Abstract
Bile acids are primarily synthesized from cholesterol in the liver and have important roles in dietary lipid absorption and cholesterol homoeostasis. Detailed roles of the orphan nuclear receptors regulating cholesterol 7α-hydroxylase (CYP7A1), the rate-limiting enzyme in bile acid synthesis, have not yet been fully elucidated. In the present study, we report that oestrogen-related receptor γ (ERRγ) is a novel transcriptional regulator of CYP7A1 expression. Activation of cannabinoid receptor type 1 (CB1 receptor) signalling induced ERRγ-mediated transcription of the CYP7A1 gene. Overexpression of ERRγ increased CYP7A1 expression in vitro and in vivo, whereas knockdown of ERRγ attenuated CYP7A1 expression. Deletion analysis of the CYP7A1 gene promoter and a ChIP assay revealed an ERRγ -binding site on the CYP7A1 gene promoter. Small heterodimer partner (SHP) inhibited the transcriptional activity of ERRγ and thus regulated CYP7A1 expression. Overexpression of ERRγ led to increased bile acid levels, whereas an inverse agonist of ERRγ, GSK5182, reduced CYP7A1 expression and bile acid synthesis. Finally, GSK5182 significantly reduced hepatic CB1 receptor-mediated induction of CYP7A1 expression and bile acid synthesis in alcohol-treated mice. These results provide the molecular mechanism linking ERRγ and bile acid metabolism.
Keywords: bile acid, cannabinoid receptors, cholesterol 7α-hydroxylase (CYP7A1), GSK5182, oestrogen-related receptor γ (ERRγ), orphan nuclear receptor, small heterodimer partner (SHP)
INTRODUCTION
Oestrogen-related receptor (ERR)-α, -β and -γ (NR3B1-3) are three members of the oestrogen-related receptor subfamily. Both ERRα and ERRγ are ubiquitously expressed, whereas ERRβ is restricted to the brain, kidney and heart [1–4]. Several synthetic compounds were characterized as ERR-specific ligands: GSK4716 is a synthetic ERRβ and ERRγ agonist, kaempferol is an ERRα and ERRγ inverse agonist and GSK5182, a tamoxifen analogue, acts as an inverse agonist of ERRγ and selectively inhibits transactivation of ERRγ [5–8]. ERRα and ERRγ play important roles in metabolism. ERRα is thought to be a useful therapeutic target for breast cancer [9] and is a key determinant of rapamycin-induced non-alcoholic fatty liver [10]. Evidence is accumulating to implicate ERRγ in the aetiology of various diseases. Our previous studies show that ERRγ is involved in regulating pyruvate dehydrogenase kinase 4 (PDK4) gene expression and is a novel transcriptional regulator of phosphatidic acid phosphatase [11, 12]. We have also shown that ERRγ regulates the expression of phosphoenolpyruvate carboxykinase 1 (PEPCK) and glucose-6-phosphatase (G6Pase), the rate-limiting enzyme in glucose production [13, 14]. However, the role of ERRγ in liver metabolism is still unclear.
Biliary secretion of cholesterol, either in the form of free cholesterol or in the form of bile acids, is the only significant route for eliminating cholesterol in mammals [15]. Bile acid synthesis from cholesterol occurs either via the classic (also called ‘neutral’) or alternative (also called ‘acidic’) bile acid biosynthetic pathway [16]. Cholesterol 7α-hydroxylase (CYP7A1) and sterol 27-hydroxylase (CYP27A1) are initial and rate-determining enzymes in the classic and alternative pathways respectively [17]. CYP7A1 gene expression is regulated mainly at the transcriptional level and is tightly controlled by nuclear receptors [15]. Liver-related homologue-1 (LRH-1, NR5A2) and COUP transcription factor 2 (COUP-TFII, NR2F2) are critical transcriptional regulators of CYP7A1 expression [18–20]. In addition, hepatocyte nuclear factor 4α (HNF4α, NR2A1) and the peroxisome proliferator-activated receptor γ coactivator 1 α (PGC-1α) are also major transcriptional regulators of CYP7A1 [18, 21]. Although bile acids are the product of this enzymatic reaction, a feedback loop exists where bile acids repress CYP7A1 expression via the farnesoid X receptor (FXR, NR1H4) [22, 23]. Activation of FXR by bile acid induces expression of the small heterodimer partner (SHP, NR0B2) and represses CYP7A1 expression [20, 24, 25]. As a negative regulator of nuclear receptors, SHP inhibits activation of HNF4α and LRH-1 in hepatocytes. Moreover, activation of FXR also increases hepatic fibroblast growth factor 19 (FGF19) expression, suggesting that hepatic FGF19 may repress CYP7A1 expression through an autocrine/paracrine mechanism in humans [24].
The endocannabinoid system, an endogenous lipid signalling pathway, has gained interest as a potential therapeutic target for various disorders, such as cancer [26] and liver metabolic disease [27]. Two G protein-coupled receptors [cannabinoid receptor type 1 (CB1 receptor) and cannabinoid receptor type 2 (CB2 receptor)] are firmly established as targets of cannabinoids [28, 29]. 2-AGE (2-arachidonyl glyceryl ether) is suggested to be an endogenous agonist of the CB1 receptor; it is a potent agonist of the CB1 receptor, but has low affinity for the CB2 receptor [30]. The hepatic CB1 receptor is a selective target to treat fatty liver, impaired glucose homoeostasis and dyslipidaemia. Moreover, hepatic CB1 receptor plays an important role in fatty acid synthesis and contributes to diet-induced obesity [31–33]. It is also reported that induction of endocannabinoids is regulated by alcohol-mediated DAGLβ (DAG-lipase β) in hepatic stellate cells, suggesting a paracrine mechanism by which hepatic stellate cell-derived endocannabinoids activate the CB1 receptor on adjacent hepatocytes [34]. Our previous work suggests that ERRγ controls hepatic CB1 receptor-mediated CYP2E1 expression and oxidative liver injury by alcohol [35]. We also found that CB1 receptor activation disrupts hepatic insulin receptor signalling via CREBH (cAMP-responsive element binding protein, hepatocyte specific)-mediated induction of the lipin1 gene expression [36]. Furthermore, our previous study demonstrates a novel regulatory mechanism of hepatic bile acid metabolism by alcohol via CB1 receptor-mediated activation of CREBH [37]. Therefore, blocking theCB1receptor signalling pathway may be beneficial in restoring hepatic metabolic homoeostasis.
In the present study, we demonstrated that ERRγ is a previously unrecognized transcriptional regulator of CYP7A1 and increases bile acid synthesis. Increase in hepatic ERRγ gene expression led to the induction of CYP7A1, whereas ablation of hepatic ERRγ gene expression abolished the induction of CYP7A1. The induction of CYP7A1 expression by ERRγ was repressed by SHP through SHP-mediated inhibition of ERRγ transcriptional activity. An inverse agonist of ERRγ reduced CYP7A1 expression and bile acid synthesis. Control of alcohol-mediated CYP7A1 level by an ERRγ-specific inverse agonist could be a novel and alternative therapeutic approach for treating cholestatic liver disease.
MATERIALS AND METHODS
Ethics statement
All animal experiments were approved by the Institutional Animal Use and Care Committee of the Korea Research Institute of Bioscience and Biotechnology.
Animal experiments
C57BL/6J mice (The Jackson Laboratory) were used. The mice were acclimatized to a 12 h light/dark cycle at 22 ± 2°C with free access to food and water in a specific pathogen-free facility. Ad-GFP and Ad-FLAG–ERRγ were injected into the tail veins of mice and the mice were killed at day 3 after injection. For the GSK5182 study, mice were divided into four groups: control, ethanol treatment, GSK5182 treatment and GSK5182/ethanol treatment, with five mice in each group. Mice received ethanol [6 g/kg body weight (BW)] by gavage or received an isocaloric maltose solution. In the GSK5182 or GSK5182/ethanol group, GSK5182 (40 mg/kg) was given by intraperitoneal (IP) injections. CB1 receptor knockout mice (CB1−/− ) were kindly provided by Dr George Kunos at the National Institute on Alcohol Abuse and Alcoholism (NIAAA)/NIH as described previously [35] and 8-week-old wild-type (WT) and CB1−/− mice were used for mouse primary hepatocyte culture. Following completion of the experiments, liver tissues and serum were collected for total RNA isolation, protein extraction and bile acid measurement.
Chemicals
GSK5182 was synthesized as described previously [7]. CDCA (chenodeoxycholic acid) was purchased from Sigma. 2-AGE (noladin ether) was purchased from Tocris Bioscience.
Plasmids
Mouse CYP7A1 gene promoter serial constructs (−3.2 kb/+234 bp, −2.6 kb/+234 bp, −1.8 kb/+234 bp, −1.5 kb/+234 bp, −1.2 kb/+234 bp and −0.7 kb/+234 bp) were cloned and ligated into the PGL3-basic vector with the XhoI/MluI enzyme site. These reporter plasmids were confirmed by DNA sequencing. Human CYP7A1-luc (−1887 bp) reporter was as described previously [38]. Expression vectors for FLAG–ERRα, FLAG–ERRβ, haemagglutinin (HA)–SHP, siSHP, HA–HNF4α, HA–LRH-1 and FLAG–ERRγ were as described previously [39, 40]. The ERR response element (ERRE)-mutated mouse CYP7A1 gene promoter was generated via site-directed mutagenesis (Stratagene) and the constructs (MT ERRE2-luc, MT ERRE1-luc and MT ERRE1 and 2-luc) were confirmed by DNA sequencing. All primers used in site-directed mutagenesis are described in Table 1.
Table 1.
List of primers, forward primer (F) and reverse primer (R), used for amplification using qPCR and site-directed mutagenesis
| Name | Sequence(5′-3′) |
|---|---|
| ERRE1 MUT F | TGCCTTCCAAGGCAATATTTCCAATCCTCTCTCCAC |
| ERRE1 MUT R | GTGGAGAGAGGATTGGAAATATTGCCTTGGAAGGCA |
| ERRE2 MUT F | CCTACTGCTTCTGCTATTTGACCTGAGAGGGTCG |
| ERRE2 MUT R | CGACCCTCTCAGGTCAAATAGCAGAAGCAGTAGG |
| m-CYP7A1 F | TCTCAAGCAAACACCATTCCT |
| m-CYP7A1 R | GGCTGCTTTCATTGCTTCA |
| m-CYP27A1 F | CCTCACCTATGGGATCTTCATC |
| m-CYP27A1 R | TTTAGGCATCCGTGTAGAGC |
| m-CYP7B1 F | AATTGGACAGCTTGGTCTGC |
| m-CYP7B1 R | TTCTCGGTGATGCTGGAGT |
| m-CYP8B1 F | GCAGCACTGAATACCCATCC |
| m-CYP8B1 R | TCTGAGAGCTGGGGAGAGG |
| h-CYP7A1 F | GCATCATAGCTCTTTACCCAC |
| h-CYP7A1 R | GGTGTTCTGCAGTCCTGTAAT |
| m-ERRγ F | GGATGGGCAAAACATATTCC |
| m-ERRγ R | ACAACGCCGAGGACTCAGA |
| m-SHP F | CACCTGCATCTCACAGCCACT |
| m-SHP R | GCCAACCCAAGCAGGAAGA |
| r-CYP7A1 F | TGCTCTGTGTTCACTTTCTG |
| r-CYP7A1 R | ACTCGGTAACAGAAGGCATA |
| r-actin F | GGCACCACACTTTCTACAAT |
| r-actin R | AGGTCTCAAACATGATCTGG |
| m-cb1R F | GGGCAAATTTCCTTGTAGCA |
| m-cb1R R | GGCTCAACGTGACTGAGAAA |
| m-LRH-1-F | TTG AGT GGG CCA GGA GTA GT |
| m-LRH-1-R | TCG GTA AAT GTG ATC GAG AAT C |
| m-HNF4α-F | GCC AAG ATT GAC AAC CTG CT |
| m-HNF4α-R | CCC ATG TGT TCT TGC ATC AG |
| m-LXRα-F | AGA GCC TCC AGG GTG AGG |
| m-LXRα-R | AGC CCT GGA CAT TAC CAA GA |
Recombinant adenovirus
Ad-GFP, Ad-FLAG–ERRγ, Ad-USi, Ad-LRH-1, Ad-HNF4α, Ad-FLAG–ERRα, Ad-shSHP, Ad-SHP and Ad-shERRγ were as described previously [40, 41]. All viruses were purified using CsCl2 or an Adeno-X maxi purification kit (Clontech). For adenoviral infections, cells were washed with PBS and left for 2–3 h in serum-free medium containing the appropriate number of viral particles (100 multiplicity of infection/virus). The medium was replaced with fresh growth medium for an additional 36–72 h before treatment.
Cell culture and transient transfection assay
293T (human embryonic kidney cell line) and HepG2 (human hepatoma cell line) cells were maintained as described previously [42]. AML12 (mouse immortalized hepatocyte) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM)/F-12 medium (Gibco-BRL) supplemented with insulin-transferrin-selenium (Gibco-BRL), dexamethasone (40 ng/ml; Sigma) and antibiotics in a humidified atmosphere containing 5% CO2 at 37°C. Transient transfections were conducted as described previously [12]. Luciferase activity was normalized to β-galactosidase activity.
Culture of primary hepatocytes
Primary rat hepatocytes were prepared from 200–250 g Sprague– Dawley rats by a collagenase perfusion method, as described previously [43]. Cells were maintained in M199 media (Cellgro) overnight for attachment and chemical treatments. Mouse primary hepatocytes were isolated and cultured from the livers of 8-weekold C57BL/6J WT and CB1 receptor knockout (CB1−/−) mice as previously described [44].
ChIP assay
The ChIP assay was performed according to the manufacturer’s protocol (Upstate Biotechnology). Immunoprecipitation was performed using ERRγ antibody or IgG (as a negative control). After recovering the DNA, quantitative PCR (qPCR) was performed using primers encompassing the mouse CYP7A1 promoter region. The primers used for PCR were as follows: (−2.3 kb/−2.1 kb) forward 5′-CTGAGGCTTTGAGAGTGATT-3′ and reverse 5′- CCACAGCCAAGTAAGTAAAG-3′ (−1.4 kb/−1.2 kb) forward 5′-CTGATGTTTTCCTTTCTCCT-3′ and reverse 5′-GGAGGATAGAAGATGGAGTT-3′.
Semi-quantitative PCR and qPCR
Total RNA from hepatocytes or liver tissues was extracted using an RNA extraction kit. cDNAs were generated with the Maxime RT PreMix Kit (Intron Biotech) and analysed by semi-quantitative PCR [1% agarose gel stained with ethidium bromide (EtBr)] and qPCR using a SYBR Green PCR kit and AB Real Time System (Applied Biosystems). All data were normalized to actin expression. All primers used in the qPCR are described in Table 1.
Co-immunoprecipitation assay and Western blot analysis
Co-immunoprecipitation (Co-IP) and Western blot analyses were performed as described previously [40]. Cell lysates were prepared from hepatocytes or liver tissues of experimental animals and Western blotting was performed using the indicated antibodies. The following primary antibodies were used for the immunoblotting assay: β-actin (AbFrontier), anti-ERRγ (Perseus Proteomics), anti-HA (Roche Applied Science), anti-FLAG M2 (Sigma Chemical Co.) and CYP7A1 (Santa Cruz Biotechnology).
Measurement of bile acid level
Total bile acids were extracted from cell culture medium with a Sep-Pak C18 cartridge (Waters Associates) followed by determination of levels using a bile acid L3K assay kit (Diazyme). Serum and tissue bile acid analysis were performed using a bile acid L3K assay kit as described previously [45].
Statistics
Values are expressed as mean ± S.E.M. Statistical significance was calculated using the unpaired Student’s t test. Differences were considered significant at P < 0.05.
RESULTS
Activation of the hepatic CB1 receptor induces CYP7A1 expression
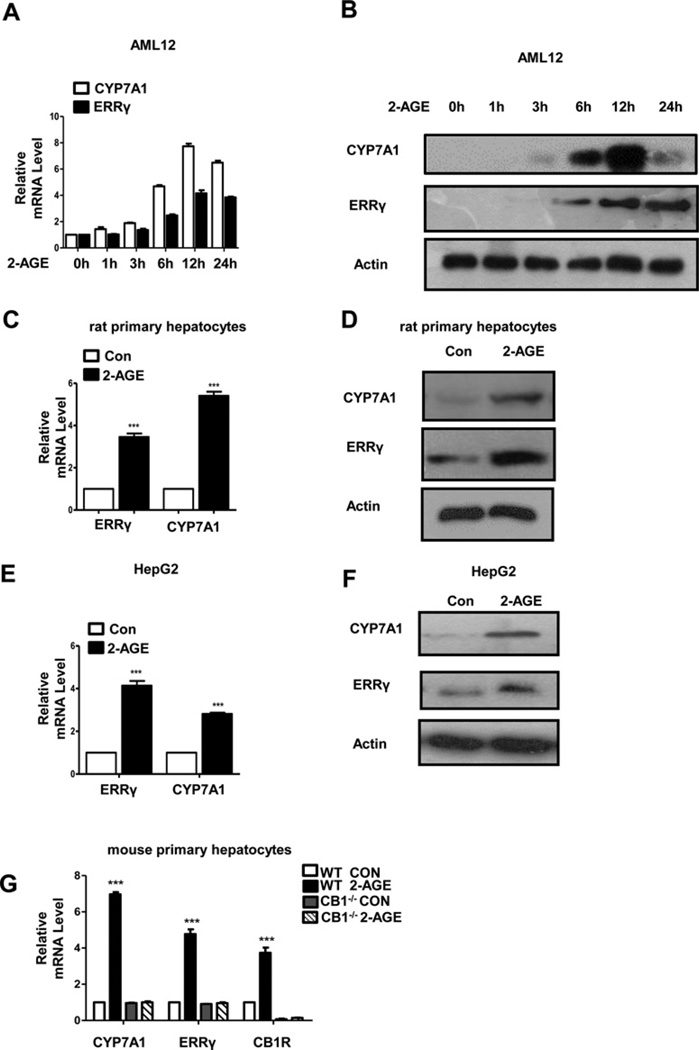
The main bile acid biosynthetic pathway is initiated by CYP7A1 [46, 47]. Our previous study showed that CB1 receptor signalling is associated with hepatic ERRγ gene expression and bile acid metabolism in response to activation of CB1 receptor signalling [35, 37]. To investigate the role of the CB1 receptor and ERRγ in bile acid metabolism, we examined the induction of CYP7A1 and ERRγ gene expression by 2-AGE, an agonist of the CB1 receptor. 2-AGE significantly increased CYP7A1 and ERRγ mRNA levels in AML12 cells from 6 h with maximal levels at 12 h (Figure 1A). Consistent with the change of mRNA levels, CYP7A1 and ERRγ protein levels were also increased during 2-AGE treatment in AML12 cells (Figure 1B). 2-AGE treatment also significantly increased ERRγ and CYP7A1 mRNA and protein levels in rat primary hepatocytes (Figures 1C and 1D). Similar increases in ERRγ and CYP7A1 mRNA and protein levels were observed in 2-AGE treated HepG2 cells (Figures 1E and 1F). To confirm whether the 2-AGE induced CYP7A1 and ERRγ expression is mainly through hepatic CB1 receptor pathway, we isolated primary hepatocytes from WT and CB1−/− mice. As expected, the treatment with 2-AGE (10 µM) significantly induced mRNA levels of CYP7A1 and ERRγ in WT hepatocytes, but not in CB1−/− hepatocytes (Figure 1G). These results demonstrate that activation of the hepatic CB1 receptor increases ERRγ and CYP7A1 mRNA and protein levels in mouse, rat and human hepatic cells.
Figure 1. 2-AGE induces ERRγ and CYP7A1 gene expression.
(A and B) AML12 cells were treated with 2-AGE (10 µM) for the indicated time period. (C and D) Rat primary hepatocytes were treated with 2-AGE (10 µM) for 12 h. ***P < 0.001. (E and F) HepG2 cells were treated with 2-AGE (10 µM) for 12 h. ***P < 0.001. ERRγ and CYP7A1 expression in (A–F) were analysed by qPCR and Western blot analysis. (G) Mouse primary hepatocytes isolated from WT and CB1−/− mice were treated with 2-AGE (10 µM) for 12 h. CYP7A1, ERRγ and CB1R gene expression were measured by qPCR analysis and normalized to actin expression. ***P < 0.001. All data are representative of at least three independent experiments. Error bars show ± S.E.M.
ERRγ mediates the induction of CYP7A1 gene expression by 2-AGE
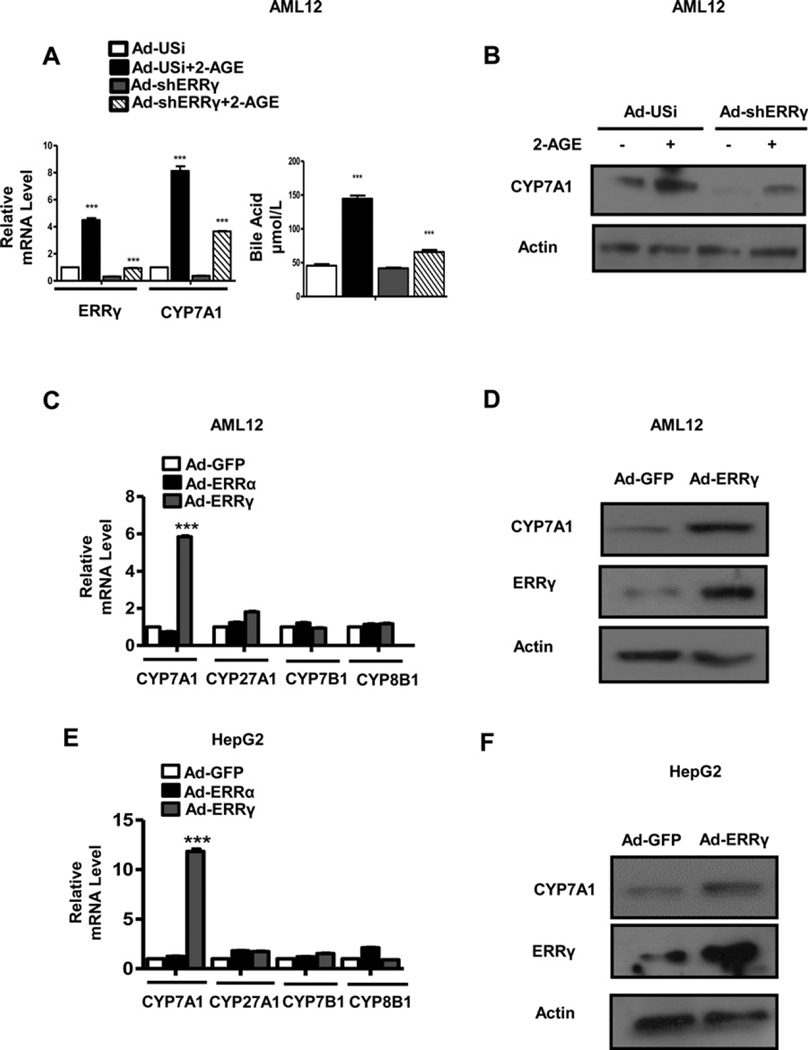
To clarify the role of ERRγ in hepatic CB1 receptor-mediated regulation of CYP7A1 gene expression, the effect of ERRγ knockdown by adenoviral overexpression of ERRγ shRNA (Ad-shERRγ) was investigated on 2-AGE-mediated CYP7A1 expression. 2-AGE treatment increased ERRγ and CYP7A1 mRNA levels, whereas expression of both genes was inhibited by Ad-shERRγ in AML12 cells. Moreover, the total bile acid levels were increased 3-fold after 2-AGE treatment in AML12 cells and this was significantly lowered after knockdown of ERRγ by Ad-shERRγ (Figure 2A). Consistent with the CYP7A1 mRNA level, the increase in CYP7A1 protein level by 2-AGE was decreased by Ad-shERRγ (Figure 2B). These results suggest that ERRγ regulates 2-AGE-mediated induction of CYP7A1 gene expression and bile acid synthesis.
Figure 2. ERRγ is involved in 2-AGE-mediated induction of CYP7A1 gene expression.
(A and B) AML12 cells were infected with adenovirus US (Ad-US) or adenovirus sh-ERRγ (Ad-shERRγ) for 48 h followed by treatment with 2-AGE (10 µM). Cell culture media were collected to determine bile acid levels (A, right panel). ***P < 0.001. (C and D) AML12 cells were infected with Ad-GFP (control), Ad-ERRα and Ad-ERRγ . ***P < 0.001. (E and F) HepG2 cells were infected with Ad-GFP (control), Ad-ERRα and Ad-ERRγ. Total RNA and protein in (A–F) were isolated and used for qPCR and Western blot analysis. ***P < 0.001. All data are representative of at least three independent experiments. Error bars show ± S.E.M.
To further confirm the direct regulation of ERRγ on CYP7A1 gene expression, we used adenoviral overexpression of ERRγ (Ad-ERRγ). Overexpression of ERRγ by Ad-ERRγ increased CYP7A1 mRNA and protein levels in AML12 and HepG2 cells (Figures 2C–2F). Conversely, Ad-ERRα failed to increase CYP7A1 mRNA levels in AML12 and HepG2 cells (Figures 2C and 2E). Moreover, overexpression of ERRα and ERRγ showed no significant effect on CYP27A1, CYP7B1 and CYP8B1 gene expression (Figures 2C and 2E). In addition, HNF4α- or LRH-1-mediated induction of CYP7A1 mRNA was significantly lower than that of ERRγ (Supplementary Figure S1A). Taken together, these results suggest that ERRγ is specifically involved in 2-AGE-mediated induction of CYP7A1 gene expression and bile acid synthesis.
ERRγ increases the activity of the CYP7A1 gene promoter
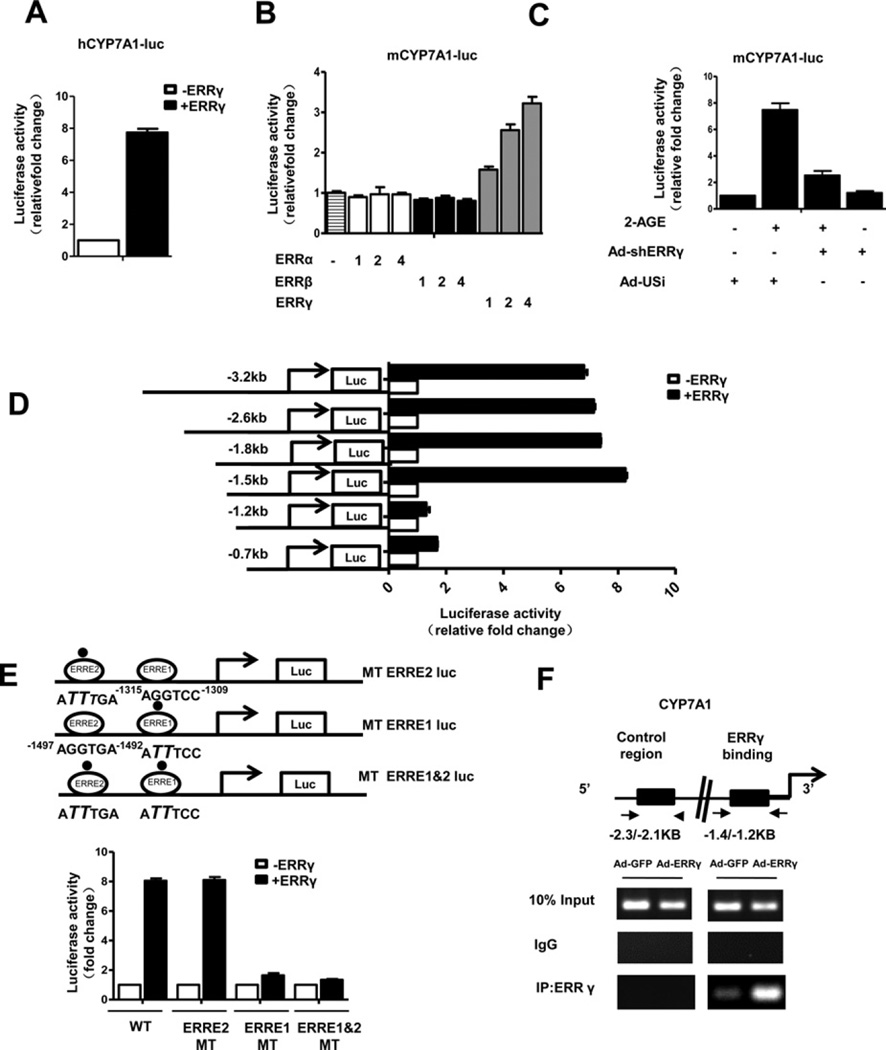
To determine the molecular mechanism by which ERRγ regulates CYP7A1 gene transcription, we performed transient transfection using ERRγ and CYP7A1-luc (CYP7A1 gene promoter luciferase reporter construct) in 293T cells. Co-transfection with ERRγ strongly induced human CYP7A1 promoter activity (Figure 3A). Then, the effect of each of the ERR subfamily on the mouse CYP7A1 promoter was examined. ERRγ specifically elevated the mouse CYP7A1 gene promoter activity in a dose-dependent manner, whereas ERRα and ERRβ showed no significant effect on the CYP7A1 gene promoter activity (Figure 3B). Moreover, HNF4α- and LRH-1-mediated induction of CYP7A1 gene promoter activity was significantly lower than that of ERRγ (Supplementary Figure S1B). Furthermore, CYP7A1 promoter activity was increased 8-fold after 2-AGE treatment. This was decreased significantly by knockdown of ERRγ using Ad-shERRγ (Figure 3C). Moreover, reporter assays of deletion constructs showed that the ERRγ -mediated activation of the CYP7A1 promoter was markedly decreased by deletion of the CYP7A1 promoter from −1.5 kb to −1.2 kb (Figure 3D). A close investigation of the CYP7A1 promoter revealed two putative binding motifs (AGGTCC, as indicated by ERRE1 and AGGTGA, as indicated by ERRE2). To further identify ERRγ-binding sites in the CYP7A1 promoter, we performed transfection assays with the WT and a point mutant of the putative ERRγ -binding sites (ERRE1 and ERRE2) in the CYP7A1 promoter. Activation of the CYP7A1 promoter by ERRγ was significantly abolished in ERRE1-mutated or the ERRE1 and 2-mutated CYP7A1 promoter, whereas no significant change in activation of the ERRE2-mutated CYP7A1 promoter was observed (Figure 3E). This result suggests that ERRγ directly regulates CYP7A1 through ERRE1. Finally, binding of ERRγ on the endogenous CYP7A1 promoter was confirmed by a ChIP assay in AML12 cells with a specific antibody against ERRγ. ERRγ was strongly recruited to the ERRE1 region of the CYP7A1 promoter whereas no significant recruitment of ERRγ was observed on the control region (Figure 3F). Overall, these results indicate the ERRγ directly binds and activates the CYP7A1 gene promoter.
Figure 3. ERRγ activates the CYP7A1 gene promoter.
(A) 293T cells were transfected with hCYP7A1-luc (−1887 bp) and ERRγ expression vector. (B) 293T cells were transfected with mCYP7A1-luc (−3.2 kb to +234 bp) and different doses of the ERR subfamily cDNA expression vectors: 1:100 ng, 2:200 ng, 4:400 ng. (C) AML12 cells were infected with Ad-US or Ad-shERRγ for 48 h then transfected with mCYP7A1-luc (−3.2 kb to +234 bp) and treated with 2-AGE (10 µM). (D) 293T cells were transfected with deletion constructs of mCYP7A1-luc and ERRγ . (E) An ERRγ -binding site point mutation was made in mCYP7A1 (−1.5 kb)-luc as described in the ‘Materials and Methods’ section. 293T cells were transiently transfected with pCDNA3 – FLAG–ERRγ, mCYP7A1 (−1.5 kb to +234 bp)-luc (WT), mCYP7A1–mtERRE2-Luc, mCYP7A1–mtERRE1-Luc and mCYP7A1–mtERRE1 and 2-Luc. The cell lysates in (A–E) were utilized for luciferase and β-galactosidase assays. Experiments in (A–E) were conducted in triplicate and data are expressed as fold activation relative to the control. (F) ChIP assay. AML12 cells were infected with Ad-GFP or Ad-ERRγ for 48 h. Input represents 10% of purified DNA in each sample. Cell extracts were immunoprecipitated with anti-ERRγ antibody and purified DNA samples were employed for PCR with primers binding to ERRE1 (−1.4 kb to −1.2 kb) and distal site (−2.3 kb to −2.1 kb) on the CYP7A1 gene promoter.
SHP inhibits ERRγ -mediated CYP7A1 gene expression
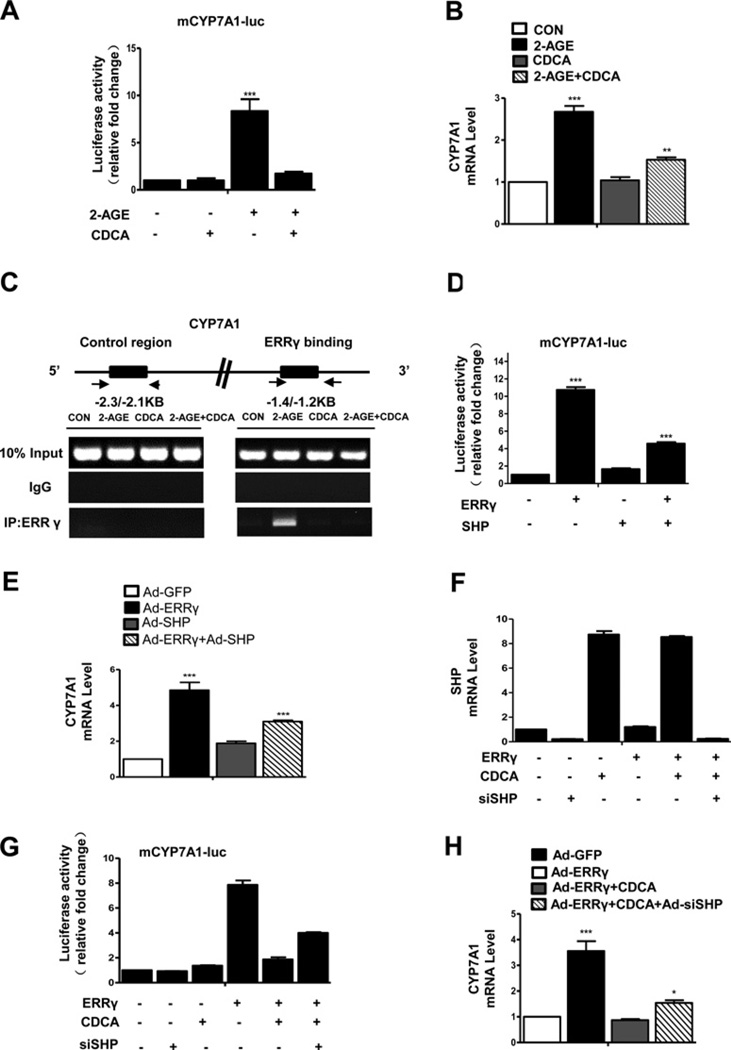
It is reported that SHP acts as transcriptional co-repressor of ERRγ [45], suggesting that SHP is involved in the regulation of ERRγ -mediated CYP7A1 expression. Moreover, the FXR/SHP pathway and the nuclear bile acid receptor FXR sense elevated hepatic bile acid levels and induce SHP gene expression [19]. A previous report also demonstrates that CDCA induces SHP gene expression in mouse AML12 cells [48]. To determine whether SHP is involved in the CB1 receptor-mediated CYP7A1 gene expression, AML12 cells were transfected with mouse CYP7A1-luc in the presence or absence of 2-AGE and CDCA. 2-AGEmediated activation of CYP7A1 promoter activity and CYP7A1 mRNA levels were significantly decreased with CDCA treatment (Figures 4A and 4B). Our ChIP assay results also indicated that 2-AGE treatment strongly enhanced ERRγ recruitment to the ERRE1 region of the endogenous CYP7A1 promoter, which was abolished by CDCA treatment (Figure 4C). Additionally, AML12 cells were infected with both Ad-ERRγ and Ad-SHP to further confirm the role of SHP in ERRγ -mediated induction of CYP7A1 gene expression. SHP significantly inhibited ERRγ - mediated induction of CYP7A1 promoter activity and CYP7A1 mRNA levels (Figures 4D and 4E). Finally, knockdown of SHP significantly abolished the inhibitory effect of CDCA on ERRγ -mediated CYP7A1 promoter activity and gene expression (Figures 4F–4H). Taken together, these results demonstrate that the induction of CYP7A1 gene transcription by ERRγ is regulated by SHP and CDCA.
Figure 4. ERRγ-mediated induction of CYP7A1 gene expression is inhibited by SHP.
(A) AML12 cells were transfected with mCYP7A1-luc (−3.2 kb to +234 bp). At 36 h post transfection, cells were treated with 2-AGE (10 µM) in the presence or absence of CDCA (25 µM). (B) AML12 cells were treated with 2-AGE (10 µM) in the presence or absence of CDCA (25 µM) and CYP7A1 expression was analysed by qPCR. **P < 0.01; ***P < 0.001. (C) ChIP assay. AML12 cells were treated with 2-AGE (10 µM) in the presence or absence of CDCA (25 µM). Input represents 10% of purified DNA in each sample. Cell extracts were immunoprecipitated with anti-ERRγ antibody and purified DNA samples were employed for PCR with primers binding to ERRE1 (−1.4 kb to −1.2 kb) and distal site (−2.3 kb to −2.1 kb) on the CYP7A1 gene promoter. (D) AML12 cells were transfected with mCYP7A1-luc (−3.2 kb to +234 bp) and co-transfected with ERRγ or SHP expression vectors. (E) AML12 cells were infected with indicated adenoviruses and CYP7A1 expression was analysed by qPCR analysis. (F and G) AML12 cells were transfected with the si-SHP expression vector and after 24 h, the cells were co-transfected with mCYP7A1-luc (−3.2 kb to +234 bp) and ERRγ expression vector in the presence or absence of CDCA (25 µM). SHP mRNA level analysed by qPCR analysis. (H) AML12 cells were infected with Ad-ERRγ and Ad-shSHP in the presence or absence of CDCA (25 µM). CYP7A1 expression was analysed by qPCR analysis. *P < 0.05; ***P < 0.001. The cell lysates in (A, D and G) were utilized for luciferase and β-galactosidase assays.
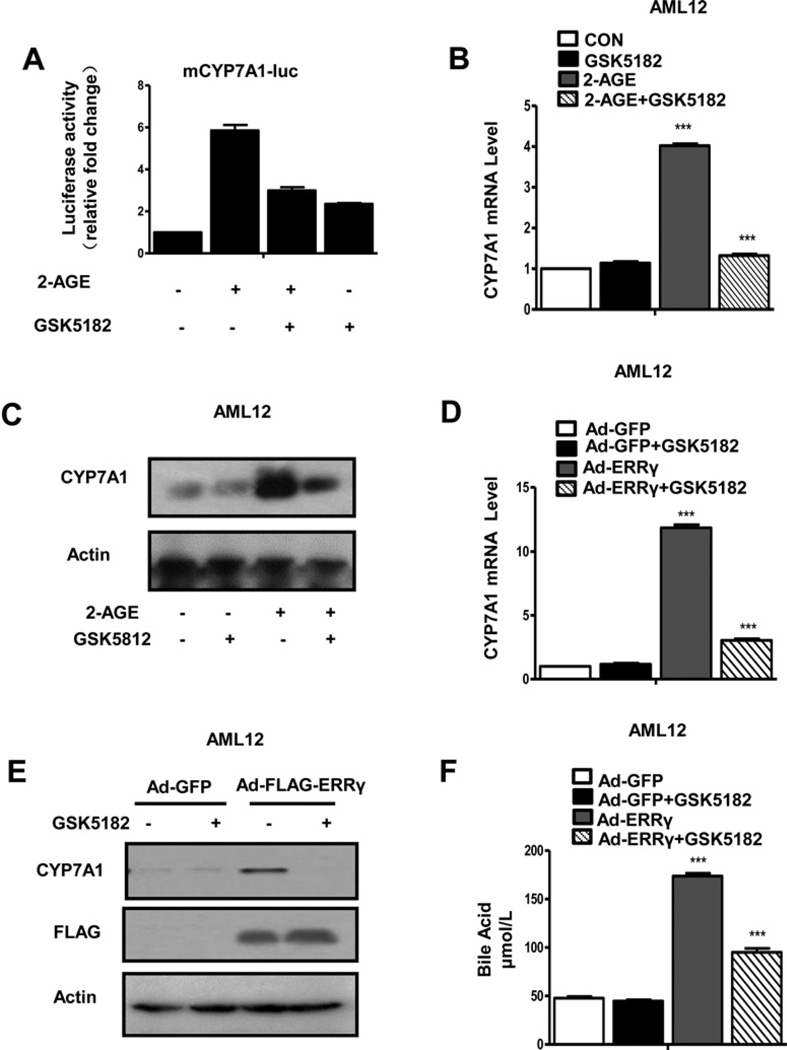
An inverse agonist of ERRγ inhibits CYP7A1 gene expression and reduces bile acid levels
GSK5182, an ERRγ inverse agonist, has been used to selectively inhibit transactivation of ERRγ [7, 12]. To further clarify the role of ERRγ in CB1 receptor-mediated induction of CYP7A1 gene expression, AML12 cells were transfected with the CYP7A1 promoter and treated with 2-AGE in the presence or absence of GSK5182. 2-AGE-activated CYP7A1 gene promoter activity and CYP7A1 gene expression were inhibited by GSK5182 (Figures 5A and 5B). Consistent with the change in CYP7A1 mRNA level, 2-AGE-induced CYP7A1 protein levels were also significantly decreased by GSK5182 (Figure 5C). The inhibitory effect of the ERRγ inverse agonist on CYP7A1 gene expression was determined by infecting AML12 cells with Ad-ERRγ and treating with GSK5182. The Ad-ERRγ - mediated increase in CYP7A1 mRNA and protein levels were significantly decreased by GSK5182 (Figures 5D and 5E). However, GSK5182 treatment did not exhibit significant effect on the recruitment of ERRγ to the region (Supplementary Figure S2A), suggesting that GSK5182 suppresses the expression of CYP7A1 gene expression by disrupting the interaction between ERRγ and its co-activator without affecting DNA-binding ability of ERRγ. Cell culture medium was collected to measure total bile acid levels. The total bile acid levels were dramatically increased after Ad-ERRγ overexpression and this increase was significantly reduced by GSK5182 (Figure 5F). In addition, to further examine the interaction between ERRγ and SHP protein, Co-IP was conducted after co-transfection of vector encoding FLAG–ERRγ or HA–SHP in AML12 cells. The results showed that ERRγ interacts with SHP and GSK5182 did not affect the interaction (Supplementary Figure S2B). Taken together, these results indicate that inactivating ERRγ with the inverse agonist GSK5182 decreases CB1 receptor-mediated CYP7A1 gene expression and bile acid synthesis.
Figure 5. GSK5182 inhibits CYP7A1 gene expression and bile acid synthesis.
(A) AML12 cells were transfected with mCYP7A1-luc (−3.2 kb to +234 bp) then treated with 2-AGE (10 µM) and GSK5182 (10 µM). Cell lysates were utilized for luciferase and β-galactosidase assays. (B and C) AML12 cells were treated with 2-AGE (10 µM) for 12 h. Then, the cell culture medium was changed and GSK5182 (10 µM) was added for the final 24 h. Total mRNAs and protein were extracted for qPCR (B) and Western blot analysis (C). ***P < 0.001. (D–F) AML12 cells were infected with Ad-GFP and Ad-ERRγ and then treated with GSK5182 (10 µM) for 24 h. CYP7A1 mRNA level was analysed by qPCR (D) and the ERRγ and CYP7A1 protein levels were measured by Western blot analysis (E), ***P < 0.001. Cell culture media were collected to determine bile acid levels (F), ***P < 0.001.
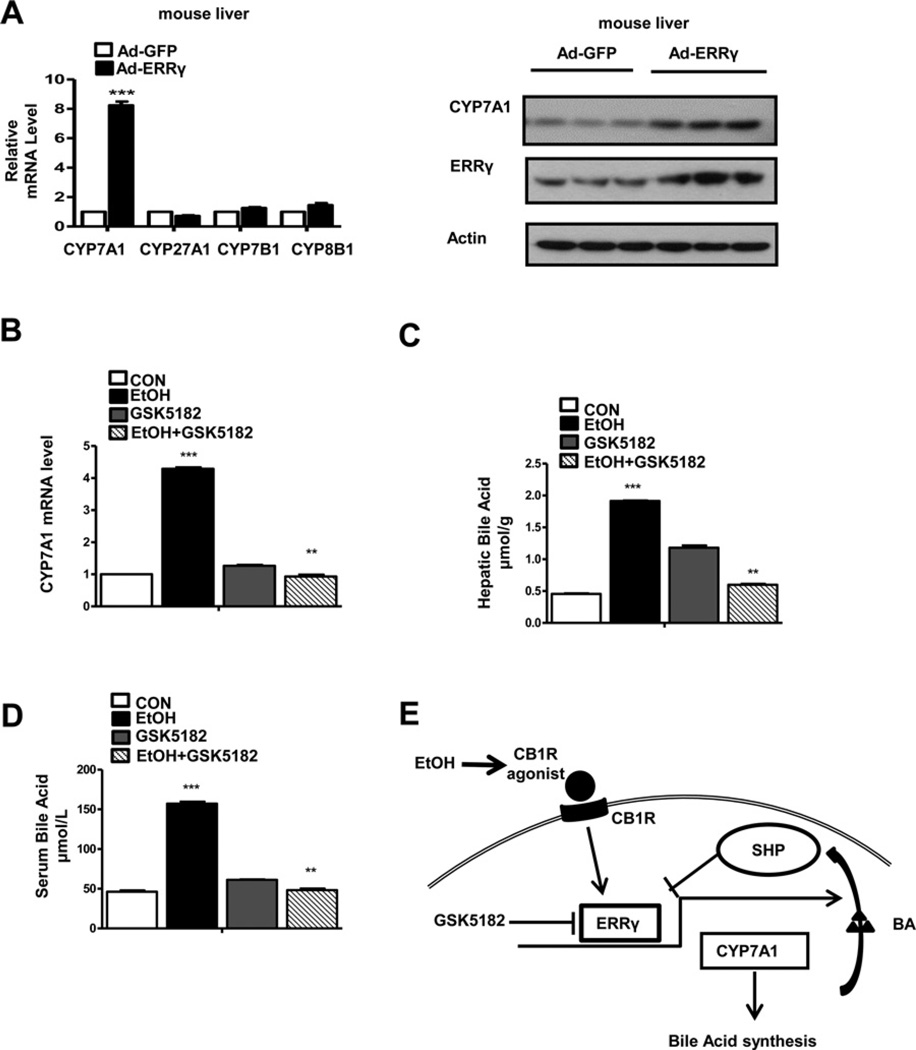
Figure 6. Hepatic CB1 receptor signalling regulates CYP7A1 gene expression via induction of ERRγ in mice.
(A) Ad-GFP or Ad-ERRγ were injected via the tail-vein into male C57BL/6J mice (n = 3–4 per group) and mice were killed at day 3. qPCR analysis and Western blot analysis were performed to measure the mRNA and protein levels in liver. ***P < 0.001. (B–D) Mice (n = 3–4) were treated with alcohol for 1 day in the presence of GSK5182. Total RNA was extracted for qPCR analyses (B). Liver tissue (C) and serum (D) were used for bile acid analysis; **P < 0.01; ***P < 0.001. (E) Proposed model for CB1 receptor-mediated induction of CYP7A1 gene expression via ERRγ. Activation of hepatic CB1 receptor increases ERRγ gene expression, which in turn increases the promoter activity and mRNA level of CYP7A1. Induction of CYP7A1 gene expression promotes bile acid synthesis and induces SHP expression. SHP inhibits ERRγ -mediated induction of CYP7A1 gene expression and promoter activity. GSK5182, an ERRγ inverse agonist, inhibits ERRγ -mediated CYP7A1 gene expression and bile acid (BA) synthesis.
An inverse agonist of ERRγ regulates CYP7A1 gene expression and bile acid synthesis in alcohol-treated mice
The hepatic endocannabinoid system is associated with the regulation of hepatic lipid metabolism, alcoholic fatty liver [49] and alcoholic liver injury [35]. We intravenously injected adenovirus ERRγ into mice to investigate the effect of ERRγ on the gene expression of hepatic bile acid metabolic enzymes in vivo. ERRγ overexpression resulted in a specific increase in CYP7A1 mRNA and protein levels but no differences in gene expression of CYP27A1CYP7B1 and CYP8B1 in mouse liver (Figure 6A). Our previous study demonstrated that hepatic ERRγ gene expression is regulated by alcohol-mediated activation of CB1 receptor signalling [35]. Based on this study, mice were administered with alcohol for 1 day in the presence or absence of GSK5182 to determine the inhibitory effect of GSK5182 on alcohol-mediated induction of CYP7A1 gene expression and bile acid levels in mice. As expected, the alcohol-mediated increase in CYP7A1 mRNA level was markedly decreased following GSK5182 treatment (Figure 6B). Notably, total bile acid levels in alcohol-treated mouse serum and liver tissue were significantly increased, whereas a significant reduction in bile acid levels was observed in mouse liver tissue and serum in the presence of GSK5182 (Figures 6C and 6D). We also found that ethanol administration increased SHP mRNA level, which was reduced by GSK5182 treatment (Supplementary Figure S3A). However, ethanol administration did not lead to significant changes in mRNA levels of other CYP7A1 regulators such as LXRα, HNF4α and LRH-1 (Supplementary Figure S3A). These results suggest that alcohol induces CYP7A1 gene expression via the hepatic CB1 receptor and that GSK5182, an inverse agonist of ERRγ, inhibits alcohol-mediated induction of CYP7A1 gene expression and bile acid synthesis.
DISCUSSION
There are several important observations in this current study. Firstly, activation of the hepatic CB1 receptor by 2-AGE induces ERRγ and CYP7A1 gene expression. Among the ERR family, ERRγ is specifically involved in the regulation of CYP7A1 gene expression. Secondly, ERRγ activates the CYP7A1 gene promoter and nuclear receptor co-repressor SHP inhibits ERRγ -mediated induction of CYP7A1 gene expression. Thirdly, GSK5182 inhibits CYP7A1 gene expression and decreases bile acid level through inhibition of ERRγ. Finally, GSK5182 regulates CYP7A1 gene expression and bile acid synthesis in alcohol-treated mice.
Several membrane receptors, such as the insulin receptor, toll-like receptor 4 and epidermal growth factor receptor, are involved in the regulation of CYP7A1 gene expression [38, 50, 51]. In the present study, we demonstrated that activation of the hepatic CB1 receptor-ERRγ signalling pathway induced CYP7A1 gene expression. A future study is to also examine a role for the hepatic CB2 receptor in the regulation of bile acid synthesis. It should be emphasized that cholesterol metabolism in rats and mice is different from that in humans and other species. For example, stimulation of CYP7A1 activity by a high cholesterol diet is only observed in rats and some inbred strains of mice [52, 53], which are highly efficient in converting cholesterol to bile acids. Our results show that activation of hepatic CB1 receptor signalling induced CYP7A1 in mouse, rat and human hepatic cells (Figure 1). Moreover, overexpression of ERRγ increases CYP7A1 mRNA and protein levels in AML12 (mouse hepatocytes) and HepG2 (human hepatoma) cells (Figure 2). These finding suggest that the ERRγ -mediated induction of CYP7A1 gene expression is conserved in mouse, rat and human.
Studies on the molecular mechanism underlying the transcriptional regulation of CYP7A1 expression have progressed greatly in recent years. CYP7A1 gene expression is under tight regulation by different signalling pathways and is regulated mainly at the transcriptional level by a number of factors, including nuclear receptors, protein kinase C activators, cytokines, growth factors and bile acids. In the present study, we found that knockdown of ERRγ did not completely inhibit 2-AGE-mediated induction of CYP7A1 gene expression (Figures 2A and 2B). This result suggests that there may be other factors involved in 2-AGE-meidated CYP7A1 expression. For example, we recently reported that CREBH, an ER-bound transcription factor, is also implicated in 2-AGEmeidated CYP7A1 expression [37]. Therefore, there may be a compensatory mechanism between ERRγ and CREBH in the CB1 pathway. In the present study, we suggest that ERRγ is a novel transcriptional regulator of CYP7A1 (Figure 3). However, ERRγ showed no significant effect on CYP27A1, CYP7B1 and CYP8B1 gene expression. Dufour et al. [54] demonstrated that ERRα and ERRγ regulate the same direct target genes [54]. However, our result shows that the CYP7A1 promoter is activated by ERRγ but not ERRα or ERRβ, showing the complexity of their function in terms of the transcriptional output of the ERR subfamily.
The transcriptional activity of ERRγ is co-regulated by PGC-1α and SHP [55, 56]. Elevated SHP protein levels result in repression of SHP promoters by an inhibitory LRH-1 and SHP heterodimeric complex [20]. Our current study indicates that SHP inhibits ERRγ -mediated induction of CYP7A1 promoter activity (Figure 4). It is reported that the transcriptional co-activator, PGC-1α, activates CYP7A1 and bile acid biosynthesis [21]. However, PGC-1α gene expression was not significantly changed in 2-AGE treated AML12 cell (result not shown). Proline-rich nuclear receptor coactivator 2 (PNRC2) and transducin-like enhancer protein 1 (TLE1) are co-activators of ERRγ by binding to its AF-1 domain [57]. Moreover, nuclear receptor coactivator 1 (NCOA1) and nuclear receptor coactivator 2 (NCOA2) are well-known AF2-dependent co-activators of ERα and other nuclear receptors, including ERRγ [58]. Therefore, we speculate that other transcriptional co-activators may enhance ERRγ transcriptional activity during CB1 receptor-mediated CYP7A1 gene transcription.
CYP7A1 overexpression results in a marked activation of the classic pathway of bile acid biosynthesis leading to the accumulation of high concentrations of bile acids and, ultimately, hepatocyte injury and impaired liver function [59]. Conversely, a role for hepatic CYP7A1 in preventing the development of obesity and diabetes has been investigated. One study shows that increases in hepatic CYP7A1 expression improve several parameters of metabolic syndrome, including obesity, hepatic steatosis and insulin resistance in mice [60]. Therefore, it is worth defining whether the modulation of CYP7A1 and bile acid synthesis through ERRγ may improve energy metabolism and insulin resistance. The hepatic manifestations of alcohol overconsumption include alcoholic fatty liver, alcoholic hepatitis, hepatic fibrosis and hepatic cirrhosis. Above all, serum bile acid levels are biomarkers for diagnosing liver diseases, diabetes and obesity. Moreover, two potent activators of human pregnane X receptor (PXR), rifampicin and ursodeoxycholic acid, were successfully used in the treatment of cholestasis through inhibition of CYP7A1 [61–63]. Our current study demonstrated that CYP7A1 gene expression and bile acid levels were decreased in a GSK5182-treated acute alcohol mouse model. This finding suggests that the inverse agonist of ERRγ, GSK5182, may be a new potential therapeutic agent for the treatment of alcohol-induced bile acid synthesis and liver injury (Figures 5 and 6).
Overall, our results reveal that ERRγ plays a significant role in the classical bile acid synthesis pathway through CB1 receptor-mediated CYP7A1 gene expression. ERRγ binds to ERRE1 and activates transcriptional activity of the CYP7A1 gene. Increases in CYP7A1 gene expression induce bile acid synthesis and SHP gene expression. Consequently, SHP inhibits the ERRγ - mediated induction of CYP7A1 gene expression and promoter activity through a regulatory loop. GSK5182, an ERRγ inverse agonist, resulted in a marked reduction in ERRγ -induced CYP7A1 transactivity (Figure 6E). When alcohol injury takes place, the CB1 receptor signalling pathway gets activated following induction and activation of ERRγ ,which plays a significant role in up-regulating the classical bile acid synthesis pathway. Therefore, targeting ERRγ can be of therapeutic potential in ameliorating alcohol-induced perturbation of bile acid homoeostasis.
Supplementary Material
Acknowledgments
We thank our laboratory members and Dr David D. Moore for cooperation and discussion related to this work.
FUNDING
This work was supported by the National Research Foundation of Korea (NRF) [grant number 20110018305]; the Korea Health Industry Development Institute [grant number A111345]; the National Institute of Health [grant numbers DK44442 and DK58379 (to J.Y.L.C.)].
Abbreviations
- 2-AGE
2-arachidonyl glyceryl ether
- CB1 receptor
cannabinoid receptor type 1
- CDCA
chenodeoxycholic acid
- Co-IP
coimmunoprecipitation
- CREBH
cAMP-responsive element binding protein, hepatocyte specific
- CYP7A1
cholesterol 7α-hydroxylase
- CYP27A1
sterol 27-hydroxylase
- ERRγ
oestrogen-related receptor γ
- ERRE
ERR response element
- FXR
farnesoid X receptor
- FGF19
fibroblast growth factor 19
- HNF4α
hepatocyte nuclear factor 4α
- LRH-1
liver-related homologue-1
- PGC-1α
peroxisome proliferator-activated receptor γ coactivator 1 α
- qPCR
quantitative PCR
- SHP
small heterodimer partner
- WT
wild-type
Footnotes
AUTHOR CONTRIBUTION
Yaochen Zhang and Don-Kyu Kim designed and performed most of the experiments. Hueng-Sik Choi and John Chiang supervised the project. In-Kyu Lee, Seong Kim and Seung Park synthesized and provided GSK5182. Chul-Ho Lee, Won-IL Jeong performed animal studies. Ji-Min Lee isolated and cultured primary hepatocytes. Yaochen Zhang, Don-Kyu Kim, John Chiang and Hueng-Sik Choi analysed data and wrote the manuscript.
REFERENCES
- 1.Giguere V, Yang N, Segui P, Evans RM. Identification of a new class of steroid hormone receptors. Nature. 1988;331:91–94. doi: 10.1038/331091a0. [DOI] [PubMed] [Google Scholar]
- 2.Yang X, Downes M, Ruth TY, Bookout AL, He W, Straume M, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 3.Luo J, Sladek R, Carrier J, Bader JA, Richard D, Giguere V. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor alpha. Mol. Cell Biol. 2003;23:7947–7956. doi: 10.1128/MCB.23.22.7947-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Razzaque MA, Masuda N, Maeda Y, Endo Y, Tsukamoto T, Osumi T. Estrogen receptor-related receptor γ has an exceptionally broad specificity of DNA sequence recognition. Gene. 2004;340:275–282. doi: 10.1016/j.gene.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 5.Busch BB, Stevens WC, Martin R, Ordentlich P, Zhou S, Sapp DW, Horlick RA, Mohan R. Identification of a selective inverse agonist for the orphan nuclear receptor estrogen-related receptor α. J. Med. Chem. 2004;47:5593–5596. doi: 10.1021/jm049334f. [DOI] [PubMed] [Google Scholar]
- 6.Mootha VK, Handschin C, Arlow D, Xie X, Pierre JS, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N. Errα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad Sci. U.S.A. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao EY, Collins JL, Gaillard S, Miller AB, Wang L, Orband-Miller LA, Nolte RT, McDonnell DP, Willson TM, Zuercher WJ. Structure-guided synthesis of tamoxifen analogs with improved selectivity for the orphan ERRγ. Bioorg. Med. Chem. Lett. 2006;16:821–824. doi: 10.1016/j.bmcl.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Fang F, Huang Z, Wang Y, Wong C. Kaempferol is an estrogen-related receptor α and γ inverse agonist. FEBS Lett. 2009;583:643–647. doi: 10.1016/j.febslet.2009.01.030. [DOI] [PubMed] [Google Scholar]
- 9.Jarzabek K, Koda M, Kozlowski L, Sulkowski S, Kottler M-L, Wolczynski S. The significance of the expression of ERRα as a potential biomarker in breast cancer. J. Steroid. Biochem. 2009;113:127–133. doi: 10.1016/j.jsbmb.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Chaveroux C, Eichner LJ, Dufour CR, Shatnawi A, Khoutorsky A, Bourque G, Sonenberg N, Giguère V. Molecular and genetic crosstalks between mTOR and ERRα are key determinants of rapamycin-induced nonalcoholic fatty liver. Cell Metab. 2013;17:586–598. doi: 10.1016/j.cmet.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Lee JH, Kim E-J, Kim D-K, Lee J-M, Park SB, Lee I-K, Harris RA, Lee M-O, Choi H-S. Hypoxia induces PDK4 gene expression through induction of the orphan nuclear receptor ERRγ. PLoS One. 2012;7:e46324. doi: 10.1371/journal.pone.0046324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim D-K, Kim JR, Koh M, Kim YD, Lee J-M, Chanda D, Park SB, Min J-J, Lee C-H, Park T-S. Estrogen-related receptor γ (ERRγ) is a novel transcriptional regulator of phosphatidic acid phosphatase, LIPIN1, and inhibits hepatic insulin signaling. J. Biol. Chem. 2011;286:38035–38042. doi: 10.1074/jbc.M111.250613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim D-K, Ryu D, Koh M, Lee M-W, Lim D, Kim M-J, Kim Y-H, Cho W-J, Lee C-H, Park SB. Orphan nuclear receptor estrogen-related receptor γ (ERRγ) is key regulator of hepatic gluconeogenesis. J. Biol. Chem. 2012;287:21628–21639. doi: 10.1074/jbc.M111.315168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim D-K, Gang G-T, Ryu D, Koh M, Kim Y-N, Kim SS, Park J, Kim Y-H, Sim T, Lee I-K. Inverse agonist of nuclear receptor ERRγ mediates anti-diabetic effect through inhibition of hepatic gluconeogenesis. Diabetes. 2013;62:3093–3102. doi: 10.2337/db12-0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiang JY. Bile acids: regulation of synthesis. J. Lipid Res. 2009;50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vlahcevic ZR, Pandak WM, Stravitz RT. Regulation of bile acid biosynthesis. Gastroenterol. Clin. N. 1999;28:1–25. doi: 10.1016/s0889-8553(05)70041-8. [DOI] [PubMed] [Google Scholar]
- 17.von Bahr S, Movin T, Papadogiannakis N, Pikuleva I, Rönnow P, Diczfalusy U, Björkhem I. Mechanism of accumulation of cholesterol and cholestanol in tendons and the role of sterol 27-hydroxylase (CYP27A1) Arterioscl. Throm. Vas. 2002;22:1129–1135. doi: 10.1161/01.atv.0000022600.61391.a5. [DOI] [PubMed] [Google Scholar]
- 18.Bhalla S, Ozalp C, Fang S, Xiang L, Kemper JK. Ligand-activated Pregnane × receptor interferes with HNF-4 signaling by targeting a common coactivator PGC-1α Functional Implications In Hepatic Cholesterol And Glucose Metabolism. J. Biol. Chem. 2004;279:45139–45147. doi: 10.1074/jbc.M405423200. [DOI] [PubMed] [Google Scholar]
- 19.Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 20.Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 21.Shin D-J, Campos JA, Gil G, Osborne TF. PGC-1α activates CYP7A1 and bile acid biosynthesis. J. Biol. Chem. 2003;278:50047–50052. doi: 10.1074/jbc.M309736200. [DOI] [PubMed] [Google Scholar]
- 22.Stravitz R, Hylemon P, Heuman D, Hagey L, Schteingart C, Ton-Nu H, Hofmann A, Vlahcevic Z. Transcriptional regulation of cholesterol 7 alpha-hydroxylase mRNA by conjugated bile acids in primary cultures of rat hepatocytes. J. Biol. Chem. 1993;268:13987–13993. [PubMed] [Google Scholar]
- 23.Chiang JY, Kimmel R, Weinberger C, Stroup D. Farnesoid X receptor responds to bile acids and represses cholesterol 7α-hydroxylase gene (CYP7A1) transcription. J. Biol. Chem. 2000;275:10918–10924. doi: 10.1074/jbc.275.15.10918. [DOI] [PubMed] [Google Scholar]
- 24.Holt JA, Luo G, Billin AN, Bisi J, McNeill YY, Kozarsky KF, Donahee M, Mansfield TA, Kliewer SA, Goodwin B. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 2003;17:1581–1591. doi: 10.1101/gad.1083503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song KH, Li T, Owsley E, Strom S, Chiang JY. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7αhydroxylase gene expression. Hepatology. 2009;49:297–305. doi: 10.1002/hep.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pisanti S, Picardi P, D’Alessandro A, Laezza C, Bifulco M. The endocannabinoid signaling system in cancer. Trends Pharmacol. Sci. 2013;34:273–282. doi: 10.1016/j.tips.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Bataller R, Gao B. Dissecting the role of CB1 receptors on chronic liver diseases. Gut. 2012;62:957–958. doi: 10.1136/gutjnl-2012-303664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuda LA, Lolait SJ, Brownstein MJ, Young AC, Bonner TI. Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature. 1990;346:561–564. doi: 10.1038/346561a0. [DOI] [PubMed] [Google Scholar]
- 29.Munro S, Thomas KL, Abu-Shaar M. Molecular characterization of a peripheral receptor for cannabinoids. Nature. 1993;365:61–65. doi: 10.1038/365061a0. [DOI] [PubMed] [Google Scholar]
- 30.Hanuš L, Abu-Lafi S, Fride E, Breuer A, Vogel Z, Shalev DE, Kustanovich I, Mechoulam R. 2-Arachidonyl glyceryl ether, an endogenous agonist of the cannabinoid CB1 receptor. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3662–3665. doi: 10.1073/pnas.061029898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tam J, Vemuri VK, Liu J, Bátkai S, Mukhopadhyay B, Godlewski G, Osei-Hyiaman D, Ohnuma S, Ambudkar SV, Pickel J. Peripheral CB1 cannabinoid receptor blockade improves cardiometabolic risk in mouse models of obesity. J. Clin. Invest. 2010;120:2953. doi: 10.1172/JCI42551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osei-Hyiaman D, DePetrillo M, Pacher P, Liu J, Radaeva S, Bátkai S, Harvey-White J, Mackie K, Offertáler L, Wang L. Endocannabinoid activation at hepatic CB1 receptors stimulates fatty acid synthesis and contributes to diet-induced obesity. J. Clin. Invest. 2005;115:1298. doi: 10.1172/JCI23057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osei-Hyiaman D, Liu J, Zhou L, Godlewski G, Harvey-White J, Jeong W-i, Bátkai S, Marsicano G, Lutz B, Buettner C. Hepatic CB1 receptor is required for development of diet-induced steatosis, dyslipidemia, and insulin and leptin resistance in mice. J. Clin. Invest. 2008;118:3160. doi: 10.1172/JCI34827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeong W-i, Osei-Hyiaman D, Park O, Liu J, Bátkai S, Mukhopadhyay P, Horiguchi N, Harvey-White J, Marsicano G, Lutz B. Paracrine activation of hepatic CB-1 receptors by stellate cell-derived endocannabinoids mediates alcoholic fatty liver. Cell Metab. 2008;7:227–235. doi: 10.1016/j.cmet.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 35.Kim D-K, Kim Y-H, Jang H-H, Park J, Kim JR, Koh M, Jeong W-I, Koo S-H, Park T-S, Yun C-H. Estrogen-related receptorγ controls hepatic CB1 receptor-mediated CYP2E1 expression and oxidative liver injury by alcohol. Gut. 2013;62:1044–1054. doi: 10.1136/gutjnl-2012-303347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryu D, Oh K-J, Jo H-Y, Hedrick S, Kim Y-N, Hwang Y-J, Park T-S, Han J-S, Choi CS, Montminy M. TORC2 regulates hepatic insulin signaling via a mammalian phosphatidic acid phosphatase, LIPIN1. Cell Metab. 2009;9:240–251. doi: 10.1016/j.cmet.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 37.Chanda D, Kim Y-H, Li T, Misra J, Kim D-K, Kim JR, Kwon J, Jeong W-I, Ahn S-H, Park T-S. Hepatic cannabinoid receptor type 1 mediates alcohol-induced regulation of bile acid enzyme genes expression via CREBH. PLoS One. 2013;8:e68845. doi: 10.1371/journal.pone.0068845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li T, Kong X, Owsley E, Ellis E, Strom S, Chiang JY. Insulin regulation of cholesterol 7α-hydroxylase expression in human hepatocytes roles of forkhead box o1 and sterol regulatory element-binding protein 1c. J. Biol. Chem. 2006;281:28745–28754. doi: 10.1074/jbc.M605815200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crestani M, Sadeghpour A, Stroup D, Galli G, Chiang JY. Transcriptional activation of the cholesterol 7α-hydroxylase gene (CYP7A) by nuclear hormone receptors. J. Lipid Res. 1998;39:2192–2200. [PubMed] [Google Scholar]
- 40.Xie Y-B, Park J-H, Kim D-K, Hwang JH, Oh S, Park SB, Shong M, Lee I-K, Choi H-S. Transcriptional corepressor SMILE recruits SIRT1 to inhibit nuclear receptor estrogen receptor-related receptor γ transactivation. J. Biol. Chem. 2009;284:28762–28774. doi: 10.1074/jbc.M109.034165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chanda D, Li T, Song K-H, Kim Y-H, Sim J, Lee CH, Chiang JY, Choi H-S. Hepatocyte growth factor family negatively regulates hepatic gluconeogenesis via induction of orphan nuclear receptor small heterodimer partner in primary hepatocytes. J. Biol. Chem. 2009;284:28510–28521. doi: 10.1074/jbc.M109.022244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chanda D, Kim Y-H, Kim D-K, Lee M-W, Lee S-Y, Park T-S, Koo S-H, Lee C-H, Choi H-S. Activation of cannabinoid receptor type 1 (Cb1r) disrupts hepatic insulin receptor signaling via cyclic AMP-response element-binding protein H (Crebh)-mediated induction of Lipin1 gene. J. Biol. Chem. 2012;287:38041–38049. doi: 10.1074/jbc.M112.377978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim YD, Park K-G, Lee Y-S, Park Y-Y, Kim D-K, Nedumaran B, Jang WG, Cho W-J, Ha J, Lee I-K. Metformin inhibits hepatic gluconeogenesis through AMP-activated protein kinase–dependent regulation of the orphan nuclear receptor SHP. Diabetes. 2008;57:306–314. doi: 10.2337/db07-0381. [DOI] [PubMed] [Google Scholar]
- 44.Lee J-M, Gang G-T, Kim D-K, Kim YD, Koo S-H, Lee C-H, Choi H-S. Ursodeoxycholic acid inhibits liver X receptor α-mediated hepatic lipogenesis via induction of the nuclear corepressor SMILE. J. Biol. Chem. 2014;289:1079–1091. doi: 10.1074/jbc.M113.491522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li T, Chanda D, Zhang Y, Choi H-S, Chiang JY. Glucose stimulates cholesterol 7α-hydroxylase gene transcription in human hepatocytes. J. Lipid Res. 2010;51:832–842. doi: 10.1194/jlr.M002782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie Y, Lee O, Nedumaran B, Seong H, Lee K, Ha H, Lee I, Yun Y, Choi H. SMILE, a new orphan nuclear receptor SHP-interacting protein, regulates SHP-repressed estrogen receptor transactivation. Biochem. J. 2008;416:463–473. doi: 10.1042/BJ20080782. [DOI] [PubMed] [Google Scholar]
- 47.Russell DW, Setchell KD. Bile acid biosynthesis. Biochemistry. 1992;31:4737–4749. doi: 10.1021/bi00135a001. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y-D, Chen W-D, Moore DD, Huang W. FXR: a metabolic regulator and cell protector. Cell Res. 2008;18:1087–1095. doi: 10.1038/cr.2008.289. [DOI] [PubMed] [Google Scholar]
- 49.Tam J, Liu J, Mukhopadhyay B, Cinar R, Godlewski G, Kunos G. Endocannabinoids in liver disease. Hepatology. 2011;53:346–355. doi: 10.1002/hep.24077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyake JH, Wang S-L, Davis RA. Bile acid induction of cytokine expression by macrophages correlates with repression of hepatic cholesterol 7α-hydroxylase. J. Biol. Chem. 2000;275:21805–21808. doi: 10.1074/jbc.C000275200. [DOI] [PubMed] [Google Scholar]
- 51.Fang Y, Han SI, Mitchell C, Gupta S, Studer E, Grant S, Hylemon PB, Dent P. Bile acids induce mitochondrial ROS, which promote activation of receptor tyrosine kinases and signaling pathways in rat hepatocytes. Hepatology. 2004;40:961–971. doi: 10.1002/hep.20385. [DOI] [PubMed] [Google Scholar]
- 52.Chiang J, Miller WF, Lin G-M. Regulation of cholesterol 7 alpha-hydroxylase in the liver. Purification of cholesterol 7 alpha-hydroxylase and the immunochemical evidence for the induction of cholesterol 7 alpha-hydroxylase by cholestyramine and circadian rhythm. J. Biol. Chem. 1990;265:3889–3897. [PubMed] [Google Scholar]
- 53.Dueland S, Drisko J, Graf L, Machleder D, Lusis A, Davis R. Effect of dietary cholesterol and taurocholate on cholesterol 7 alpha-hydroxylase and hepatic LDL receptors in inbred mice. J. Lipid Res. 1993;34:923–931. [PubMed] [Google Scholar]
- 54.Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, Evans RM, Blanchette M, Giguere V. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRα and γ. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 55.Sanyal S, Kim J-Y, Kim H-J, Takeda J, Lee Y-K, Moore DD, Choi H-S. Differential regulation of the orphan nuclear receptorsmall heterodimer partner (SHP) gene promoter by orphan nuclear receptor ERR isoforms. J. Biol. Chem. 2002;277:1739–1748. doi: 10.1074/jbc.M106140200. [DOI] [PubMed] [Google Scholar]
- 56.Hentschke M, Süsens U, Borgmeyer U. PGC-1 and PERC, coactivators of the estrogen receptor-related receptor γ. Biochem. Biophys. Res. Commun. 2002;299:872–879. doi: 10.1016/s0006-291x(02)02753-5. [DOI] [PubMed] [Google Scholar]
- 57.Hentschke M, Borgmeyer U. Identification of PNRC2 and TLE1 as activation function-1 cofactors of the orphan nuclear receptor ERRγ. Biochem. Biophys. Res. Commun. 2003;312:975–982. doi: 10.1016/j.bbrc.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 58.Julie M, Hall J, McDonnell D. Coregulators in nuclear estrogen receptor action: from concept to therapeutic targeting. [J]. Mol. Interv. 2005;5:343–357. doi: 10.1124/mi.5.6.7. [DOI] [PubMed] [Google Scholar]
- 59.Aranha MM, Cortez-Pinto H, Costa A, da Silva IBM, Camilo ME, de Moura MC, Rodrigues CM. Bile acid levels are increased in the liver of patients with steatohepatitis. Eur. J. Gastroen. Hepat. 2008;20:519–525. doi: 10.1097/MEG.0b013e3282f4710a. [DOI] [PubMed] [Google Scholar]
- 60.Liu T, Owsley E, Matozel M, Hsu P, Chiang JY. Transgenic expression of CYP7A1 in the liver prevents high fat diet-induced obesity and insulin resistance in mice. FASEB J. 2010;24:570.574. doi: 10.1002/hep.23721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie W, Radominska-Pandya A, Shi Y, Simon CM, Nelson MC, Ong ES, Waxman DJ, Evans RM. An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc. Natl. Acad. Sci. 2001;98:3375–3380. doi: 10.1073/pnas.051014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Staudinger J, Liu Y, Madan A, Habeebu S, Klaassen CD. Coordinate regulation of xenobiotic and bile acid homeostasis by pregnane X receptor. Drug Metab. Dispos. 2001;29:1467–1472. [PubMed] [Google Scholar]
- 63.Schuetz EG, Strom S, Yasuda K, Lecureur V, Assem M, Brimer C, Lamba J, Kim RB, Ramachandran V, Komoroski BJ. Disrupted bile acid homeostasis reveals an unexpected interaction among nuclear hormone receptors, transporters, and cytochrome P450. J. Biol. Chem. 2001;276:39411–39418. doi: 10.1074/jbc.M106340200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.