinations. Dev. Biol. 1978;65:58–68. doi: 10.1016/0012-1606(78)90179-3. pmid: 680361. [DOI] [PubMed] [Google Scholar]
- 9.Ferraris C, Chevalier G, Favier B, Jahoda CA, Dhouailly D. Adult corneal epithelium basal cells possess the capacity to activate epidermal, pilosebaceous and sweat gland genetic programs in response to embryonic dermal stimuli. Development. 2000;127:5487–5495. doi: 10.1242/dev.127.24.5487. pmid: 11076768. [DOI] [PubMed] [Google Scholar]
- 10.Rhee H, Polak L, Fuchs E. Lhx2 maintains stem cell character in hair follicles. Science. 2006;312:1946–1949. doi: 10.1126/science.1128004. pmid: 16809539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rendl M, Polak L, Fuchs E. BMP signaling in dermal papilla cells is required for their hair follicle-inductive properties. Genes Dev. 2008;22:543–557. doi: 10.1101/gad.1614408. pmid: 18281466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beronja S, Livshits G, Williams S, Fuchs E. Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nat. Med. 2010;16:821–827. doi: 10.1038/nm.2167. pmid: 20526348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Botchkarev VA, et al. Noggin is a mesenchymally derived stimulator of hair-follicle induction. Nat. Cell Biol. 1999;1:158–164. doi: 10.1038/11078. pmid: 10559902. [DOI] [PubMed] [Google Scholar]
- 14.Plikus M, et al. Morpho-regulation of ectodermal organs: Integument pathology and phenotypic variations in K14-Noggin engineered mice through modulation of bone morphogenic protein pathway. Am. J. Pathol. 2004;164:1099–1114. doi: 10.1016/S0002-9440(10)63197-5. pmid: 14982863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Genderen C, et al. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. pmid: 7958926. [DOI] [PubMed] [Google Scholar]
- 16.Andl T, Reddy ST, Gaddapara T, Millar SE. WNT signals are required for the initiation of hair follicle development. Dev. Cell. 2002;2:643–653. doi: 10.1016/s1534-5807(02)00167-3. pmid: 12015971. [DOI] [PubMed] [Google Scholar]
- 17.Ahtiainen L, et al. Directional cell migration, but not proliferation, drives hair placode morphogenesis. Dev. Cell. 2014;28:588–602. doi: 10.1016/j.devcel.2014.02.003. pmid: 24636260. [DOI] [PubMed] [Google Scholar]
- 18.Cui C-Y, et al. Involvement of Wnt, Eda and Shh at defined stages of sweat gland development. Development. 2014;141:3752–3760. doi: 10.1242/dev.109231. pmid: 25249463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. pmid: 10498690. [DOI] [PubMed] [Google Scholar]
- 20.Jamora C, DasGupta R, Kocieniewski P, Fuchs E. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature. 2003;422:317–322. doi: 10.1038/nature01458. pmid: 12646922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ouspenskaia T, Matos I, Mertz AF, Fiore VF, Fuchs E. WNT-SHH antagonism specifies and expands stem cells prior to niche formation. Cell. 2016;164:156–169. doi: 10.1016/j.cell.2015.11.058. pmid: 26771489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kratochwil K, Dull M, Farinas I, Galceran J, Grosschedl R. Lef1 expression is activated by BMP-4 and regulates inductive tissue interactions in tooth and hair development. Genes Dev. 1996;10:1382–1394. doi: 10.1101/gad.10.11.1382. pmid: 8647435. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, et al. Reciprocal requirements for EDA/EDAR/NF-kappaB and Wnt/β-catenin signaling pathways in hair follicle induction. Dev. Cell. 2009;17:49–61. doi: 10.1016/j.devcel.2009.05.011. pmid: 19619491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rinn JL, et al. A dermal HOX transcriptional program regulates site-specific epidermal fate. Genes Dev. 2008;22:303–307. doi: 10.1101/gad.1610508. pmid: 18245445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plaisancié J, et al. Mutations in WNT10A are frequently involved in oligodontia associated with minor signs of ectodermal dysplasia. Am. J. Med. Genet. A. 2013;161A:671–678. doi: 10.1002/ajmg.a.35747. pmid: 23401279. [DOI] [PubMed] [Google Scholar]
- 26.Vincent JP, Lawrence PA. Drosophila wingless sustains engrailed expression only in adjoining cells: Evidence from mosaic embryos. Cell. 1994;77:909–915. doi: 10.1016/0092-8674(94)90139-2. pmid: 8004677. [DOI] [PubMed] [Google Scholar]
- 27.Loomis CA, et al. The mouse Engrailed-1 gene and ventral limb patterning. Nature. 1996;382:360–363. doi: 10.1038/382360a0. pmid: 8684466. [DOI] [PubMed] [Google Scholar]
- 28.Kamberov YG, et al. A genetic basis of variation in eccrine sweat gland and hair follicle density. Proc. Natl. Acad. Sci. U.S.A. 2015;112:9932–9937. doi: 10.1073/pnas.1511680112. pmid: 26195765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woo W-M, Zhen HH, Oro AE. Shh maintains dermal papilla identity and hair morphogenesis via a Noggin-Shh regulatory loop. Genes Dev. 2012;26:1235–1246. doi: 10.1101/gad.187401.112. pmid: 22661232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu Y-C, Li L, Fuchs E. Transit-amplifying cells orchestrate stem cell activity and tissue regeneration. Cell. 2014;157:935–949. doi: 10.1016/j.cell.2014.02.057. pmid: 24813615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dahmann C, Basler K. Opposing transcriptional outputs of Hedgehog signaling and engrailed control compartmental cell sorting at the Drosophila A/P boundary. Cell. 2000;100:411–422. doi: 10.1016/s0092-8674(00)80677-7. pmid: 10693758. [DOI] [PubMed] [Google Scholar]
- 32.Prin F, Dhouailly D. How and when the regional competence of chick epidermis is established: Feathers vs. scutate and reticulate scales, a problem en route to a solution. Int. J. Dev. Biol. 2004;48:137–148. doi: 10.1387/ijdb.15272378. pmid: 15272378. [DOI] [PubMed] [Google Scholar]
- 33.Basler K, Struhl G. Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature. 1994;368:208–214. doi: 10.1038/368208a0. pmid: 8145818. [DOI] [PubMed] [Google Scholar]
- 34.Alexandre C, Vincent J-P. Requirements for transcriptional repression and activation by Engrailed in Drosophila embryos. Development. 2003;130:729–739. doi: 10.1242/dev.00286. pmid: 12506003. [DOI] [PubMed] [Google Scholar]
- 35.Youssef KK, et al. Adult interfollicular tumour-initiating cells are reprogrammed into an embryonic hair follicle progenitor-like fate during basal cell carcinoma initiation. Nat. Cell Biol. 2012;14:1282–1294. doi: 10.1038/ncb2628. pmid: 23178882. [DOI] [PubMed] [Google Scholar]
- 36.Zou H, Niswander L. Requirement for BMP signaling in interdigital apoptosis and scale formation. Science. 1996;272:738–741. doi: 10.1126/science.272.5262.738. pmid: 8614838. [DOI] [PubMed] [Google Scholar]
- 37.Noramly S, Morgan BA. BMPs mediate lateral inhibition at successive stages in feather tract development. Development. 1998;125:3775–3787. doi: 10.1242/dev.125.19.3775. pmid: 9729486. [DOI] [PubMed] [Google Scholar]
- 38.Roberts DJ, Smith DM, Goff DJ, Tabin CJ. Epithelialmesenchymal signaling during the regionalization of the chick gut. Development. 1998;125:2791–2801. doi: 10.1242/dev.125.15.2791. pmid: 9655802. [DOI] [PubMed] [Google Scholar]
- 39.Narita T, et al. BMPs are necessary for stomach gland formation in the chicken embryo: A study using virally induced BMP-2 and Noggin expression. Development. 2000;127:981–988. doi: 10.1242/dev.127.5.981. pmid: 10662637. [DOI] [PubMed] [Google Scholar]
- 40.Mayer JA, Foley J, De La Cruz D, Chuong C-M, Widelitz R. Conversion of the nipple to hair-bearing epithelia by lowering bone morphogenetic protein pathway activity at the dermal-epidermal interface. Am. J. Pathol. 2008;173:1339–1348. doi: 10.2353/ajpath.2008.070920. pmid: 18832580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang J, et al. FGF-regulated BMP signaling is required for eyelid closure and to specify conjunctival epithelial cell fate. Development. 2009;136:1741–1750. doi: 10.1242/dev.034082. pmid: 19369394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: Genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl. Acad. Sci. U.S.A. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. pmid: 10411913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mishina Y, Hanks MC, Miura S, Tallquist MD, Behringer RR. Generation of Bmpr/Alk3 conditional knockout mice. Genesis. 2002;32:69–72. doi: 10.1002/gene.10038. pmid: 11857780. [DOI] [PubMed] [Google Scholar]
- 44.King JA, Marker PC, Seung KJ, Kingsley DM. BMP5 and the molecular, skeletal, and soft-tissue alterations in short ear mice. Dev. Biol. 1994;166:112–122. doi: 10.1006/dbio.1994.1300. pmid: 7958439. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen H, Rendl M, Fuchs E. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 2006;127:171–183. doi: 10.1016/j.cell.2006.07.036. pmid: 17018284. [DOI] [PubMed] [Google Scholar]
- 46.Long F, Zhang XM, Karp S, Yang Y, McMahon AP. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development. 2001;128:5099–5108. doi: 10.1242/dev.128.24.5099. pmid: 11748145. [DOI] [PubMed] [Google Scholar]
- 47.Mao J, et al. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res. 2006;66:10171–10178. doi: 10.1158/0008-5472.CAN-06-0657. pmid: 17047082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. pmid: 11299042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee H-Y, et al. Instructive role of Wnt/β-catenin in sensory fate specification in neural crest stem cells. Science. 2004;303:1020–1023. doi: 10.1126/science.1091611. pmid: 14716020. [DOI] [PubMed] [Google Scholar]
- 50.Lustig B, et al. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell. Biol. 2002;22:1184–1193. doi: 10.1128/MCB.22.4.1184-1193.2002. pmid: 11809809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pummila M, et al. Ectodysplasin has a dual role in ectodermal organogenesis: Inhibition of Bmp activity and induction of Shh expression. Development. 2007;134:117–125. doi: 10.1242/dev.02708. pmid: 17164417. [DOI] [PubMed] [Google Scholar]
- 52.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. pmid: 20436464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. pmid: 22383036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.