Abstract
We previously demonstrated that an αvβ5 integrin/FAK- mediated pathway regulated the phagocytic properties of human trabecular meshwork (HTM) cells. Here we demonstrate that this process is mediated by Rac-1 and a previously unreported signaling pathway that utilizes the Tiam1 as well as a novel ILK/RhoG/ELMO2 signaling pathway. Phagocytosis in both a TM-1 cell line and normal HTM cells was mediated by Rac1 and could be significantly decreased by >75% using the Rac1 inhibitor EHop-016. Knockdown of Rac1 in TM-1 cells also inhibited phagocytosis by 40% whereas overexpression of a constitutively active Rac1 or stimulation with PDGF increased phagocytosis by 83% and 32% respectively. Tiam1 was involved in regulating phagocytosis. Knockdown of Tiam1 inhibited phagocytosis by 72% while overexpression of Tiam1 C1199 increased phagocytosis by 75%. Other upstream effectors of Rac1 found to be involved included ELMO2, RhoG, and ILK. Knockdowns of ELMO2, ILK, and RhoG caused a reduction in phagocytosis by 51%, 55% and 46% respectively. In contrast, knockdown of Vav2 and Dock1 or overexpression of Vav2 Y159/172F did not cause a significant change in phagocytosis. These data suggest a novel link between Tiam1 and RhoG/ILK/ELMO2 pathway as upstream effectors of the Rac1-mediated phagocytic process in TM cells.
Keywords: phagocytosis, trabecular meshwork, glaucoma, Rac1, RhoG, Dock180, Vav2, Tiam1, ILK, ELMO2
INTRODUCTION
Glaucoma is a heterogenous eye disease that leads to irreversible blindness. The pathogenesis of glaucoma is multifactorial but it is frequently associated with an elevated intraocular pressure (IOP). This pressure increase occurs when aqueous humor outflow through the trabecular meshwork (TM)-Schlemm’s canal system is restricted. The phagocytic properties of TM cells are thought to play an important role in maintaining the outflow pathway by clearing cellular debris and degraded extracellular matrix proteins that can restrict the outflow pathway (Buller et al., 1990). In both POAG and steroid-induced glaucoma, phagocytosis is believed to be impaired thus possibly contributing to the reduction in outflow facility.
Recent studies show that phagocytosis in TM cells utilizes an αvβ5 integrin/FAK signaling pathway and that this process is inhibited when the αvβ3 integrin is activated (Gagen et al., 2013), thereby establishing a functional role for integrins in maintaining TM homeostasis. The downstream modulators of αvβ5 integrin-mediated phagocytosis in the TM are still unknown. It is also unknown how αvβ3 integrin signaling inhibits this process.
To date, a number of other integrins besides αvβ5 integrin have been shown to participate in phagocytosis. These integrins include αvβ3 integrin in microglial cells, α2β1 integrin in fibroblasts and αMβ2 in macrophages (Dupuy and Caron, 2008). These integrins have been associated with a wide range of phagocytic processes including the uptake of apoptotic cells, pathogens and matrix proteins such as collagen fibrils. They usually work with co-receptors such as Mer and BAI-1 to bind and engulf the cell or matrix protein. However, in some instances, an opsonin such as MGF-E8 bound to the integrin is used for the efficient removal of apoptotic cells (Akakura et al., 2004). Together, these integrin–mediated complexes trigger the signaling pathway(s) that coordinate the reorganization of the actin cytoskeleton and the extension of the membrane into a phagocytic cup.
Several different signal transduction pathways have been linked to integrin-mediated phagocytosis and the precise pathway(s) may be cell-type specific or depend on what is being engulfed. One common thread is that all these pathways appear to converge on various members of the Rho GTPase family, particularly Rac1, which are needed to coordinate the reorganization of the actomyosin cytoskeleton during the formation of the phagocytic cup (Dupuy and Caron, 2008, Sayedyahossein and Dagnino, 2013). Integrins are believed to play an important role in coordinating the spatial and temporal recruitment of active Rac1 to the membrane site of phagocytosis.
Rac1 activation requires specific guanine nucleotide exchange factors (GEFs) (Goicoechea et al., 2014, Rossman et al., 2005, Schmidt and Hall, 2002) that mediate the exchange of GDP for GTP. Although there are at least 30 GEFs that can regulate Rac1 activity (Bai et al., 2015), only a few have been shown to be involved in phagocytosis. The best characterized GEF known to facilitate Rac1 activation during αvβ5 integrin-mediated phagocytosis (Akakura et al., 2004, Albert et al., 2000) is the atypical GEF DOCK1 (Dedicator of cytokinesis1). By itself DOCK1 cannot activate Rac1 and needs to form a bipartite complex with the adapter protein ELMO1 (Engulfment and cell motility) in order to activate Rac1 (Cote and Vuori, 2007, deBakker et al., 2004, Laurin and Cote, 2014). To date only ELMO1 has been shown to functionally cooperate with DOCK1 during phagocytosis (Gumienny et al., 2001). ELMO2, another member of the ELMO family, is involved in Rac1 activation but appears to bind integrin linked kinase (ILK) (Ho et al., 2009) and has never been shown to play a role in phagocytosis. In some cases, DOCK1 can activate Rac1 in phagocytosis using an alternative mechanism that involves RhoG which is another member of the Rac family of GTPases (Cote and Vuori, 2007, deBakker et al., 2004)
The Rac1/DOCK1/ELMO1 pathway, however, is not the only relevant Rac-GEF pathway involved in phagocytosis. Members of the Vav family of proteins (Vav1-3) have also been implicated to have a role in phagocytosis. Vav1 is exclusively restricted to hematopoietic cells, whereas Vav2 and Vav3 are widely expressed (Hornstein et al., 2004). Vav2 has been shown to regulate collagen phagocytosis in fibroblasts in vitro (Arora et al., 2008) and to be involved in Rac1 activation (Sauzeau et al., 2010). Interestingly, Vav2/Vav3-deficient mice display characteristics of a glaucomatous phenotype including elevated IOP and loss of inner retinal cells (Fujikawa et al., 2010). Tiam1 (T-Cell Lymphoma Invasion And Metastasis 1), which is the best characterized GEF known to activate Rac1 has, to date, not been shown to play a major role in integrin-mediated phagocytosis.
In the current study, we investigated the signaling components involved in phagocytosis by TM cells downstream of αvβ5 integrin/FAK signaling. Here we demonstrate that αvβ5 integrin/FAK-mediated phagocytosis by TM cells is regulated by the GTPases Rac1 and RhoG. Activation of this pathway utilizes ELMO2, ILK, and Tiam1. A role for Dock1 or Vav2, however, could not be established. Together these studies indicate that, although phagocytosis in TM cells uses some of the same regulatory mechanisms found in other phagocytic cells, TM cells also utilize some unique components to control phagocytosis. Finally, these studies suggest there may be a differential use of GEFs by integrins in the TM to control phagocytosis. Understanding how integrin-mediated mechanisms regulate phagocytosis in TM cells should provide insight into novel approaches and therapies to manage signaling pathways governing normal TM function.
METHODS
Materials
The monoclonal antibodies (mAb) EP1067Y (rabbit-anti-Vav2) and 0.T.127 (mouse-anti-Rac1) were purchased from Abcam (Cambridge, MA). IRDye800-conjugated secondary goat anti-rabbit and IRDye700-conjuagted secondary goat anti-mouse antibodies were purchased from Li-Cor Biosciences (Lincoln, NE). pHrodo® Red S. aureus bioparticles, Hoescht 33342 nuclear stain and CellMask Green were purchased from Invitrogen Life Technologies (Carlsbad, CA). siRNA against human Rac1, Vav2, DOCK180, ELMO2, Tiam1, and RhoG (ON-TARGETplus SMARTpool, Human ITGB5) and non-targeting siRNA (ON-TARGETplus Non-targeting siRNA#1) were purchased from Dharmacon (Lafayette, CO). Rac1 inhibitors NSC23766 and EHop-016 were purchased from EMD Millipore (Billerica, MA). The Rac1 inhibitor EHT 1864 was purchased from Tocris Bioscience (Bristol, UK). Both the Vav2 and pc.HA plasmids were provided by Dr. Joan Brugge (Moores et al., 2000) through Addgene (plasmid #14554; Cambridge, MA). The Tiam1 plasmids were gifts from Dr. John Collard (Stam et al., 1997). The Rac1 plasmids constructed by Subauste et al (Subauste et al., 2000) were provided by Dr. Patricia Keely (University of Wisconsin).
Cell Culture
The immortalized human TM-1 cell line was established as previously described (Filla et al., 2002). Cells were grown in low-glucose Dulbecco’s modified Eagle’s medium (DMEM, Sigma-Aldrich), 2 mM L-glutamine (Sigma-Aldrich), 1% amphotericin B (Mediatech, Herndon, VA), and 0.05% gentamicin (Mediatech) in the presence of 10% fetal bovine serum (FBS, Atlanta Biologicals, Flowery Branch, GA). The normal human TM (HTM) cell strain N25TM-8 was isolated from a corneal rim obtained from a 25-year old donor eye with no known history of ocular disease and characterized as previously described (Filla et al., 2004). N25TM-8 cells were cultured in low glucose Dulbecco’s modified Eagle’s medium (DMEM; Sigma, St. Louis, MO), 15% fetal bovine serum (Atlanta Biologicals, Atlanta, GA), 2 mM L-glutamine (Sigma), 1% amphoteracin B (Mediatech, Herndon, VA), 0.05% gentamicin (Mediatech) and 1 ng/mL FGF-2 (Peprotech, Rocky Hill, NJ).
siRNA and Plasmid Transfections
For the mRNA knock-down experiments, TM-1 cell lines were transfected 48 h prior to the phagocytosis assay with 100nM siRNA against human Rac1, Vav2, ELMO2, Tiam1, RhoG, ILK, or Dock1 (Dharmacon, Lafayette, CO) respectively using the Mirus siQuest transfection reagent (Mirus, Madison, WI) according to the manufacturer’s instructions. siRNA concentrations were determined empirically. Non-targeting siRNA (100 nM) was used as a negative control. qPCR was used to validate knockdown of gene expression.
In experiments where Rac1, Tiam1 or Vav2 were overexpressed, TM-1 cells were transfected with 500 ng of plasmid DNA containing an active Tiam1 fragment (C1199), an inactive Tiam1 fragment (Tiam1-PhN-CCEx), constitutively active Rac1-61L, constitutively inactive Rac1-17N, pcDNA3 (empty vector control for Tiam1 and Rac1 constructs), constitutively active Vav2 (CA-Vav2), or pC.HA (empty vector control for CA-Vav2). Mock transfections, receiving no DNA, were also used as negative controls. Transfections were done using Mirus TransIT-LT1 transfection reagent (Mirus, Madison, WI). DNA used in the transfections was obtained using a Qiagen EndoFree Plasmid Maxi Kit (Qiagen, Germany). The CA-Vav2 Y159/172F construct (Moon and Gomez, 2010) was made was using the Quik-change II XL Kit (Qiagen) according to the manufacturer’s instructions. The primers used to make the CA-Vav2 Y159F mutation were: forward 5′-GGGGAGGAC ATCTTCGACTGCGTCCCG-′3 and reverse 5′-CCC CTCCTGTAGAAGCTGACGCAGGGC-′3. The primers used to introduce the Y172F mutation were: forward 5′-GGGGACGACATCTTCGAGGACATCATC and reverse CCCCTGCTGTAGAAG CTCCTGTAGTAG-′3.
qPCR
The RNeasy ® Mini Kit (Qiagen, Valencia, CA) was used to isolate total RNA from transfected or untransfected cells. cDNA was generated using High Capacity cDNA Reverse Transcription Kit as per manufacturers’ instructions (Invitrogen). Quantitative RT-PCR (qRT-PCR, 7300 Real Time PCR System, Applied Biosciences) was used to evaluate mRNA levels using specific primers for Rac1, Vav2, Dock1, RhoG, Tiam1, Tiam2, and ELMO1, 2, and 3 made at the UW Biotechnology Center, Madison, WI. Results were normalized to the housekeeping gene succinate dehydrogenase complex subunit (SDHA). The primers used for the qPCR reactions are listed in Table 1 in the Supplemental Data.
Phagocytosis of S. aureus bioparticles
TM-1 or HTM cells were grown to confluence in 6-well tissue culture-treated plates or on sterile glass cover slips (Bellco, Vineland, NJ). Cultures were then challenged with pHrodo® Red S. aureus bioparticle conjugates (pHrodo bioparticles; Invitrogen) for 4 h as previously described (Gagen et al., 2013). In some experiments, cells were pre-incubated for 30 min with a Rac1 inhibitor (NSC23766, EHop-016 or EHT 1864) followed by the 4 h incubation with pHrodo bioparticles in the presence of the inhibitor. Following the phagocytic challenge, control, Rac1 inhibitor-treated and siRNA-transfected TM-1 cells were prepared for FACS or fixed in 1.0% paraformaldehyde, in phosphate buffered saline (PBS) for 20 min at room temperature and stained with Hoescht 33342 for fluorescence microscopy. An epifluorescence microscope (Axioplan 2, Zeiss, Thornwood, NY) and image acquisition software (Axiovision 4.8) was used to image pHrodo™ bioparticles fluorescence and cell nuclei.
For all other experiments, cells were seeded onto 4-well chambered glass coverslips (ibidi USA, Madison, WI) at a density of 1 × 105 cells/ml for 24 h prior to being transfected. Forty-eight hours post-transfection, cells were placed in media containing 1% FBS for an additional 24 h prior and then exposed to the pHrodo bioparticles for 4 h. The level of phagocytosis was determined using a fluorescence microscopy assay. It was necessary to use this assay for these studies, because the transfection reagent needed for the knockdowns impaired the ability to process the cells for FACS. The phagocytic activity was calculated by determining the ratio of cells containing pHrodo bioparticles versus total number of cells from 3–4 separate randomly chosen areas per coverslip. Cells were considered positive if they had engulfed ≥3 bioparticles and designated as being a bioparticle-positive cell (BPC). Each experiment was independently repeated at least 3 times and between 200–740 cells per coverslip were counted. Cells were stained with CellMask Green (1/1000 dilution) to identify the cells. All cells were viewed unfixed using an inverted fluorescence microscope (Axiovert 200m, Zeiss, Thornwood, NY) and image acquisition software (Axiovision 4.8) to image pHrodo™ bioparticles fluorescence and stained cells.
Fluorescence-activated cell-sorting (FACS) analysis
FACS analyses were performed as previously described (Gagen et al., 2013). Cells were lifted using either a 0.05% Trypsin/0.25mM EDTA solution or a Cell Dissociation Solution (Sigma-Aldrich, St Louis, MO) and then washed in complete medium containing serum followed by warm PBS. Cells were then resuspended in 1% paraformaldehyde in buffered saline and the fluorescence intensity of the pHrodo bioparticles was analyzed using the BD FACSCalibur System (BD Biosciences). Quantification of phagocytosis was done by comparing the geometric mean fluorescence intensity (MFI) of phagocytosed pHrodo bioparticles between the different treatment groups and their respective controls, as previously described (Gagen et al., 2013). The ratio between the treated MFI and untreated MFI × 100% was used to calculate % phagocytic activity relative to control.
Immunoblotting
TM-1 cells were lysed with RIPA lysis buffer (25 mM Hepes, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM NaF, 1% NP-40, 0.25% deoxycholate) containing HALT phosphatase inhibitor cocktail and HALT protease inhibitor cocktail (Thermo Scientific, Rockford, IL). Cells were incubated for 10 min at 4 °C and debris was removed by centrifugation at 10,000 × g. Protein concentration was determined using a BCA assay (Thermo Scientific). The cell lysate (10 μg) was loaded onto a 10% SDS-PAGE gel and transferred to Immobilin-FL for imaging on an Odyssey CLx infrared reader (Li-Cor) using Image Studio ver 5.0 imaging software (Li-Cor). Membranes were blocked in 3% BSA/TBS for 1 h and then incubated with a primary antibody diluted in 1% BSA/TBS/0.1% TX-100 for 1 h. Membranes were washed with TBS/0.1% TX-100 and incubated for 1 h with anti-mouse or anti-rabbit IRDye® 800CW-conjugated secondary Ab for infrared imaging.
Statistical analysis
Statistical analyses were performed using a Student’s t-test or one-way ANOVA followed by a Tukey HSD post-test. A p value < 0.05 was considered significant. FACs data are reported as mean percentage of fluorescence intensity (%MFI) ± SEM.
RESULTS
The Rac1 inhibitor EHop-016 selectively impairs phagocytosis in TM-1 cells
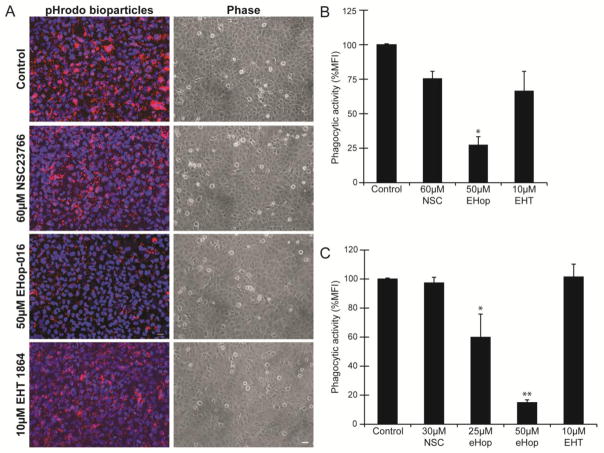
To determine if TM cells utilized a Rac1 signaling pathway to regulate phagocytosis, TM-1 cells were pretreated with the Rac1 inhibitors NSC23766, EHT 1864, and EHop-016 prior to being challenged with the pHrodo bioparticles. NSC23766 and EHT 1864 both prevent Rac1 activation by the Rac1-specific GEFs Trio and Tiam1 (Gao et al., 2004, Shutes et al., 2007), while EHop-016 prevents Rac1 activation via the GEFs Vav1 and, to a lesser extent, Tiam1 (Montalvo-Ortiz et al., 2012). By phase microscopy, none of the inhibitors had any effect on cell morphology at the concentrations used compared to vehicle-treated control TM-1 cells (Fig 1A). However, the phagocytic activity of the cells differed depending on which inhibitor was used. As shown in Figure 1A, TM-1 cells treated with 50 μM EHop-016 demonstrated a clear reduction in the uptake of pHrodo bioparticles compared to control cells or cells treated with 60 μM NSC23766 or 10 μM EHT 1864. In contrast, NSC2366 and EHT 1864 caused only a slight decrease in the uptake of pHrodo bioparticles compared to the vehicle-treated control.
Figure 1. The Rac1 inhibitor EHop-016 impairs phagocytosis in TM-1 cells.
(A) Following a 4 h challenge with pHrodo bioparticles, TM-1 cells treated with 50μM EHop-016 demonstrated a visible reduction in the uptake of the bioparticles compared to vehicle-treated control TM-1 cells or cells treated with the other inhibitors. By phase microscopy the treatment of TM-1 cells with the various inhibitors did not appear to alter the morphology of cells compared to control TM-1 cells. Scale bar = 20μm. Images are representative of results from 3 independent experiments. (B) Quantification of the uptake of the bioparticles by FACS showed that phagocytosis was significantly reduced (*) by 75% (p<0.05) in cells treated with EHop-016 compared to untreated cells. In contrast, uptake of the bioparticles was not significantly reduced compared to the control cells following treatment with the other inhibitors. Phagocytic activity was determined by measuring the mean fluorescence intensity (%MFI) ± SEM of pHrodo particles in 4 independent experiments. (C) HTM cells treated with 25 μM EHop-16 showed a 49% (*p<0.05) reduction in phagocytosis compared to untreated and cells treated with 50 μM EHop-16 showed a significant reduction (**) of 81% (p<0.01) compared to untreated in 3 independent experiments done in triplicates. The other inhibitors, NSC23766 and EHT 1864, did not show a significant reduction in phagocytosis compared to the untreated cells. The lower concentration (30μM) of NSC23766 was used in the study with the normal HTM cells, because the higher concentration (60μM) proved toxic (data not shown).
Quantification of the phagocytic activity using FACS verified the observations from the fluorescence microscopy study. As shown in Figure 1B, the percent mean fluorescent intensity (% MFI) in TM-1 cells treated with the EHop-016 inhibitor was significantly reduced by 75% compared to control cells (p<0.05). In contrast, the % MFI of NSC23766 inhibitor-treated or EHT 1864 inhibitor-treated cells was not statistically different from untreated cells. A similar result was observed in a normal HTM cell strain (Fig 1C). Neither the NSC2366 nor the EHT 1864 inhibitor reduced the phagocytic activity of HTM cells, while the EHop-016 inhibitor showed a dose dependent reduction in the phagocytic activity of the HTM cells compared to the no treatment control cells. At 25 μM, EHop-16 showed a 49% (*p<0.05) reduction in phagocytosis compared to untreated and cells treated with 50 μM EHop-16 showed a significant reduction (**) of 81% (p<0.01). Thus, phagocytosis in both normal and transformed TM cells appears to utilize a Rac1-dependent pathway.
Knockdown of Rac1 inhibits phagocytosis in TM-1 cells
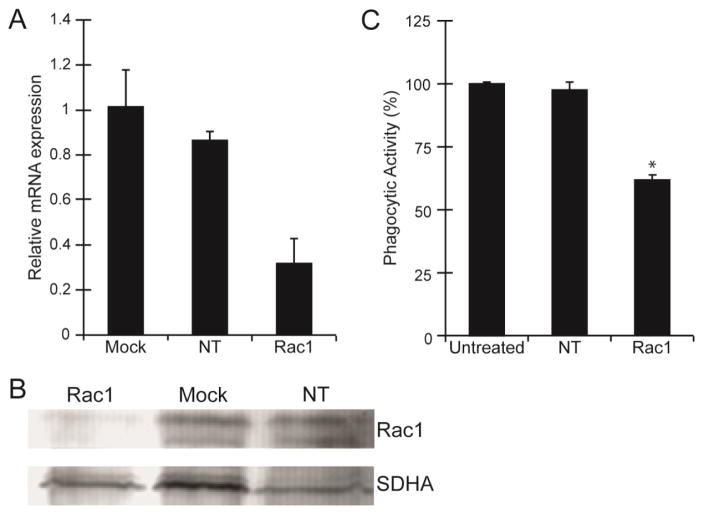
To further demonstrate that phagocytosis was a Rac1–mediated process, siRNA was used to knock down Rac1 expression in the immortalized TM-1 cell line. By qPCR analysis 100 nM Rac1 siRNA knocked down Rac1 gene expression by ~68% compared to mock transfected cells and 65% compared to cells transfected with 100nM non-targeting (NT) siRNA (Fig 2A). Immunoblot analysis confirmed that protein levels of Rac1 were reduced following transfection with Rac1 siRNA (Fig 2B).
Figure 2. Phagocytosis was inhibited in TM-1 cells transfected with Rac1 siRNA.
(A) Level of Rac1 mRNA expression in TM-1 cells transfected with Rac1 siRNA was reduced compared to mock transfected cells or cells transfected with a NT siRNA. At least 3 independent experiments were done in triplicates. Data was normalized to SDHA and expressed relative to the untransfected control. (B) Western blot analysis of total Rac1 following siRNA transfections showed that Rac1 protein levels were also reduced compared to mock transfected cells and cells transfected with a NT siRNA. (C) Rac1 siRNA-transfected cells showed a statistically significant (*) reduction in phagocytic activity by ~40% compared to cells transfected with NT siRNA or untreated control cells. Phagocytic activity of TM-1 cells was assessed using FACS analysis following the phagocytic challenge with pHrodo bioparticles and expressed relative to mock transfected control. Each experiment was done using triplicate determinations. All results (A–C) were verified in at least three independent experiments.
When Rac1 siRNA transfected TM-1 cells were challenged with pHrodo bioparticles to determine their level of phagocytic activity, FACS analysis showed that the phagocytic activity was reduced by 40% (p<0.05) compared to mock transfected cells or NT siRNA-transfected cells (Fig 2C). In contrast, there was no difference in the phagocytic activity in NT siRNA-transfected cells compared to untreated controls.
PDGF and expression of a constitutively active form of Rac1 (Rac-61L) stimulates phagocytosis
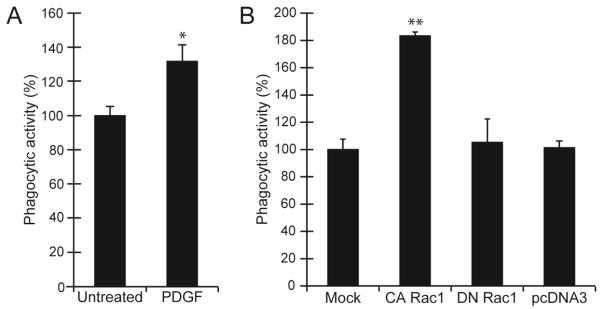
Finally, to confirm that Rac1 is necessary for the efficient engulfment of the pHrodo bioparticles, TM-1 cells were treated with the growth factor PDGF which is known to activate Rac1 or transfected with a constitutively active (CA) form of Rac1 (Rac-61L). As shown in Figure 3A, addition of 60 ng/ml PDGF to the media during the phagocytic assay caused a statistically significant (p<0.05) increase in the levels of phagocytosis by 32% compared to untreated TM-1 cells. Overexpression of Rac-61L in TM-1 cells also caused a significant increase in phagocytosis by 83% (p< 0.01) compared to mock transfected cells, cells transfected with a dominant negative (DN) Rac1-17N or cells transfected with the empty vector (Figure 3B). Collectively, these data suggest that phagocytosis in TM cells is mediated by Rac1.
Figure 3. Phagocytosis was enhanced in cells treated with PDGF or transfected with Rac1-61L.
(A) TM-1 cells treated with 60ng/ml PDGF during the duration of the phagocytic assay showed a statistically significant (* p<0.05) increase in the number of cells that took up the pHrodo bioparticles compared to untreated control cells. (B) Overexpression of a CA-Rac1-61L also caused a significant increase (**p<0.01) in the uptake of pHrodo bioparticles compared to mock transfected cells, cells transfected with DN-Rac1-N17, or cells transfected with an empty pcDNA3 vector. Phagocytic activity of TM-1 cells was measured after 4 hr challenge with the pHrodo bioparticles in the presence of 60ng/ml PDGF and assessed by counting the number of cells containing ≥3 pHrodo bioparticles. The phagocytic activity is expressed as the percentage of bioparticle positive cells (BPC) relative to mock transfected control. Histograms are representative of the results obtained in three independent experiments as described in materials and methods.
Tiam1 and not Vav2 plays a role in phagocytosis
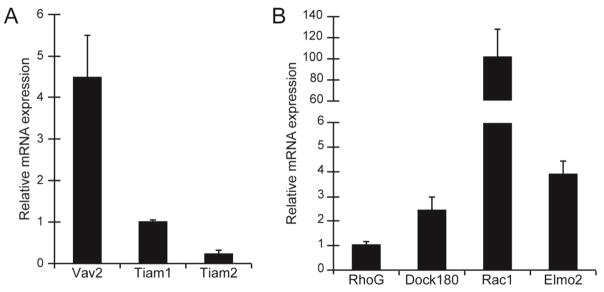
These data not only indicated that phagocytosis is a Rac1-mediated process, but they suggested that a specific GEF may be involved in αvβ5 integrin-mediated phagocytosis by TM cells since only EHop-016 showed a significant knockdown of phagocytosis. Since the EHop-016 inhibitor was developed to block the association of Vav1 with Rac1 and, to a lesser extent, the activation of Rac1 by Tiam1, we wanted to determine if either of these GEFs was involved in regulating phagocytosis in TM-1 cells. Of the three members in the Vav family, qPCR showed that TM-1 cells (Fig 4A) and HTM cells both expressed Vav2 mRNA (data not shown). Vav3 mRNA was not detected in TM-1 cells and HTM cells only expressed trace levels of Vav3 mRNA (data not shown). Vav1 mRNA was also not expressed by the immortalized TM-1 cell line which was to be expected since Vav1 expression is restricted to hematopoietic cells (Hornstein et al., 2004). Both TM-1 (Fig 4A) and HTM cells (data not shown) were also found to express Tiam1 and, to a lesser extent, Tiam2 mRNA. In addition, to Vav2 and Tiam1 and 2, TM-1 cells were found to express Dock1, RhoG, and ELMO2 mRNA (Fig 4B). All of these proteins with the exception of ELMO2 have been implicated in playing a role in phagocytosis.
Figure 4. Expression of factors involved in Rac1 activation in TM-1 cells.
(A) RT-PCR analysis showed that TM-1 cells expressed mRNA for the GEFs Vav2, Tiam1 and Tiam2. RNA was normalized to SDHA and expressed relative to Tiam1 mRNA levels. (B) RT-PCR showed that RhoG, Dock 180 and ELMO2 mRNA could be detected in TM-1 cells. Neither ELMO1 nor ELMO3 mRNA could be detected (not shown). Level of Rac1 mRNA was used as a positive control. RNA was normalized to SDHA and expressed relative to RhoG mRNA levels. All experiments were done using triplicate determinations and the histograms shown are representative of three independent experiments.
To determine if Vav2 was involved in αvβ5 integrin mediated phagocytosis, TM-1 cells were transfected with human Vav2 siRNA to knockdown Vav2 expression. TM-1 cells were used for the remainder of the experiments instead of normal HTM cells because HTM express both αvβ3 and αvβ5 integrins whereas TM-1 cells express only the αvβ5 integrin and not the αvβ3 integrin (Filla et al., 2011). Thus by using TM-1 cells we could study αvβ5 integrin mediated phagocytosis in the absence of αvβ3 integrins which had previously been shown to affect phagocytosis in TM cells (Gagen et al., 2013).
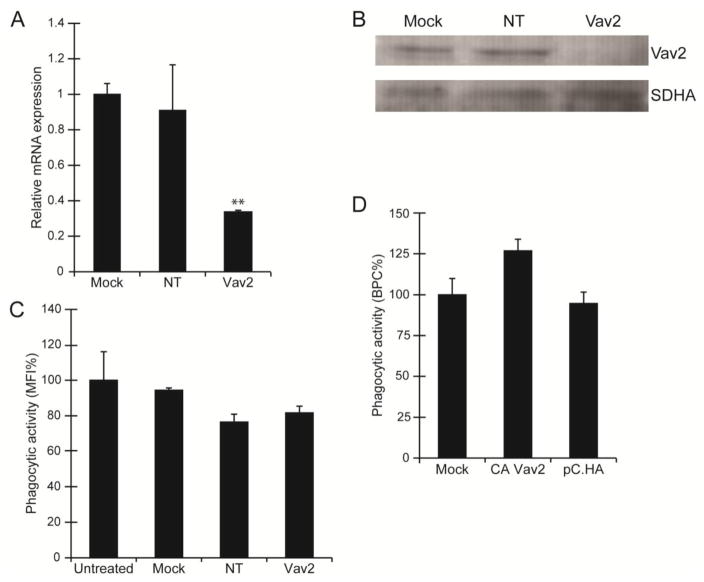
Forty-eight hours post-transfection, qPCR showed that Vav2 mRNA was reduced in cells treated with 100 nM Vav2 siRNA compared to mock transfected cells or cells treated with 100 nM NT siRNA (Fig 5A). Furthermore, protein levels of Vav2 were also reduced in cells transfected with Vav2 siRNA (Fig 5B). Yet, despite the knockdown of both Vav2 mRNA and protein levels, FACs analysis failed to show a statistically significant decrease in phagocytosis compared to the untreated controls (Fig 5C). To help determine if Vav2 was involved, we then overexpressed a constitutively active form of Vav2 (CA-Vav2 Y159/172F) in TM-1 cells. As shown in Figure 5D, overexpression of the CA-Vav2 Y159/172F also failed to show a statistically significant increase in phagocytosis. This data suggested that Vav2 may not be the primary GEF involved in the activation of Rac1. Given that EHop-016 can also block Rac1 activation by Tiam1, we sought to determine if this GEF might be involved in phagocytosis.
Figure 5. Phagocytosis was not significantly changed by Vav2 siRNA or overexpression of CA-Vav2Y159/172F.
(A) Levels of Vav2 mRNA expression was reduced in TM-1 cells transfected with Vav2 siRNA compared to mock transfected cells or cells transfected with a NT siRNA. RNA was normalized to SDHA and expressed relative to Vav2 mRNA level in mock treated cells. All experiments were done using triplicate determinations and the data shown here are representative of 3 independent experiments. (B) Western blot analysis of total Vav2 following siRNA transfections showed that protein levels for Vav2 were reduced in cells transfected with the Vav2 siRNA compared to mock transfected cells and cells transfected with a NT siRNA. SDHA was used as the loading control. The western blot shown is representative of 3 independent experiments. (C) Quantification of the phagocytic activity of Vav2 siRNA transfected TM-1 cells was assessed using FACS analysis following the 4 hr phagocytic challenge with pHrodo bioparticles and expressed as %MFI relative to untransfected controls. Despite the knockdown of Vav2 mRNA and protein levels, no effect on phagocytosis was measured. The histogram is representative of the results obtained in three independent experiments as described in materials and methods. (D) Overexpression of a CA-Vav2Y159/172F also failed to induce a statistically significant increase in phagocytosis compared to mock transfected control cells or cells transfected with the empty vector (pC.HA). Data analysis is based on two independent experiments done as described in materials and methods and expressed as %BPC relative to mock control.
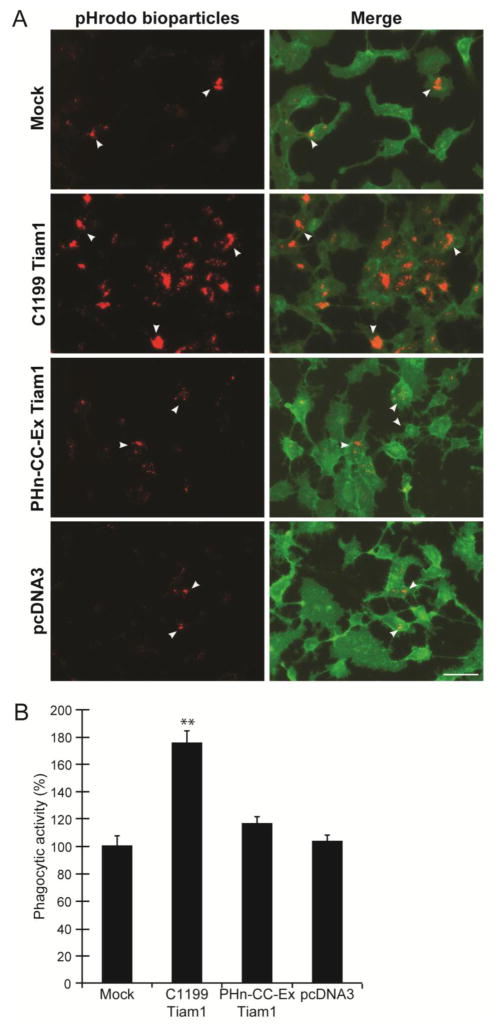
To determine if Tiam1 was involved in regulating Rac1-mediated phagocytosis, a truncated, but active, fragment (C1199) of Tiam1 was expressed in TM-1 cells (Stam et al., 1997). A smaller inactive Tiam1 fragment (Phn-cc-Ex) that lacked the Rac1 activation domain (Baumeister et al., 2003) and an empty vector were used as negative controls. By fluorescent microscopy, TM-1 cells transfected with C1199 Tiam1 appeared to engulf a larger number of pHrodo bioparticles compared to the mock transfected controls (Fig 6A). Quantification of the images verified this observation and showed that overexpression of Tiam1 C1199 resulted in a statistically significant increase by 75% in phagocytosis (p<0.01) compared to mock transfected cells (Fig 6B). In contrast, overexpression of Phn-cc-Ex Tiam1 or the empty vector pcDNA3 had no effect (Fig 6B). Furthermore, siRNA knockdown of Tiam1 resulted in a 72% decrease (p<0.01) in phagocytosis (Fig 7) compared to NT siRNA controls. Together these data suggest that Tiam1 appears to be the GEF primarily responsible for the activation of Rac1 during phagocytosis.
Figure 6. Overexpression of Tiam1 C1199 increased phagocytosis.
(A) Fluorescence micrographs showed a significant increase in the uptake of pHrodo bioparticles (red) in cells transfected with Tiam1 C1199 compared to mock transfected cells, cells transfected with the Tiam1 pHn-CC-Ex construct or the empty pcDNA3 vector. Cells were counterstained with CellMask Green to visualize the cells. Scale bar = 50 μm. (B) Quantification of the number of cells taking up pHrodo™ bioparticles showed that expression of the active Tiam1 construct C1199 caused a significant increase (**, p<0.01) in the number of cells taking up the pHrodo bioparticles compared to mock transfected control cells. Phagocytosis was quantified by counting the number of cells that had taken up ≥3 pHrodo bioparticles and then expressed relative to level of phagocytosis in mock transfected cells as %BPC ±SEM. All results are representative of the data from three independent experiments.
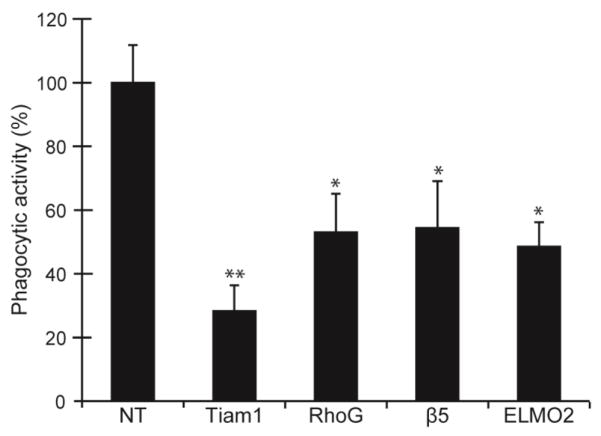
Figure 7. siRNA knockdown of Tiam1, RhoG, and ELMO2 inhibit phagocytosis.
Level of phagocytosis was determined in cells transfected with siRNA to Tiam1, RhoG, ELMO2, or β5 integrin subunit. A NT siRNA was used as a negative control. Cells transfected with siRNA to Tiam1, RhoG, β5 integrin subunit, or ELMO2 showed a statistically significant reduction in the phagocytic properties of the cells compared to cells transfected with NT siRNA. The histogram shown is representative of the results obtained from three independent experiments. All experiments were done as described for Figure 6 using triplicate determinations. ** p<0.01. *p<0.05.
TM-1 cells utilize RhoG and a novel ELMO2 and ILK protein complex during phagocytosis
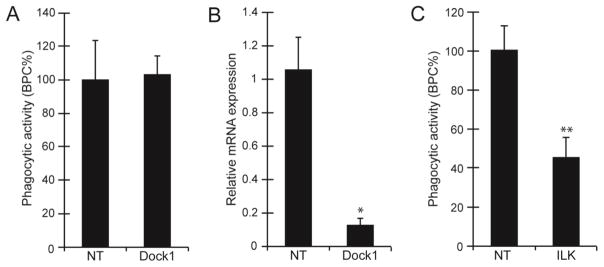
Finally, we examined whether the unconventional ELMO/Dock1 complex which has been implicated in αvβ5 integrin mediated phagocytosis in other phagocytic cells (Akakura et al., 2004) was involved in the activation of Rac1 during phagocytosis in TM-1 cells. As shown earlier in Figure 4B, TM-1 cells express low levels of DOCK1 mRNA and, surprisingly, only ELMO2 mRNA. Neither ELMO1 nor ELMO3 mRNA could be detected in TM-1 cells. TM-1 cells were also found to contain mRNA for another member of the Rac family known as RhoG which has recently been identified as a binding partner for the ELMO1/DOCK1 complex (deBakker et al., 2004, Kim et al., 2011) and can participate in regulating phagocytosis (Tzircotis et al., 2011). To determine if any of these proteins could be involved in phagocytosis, the expression of these proteins was knocked down using siRNAs to DOCK1, ELMO2, or RhoG, respectively. Knockdown of the β5 integrin subunit was used as a positive control. As shown in Figure 7, knockdown of both ELMO2 and RhoG produced a statistically significant reduction in phagocytosis by 51% (p<0.05) and 46% (p<0.05) respectively compared to NT siRNA controls suggesting that ELMO2 and RhoG may also play a role in regulating phagocytosis. In contrast, knockdown of Dock1 expression did not show a statistically significant reduction in phagocytosis compared to NT siRNA-transfected cells (Fig 8A). This was not due to an inability to knock down Dock1 mRNA, since cells transfected with DOCK1 siRNA showed a 91% reduction in DOCK1 mRNA expression compared to cells transfected with NT siRNA (Fig 8B).
Figure 8. Knock down of ILK impairs phagocytosis.
(A) Quantification of the number of cells transfected with Dock1 siRNA that took up pHrodo bioparticles. (B) The level of DOCK1 mRNA expression was significantly reduced in TM-1 cells transfected with Dock1 siRNA compared to cells transfected with a NT siRNA (*p<0.05). RNA was normalized to SDHA and expressed relative to Dock1 mRNA level in Mock transfected cells. The histogram is representative of the results obtained from three independent experiments. All experiments were done as described for Figure 6 using triplicate determinations. (C) The number of cells that took up pHrodo bioparticles was reduced in cells transfected with siRNA to ILK compared to cells transfected with non-targeting siRNA. The histograms shown (A & C) are representative of the results obtained from three independent experiments. Phagocytosis was quantified by counting the number of cells that had taken up ≥3 pHrodo bioparticles and expressed as %BPC ± SEM relative to NT control as described for Figure 6.
Since Dock1 did not appear to be involved, we looked for another binding partner for ELMO2. ILK is a pseudokinase that has recently been shown to associate with ELMO2 (Ho et al., 2009) and participate in phagocytosis (Sayedyahossein et al., 2012) and is found in HTM cells (Faralli et al., 2011). As shown in Figure 8C, knockdown of ILK with siRNA also resulted in a statistically significant decrease in phagocytosis by 55% (p<0.01) compared to NT siRNA control cells. This suggests that Rac1-mediated phagocytosis in TM-1 cells is regulated by Tiam1, RhoG and the newly discovered ELMO2/ILK pathway.
DISCUSSION
Previous studies showed that both normal and immortalized TM cells can use an αvβ5 integrin/FAK-mediated pathway to regulate phagocytosis (Gagen et al., 2013). Here, we extend those studies to demonstrate that αvβ5 integrin-mediated phagocytosis in immortalized TM-1 and HTM cells is a Rac1-mediated process that involves a number of upstream effectors known to activate Rac1 including RhoG, Tiam1, ELMO2 and ILK (Fig 9). Although it is not surprising that phagocytosis is a Rac1-mediated process, many of the upstream effectors involved in the activation of Rac1 for phagocytosis in TM cells are novel. This study demonstrated for the first time that Tiam1 as well as ELMO2 and ILK play a role in mediating phagocytosis. Another unusual finding was that DOCK1 and ELMO1 which had been shown to activate Rac1 in the αvβ5 integrin-mediated phagocytic process in phagocytic cells (Akakura et al., 2004), did not appear to play a role in the phagocytic process in TM-1 cells. Whether all these GEFs are also active in normal HTM cells remains to be determined. However, both TM-1 and HTM cells express Vav2, Tiam1, and ILK suggesting that the αvβ5 integrin-mediated phagocytic pathway may be the conserved in both cell types.
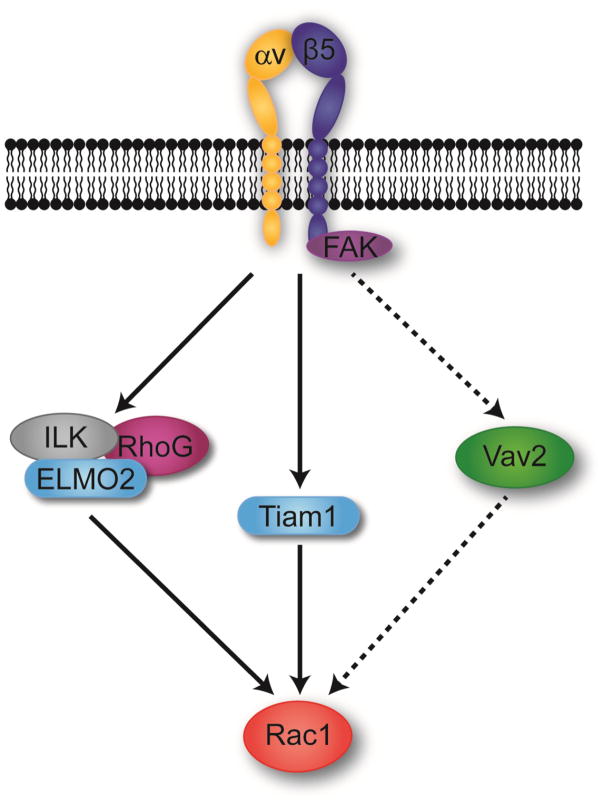
Figure 9. Schematic of the αvβ5 integrin-mediated phagocytic pathway in TM-1 cells.
The diagram illustrates that at least two major GEF mediated pathways may be used by TM-1 cells to activate Rac1 during αvβ5-mediated phagocytosis. The Tiam1 and ILK/ELMO2/RhoG pathways were drawn as separate pathways to illustrate that we do not know how these GEFs cooperate to regulate Rac1 activity. In addition, a Vav2-mediated pathway was drawn with a dotted line to indicate that its role in activating Rac1 is unclear.
The reason for a need for multiple ways to activate Rac1 is not clear. Phagocytosis is a multistep process that necessitates the remodeling of the actin cytoskeleton to drive the engulfment of the particle. Each step requires the precise spatial and temporal regulation of multiple proteins to the membrane site of phagocytosis and the remodeling of the cytoskeleton network (Freeman and Grinstein, 2014). Among the first steps involved in the remodeling of the cytoskeleton is the depolymerization of cortical actin to increase the mobility of integrins and their co-receptors to the phagocytic site. Subsequent steps involve the polymerization of a branched actin and myosin network to facilitate the formation of the phagocytic cup and its eventual closure. Rac1 activation is involved in both the depolymerization of cortical actin and the formation of a branched actomyosin network whereas RhoA is involved in closure of the phagocytic cup. Hence the use of the different upstream effectors in Rac1 activation may represent a way to control the different spatial and temporal roles of Rac1 during the phagocytic process.
Rac1-mediated phagocytosis is not an unusual finding, as many cell types utilize this signaling pathway to drive actin cytoskeleton reorganization into phagocytic cups (Dupuy and Caron, 2008, Freeman and Grinstein, 2014). What is novel is the use of both Tiam1 and ELMO2 to control phagocytosis in TM-1 cells. Although Tiam1 is known to be a Rac1 specific GEF, its activity has never been associated with phagocytosis before. Yet in TM-1 cells, Tiam1 seems to be a major player in phagocytosis as knockdown of this GEF resulted in nearly complete inhibition of phagocytosis. The most likely role for Tiam1 in phagocytosis could be the GTP loading of Rac1. However, recent studies have suggested that Tiam1 can also function as an adaptor protein that directs the localized activation of Rac1 at the plasma membrane (Liu et al., 2013). This use of Tiam1 as an adaptor protein rather than a GTP loading factor could explain why so many GEFs are involved in regulating Rac1-mediated phagocytosis.
Both RhoG (Tzircotis et al., 2011) and ILK (Sayedyahossein and Dagnino, 2013, Sayedyahossein et al., 2012) have previously been demonstrated to play a role in phagocytosis. RhoG has recently been implicated to form a complex with ELMO1 and Dock1 during phagocytosis whereas ILK has been shown to mediate phagocytosis of pathogens in epithelial cells. This raises the question of whether a RhoG/ILK/ELMO2 complex may substitute for the role that an ELMO1/DOCK1 complex normally plays in phagocytosis. ILK has been recently shown to complex with ELMO2 and since ILK and RhoG do not bind each other, ELMO2 is thought to be an obligatory bridge in a RhoG/ILK complex (Ho et al., 2009). Furthermore an ELMO2/RhoG/ILK complex has recently been shown to regulate microtubule dynamics downstream of Rac1 activation (Jackson et al., 2015). Thus, it seems plausible that an ELMO2/RhoG/ILK complex may be involved in regulating Rac1 activation in phagocytosis. Clearly additional biochemical studies are needed to verify this.
It is not exactly clear why EHop-016 inhibited phagocytosis in these initial studies and the NSC23766 and EHT 1864 inhibitors did not since all these inhibitors have been reported to inhibit Tiam1-Rac1 mediated processes (Gao et al., 2004, Montalvo-Ortiz et al., 2012, Shutes et al., 2007). The answer may lie in the specific mechanism of action for each of these inhibitors. EHT 1864, in contrast to the other inhibitors, does not block binding of Tiam1 to Rac1, but rather interferes with the process of nucleotide exchange and the binding of certain downstream effectors associated with Rac1 (Shutes et al., 2007). So in this case the downstream effectors used by Rac1 to mediate phagocytosis may not be affected by EHT 1864. EHop-016 and its parent compound NSC23766, on the other hand, both bind to the docking groove in Rac1 that is used by various GEFs including Vav1 and Tiam1. NSC23766 does not prevent Vav1 binding to Rac1, whereas EHop-016 does. Furthermore, EHop-016 is a more potent inhibitor than NSC23766 by 100-fold. Presumably the difference lies in how these two different inhibitors fit into the docking groove of Rac1. EHop-016 is able to fit deeper into the groove than NSC23766. Theoretically this would mean that higher and potentially more toxic concentrations of NSC23766 might be able to inhibit phagocytosis.
Finally, it is interesting that a previous study by Filla, et al. (Filla et al., 2009) showed that activation of the αvβ3 integrin also activated Rac1 in TM cells, but that this activation resulted in a decrease in phagocytosis (Gagen et al., 2013). At first this finding seems to be a contradiction, especially since αvβ3 integrin has been shown to regulate phagocytosis (Sayedyahossein and Dagnino, 2013). However, when αvβ3 integrin was activated in TM cells it used another GEF called TRIO to activate Rac1 which resulted in the formation of cross-linked actin networks (CLANs) (Filla et al., 2006, Filla et al., 2009) that are prevalent in glaucomatous TM (Clark et al., 2005, Hoare et al., 2009) and lamina cribosa cells (Job et al., 2010). This suggests that there is a differential use of GEFs by integrins which is responsible for mediating the various cytoskeleton functions of the TM and not just those involved in phagocytosis. If that is the case, targeting GEFS rather than the Rho GTPases may represent a viable therapeutic target to treat glaucoma without interfering with all the normal cytoskeleton functions of TM cells. The recent finding that SNPs identified in ARHGEF12 may influence the risk of developing glaucoma supports this idea (Springelkamp et al., 2015).
Supplementary Material
Highlights.
Rac1-mediated phagocytosis involves the guanine exchange factor, Tiam1
Identification of ELMO2 as a downstream effector of αvβ5 integrin-mediated phagocytosis
Both ILK and RhoG participate in the αvβ5 integrin-mediated phagocytic pathway
αvβ5 integrin-mediated Rac1 activation is dependent upon multiple guanine exchange factors
Acknowledgments
The authors would like to thank Drs. Jennifer Faralli and Mark S. Filla for their assistance in editing the text and their thoughtful comments. They would also like to thank Ralph Moeller Trane who is the biostatistician in the UW Vision Core for his assistance with the statistical analysis.
Grant Support: This work was supported by NEI grants EY06665-01, EY017006, EY020490 (D.M.P.), and a Core grant to the Department of Ophthalmology and Visual Sciences (P30 EY016665),
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akakura S, Singh S, Spataro M, Akakura R, Kim JI, Albert ML, Birge RB. The opsonin MFG-E8 is a ligand for the alphavbeta5 integrin and triggers DOCK180-dependent Rac1 activation for the phagocytosis of apoptotic cells. Exp Cell Res. 2004;292:403–416. doi: 10.1016/j.yexcr.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Albert ML, Kim JI, Birge RB. alphavbeta5 integrin recruits the CrkII-Dock180-rac1 complex for phagocytosis of apoptotic cells. Nat Cell Biol. 2000;2:899–905. doi: 10.1038/35046549. [DOI] [PubMed] [Google Scholar]
- Arora PD, Marignani PA, McCulloch CA. Collagen phagocytosis is regulated by the guanine nucleotide exchange factor Vav2. Am J Physiol Cell Physiol. 2008;295:C130–137. doi: 10.1152/ajpcell.00168.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Xiang X, Chunmei L, Shi L. Regulating Rac in the nervous system: molecular functions and disease implications of Rac GEFs and GAPs. Biomed Research International. 2015 doi: 10.1155/2015/632450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister MA, Martinu L, Rossman KL, Sondek J, Lemmon MA, Chou MM. Loss of phosphatidylinositol 3-phosphate binding by the C-terminal Tiam-1 pleckstrin homology domain prevents in vivo Rac1 activation without affecting membrane targeting. J Biol Chem. 2003;278:11457–11464. doi: 10.1074/jbc.M211901200. [DOI] [PubMed] [Google Scholar]
- Buller C, Johnson DH, Tschumper RC. Human trabecular meshwork phagocytosis. Observations in an organ culture system. Invest. Ophthalmol. Vis Sci. 1990;31:2156–2163. [PubMed] [Google Scholar]
- Clark AF, Brotchie D, Read AT, Hellberg P, English-Wright S, Pang IH, Ethier CR, Grierson I. Dexamethasone alters F-actin architecture and promotes cross-linked actin network formation in human trabecular meshwork tissue. Cell Motil Cytoskeleton. 2005;60:83–95. doi: 10.1002/cm.20049. [DOI] [PubMed] [Google Scholar]
- Cote JF, Vuori K. GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends in Cell Biology. 2007;17:383–393. doi: 10.1016/j.tcb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deBakker CD, Haney LB, Kinchen JM, Grimsley C, Lu M, Klingele D, Hsu PK, Chou BK, Cheng LC, Blangy A, Sondek J, Hengartner MO, Wu YC, Ravichandran KS. Phagocytosis of apoptotic cells is regulated by a UNC-73/TRIO-MIG-2/RhoG signaling module and armadillo repeats of CED-12/ELMO. Curr Biol. 2004;14:2208–2216. doi: 10.1016/j.cub.2004.12.029. [DOI] [PubMed] [Google Scholar]
- Dupuy AG, Caron E. Integrin-dependent phagocytosis: spreading from microadhesion to new concepts. J Cell Sci. 2008;121:1773–1783. doi: 10.1242/jcs.018036. [DOI] [PubMed] [Google Scholar]
- Faralli JA, Newman JR, Sheibani N, Dedhar S, Peters DM. Integrin linked kinase regulates integrin signaling in human trabecular meshwork cells. Invest Ophthalmol Vis Sci. 2011;52:1684–1692. doi: 10.1167/iovs.10-6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filla M, Woods A, Kaufman PL, Peters DMP. β1 and β3 integrins cooperate to induce syndecan-4 containing cross-linked actin networks (CLANs) in human trabecular meshwork (HTM) cells. Invest Ophthal Vis Sci. 2006;47:1956–1967. doi: 10.1167/iovs.05-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filla M, David G, Weinreb RN, Kaufman PL, Peters DM. Distribution of syndecans 1–4 within the anterior segment of the human eye: expression of a variant syndecan-3 and matrix-associated syndecan-2. Exp Eye Res. 2004;79:61–74. doi: 10.1016/j.exer.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Filla M, Schwinn MK, Sheibani N, Kaufman PL, Peters DM. Regulation of cross-linked actin network (CLAN) formation in human trabecular meshwork (HTM) cells by convergence of distinct β1 and β3 integrin pathways. Invest Ophthal Vis Sci. 2009;50:5723–5731. doi: 10.1167/iovs.08-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filla M, Schwinn MK, Nosie AK, Clark RW, Peters DM. Dexamethasone-associated cross-linked actin network (CLAN) formation in human trabecular meshwork (HTM) cells involves β3 integrin signaling. Invest Ophthal Vis Sci. 2011;52:2952–2959. doi: 10.1167/iovs.10-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filla MS, Lui X, Nguyen TD, Polansky JR, Brandt BR, Kaufman PL, Peters DM. In vitro localization of TIGR/MYOC in trabecular meshwork extracellular matrix and binding to fibronectin. Invest Ophthal Vis Sci. 2002;43:151–161. [PubMed] [Google Scholar]
- Freeman SA, Grinstein S. Phagocytosis: recptors, signal integration, and the cytoskeleton. Immunol Rev. 2014;262:193–215. doi: 10.1111/imr.12212. [DOI] [PubMed] [Google Scholar]
- Fujikawa K, Iwata T, Inoue K, Akahori M, Kadotani H, Fukaya M, Watanabe M, Chang Q, Barnett EM, Swat W. VAV2 and VAV3 as candidate disease genes for spontaneous glaucoma in mice and humans. PLoS ONE. 2010;5:e9050. doi: 10.1371/journal.pone.0009050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagen D, Filla M, Clark R, Litton P, Peters DM. Activated αvβ3 integrin regulates αvβ5 integrin-mediated phagocytosis in trabecular meshwork cells. Invest Ophthal Vis Sci. 2013;54:5000–5011. doi: 10.1167/iovs.13-12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Dikerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci USA. 2004;101:7618–7623. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goicoechea S, Awadia S, Garcia-Mata R. I’m coming to GEF you. Regulation of RhoGEFs during Cell Migration. Cell Adh Migr. 2014;8:535–549. doi: 10.4161/cam.28721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny TL, Brugnera E, Tosello-Trampont AC, Kinchen JM, Haney LB, Nishiwaki K, Walk SF, Nemergut ME, Macara IG, Francis R, Schedl T, Qin Y, Van Aelst L, Hengartner MO, Ravichandran KS. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 2001;107:27–41. doi: 10.1016/s0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- Ho E, Irvine T, Vilk GJ, Lajoie G, Ravichandran KS, D’Souza SJ, Dagnino L. Integrin-linked kinase interactions with ELMO2 modulate cell polarity. Mol Cell Biol. 2009;20:3033–3043. doi: 10.1091/mbc.E09-01-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoare MJ, Grierson I, Brotchie D, Pollock N, Cracknell K, Clark AF. Cross-linked actin networks (CLANs) in the trabecular meshwork of the normal and glaucomatous human eye in situ. Invest Ophthal Vis Sci. 2009;50:1255–1263. doi: 10.1167/iovs.08-2706. [DOI] [PubMed] [Google Scholar]
- Hornstein I, Alcover A, Katzav S. Vav proteins, masters of the world of cytoskeleton organization. Cell Signaling. 2004;16:1–11. doi: 10.1016/s0898-6568(03)00110-4. [DOI] [PubMed] [Google Scholar]
- Jackson BC, Ivanova IA, Dagino L. An ELMO2-RhoG-ILK network modulates microtubule dynamics. Mol Biol Cell. 2015;26:2712–2725. doi: 10.1091/mbc.E14-10-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job R, Raja V, Grierson I, Currie L, O’Reilly S, Pollock N, Knight E, Clark AF. Cross-linked actin networks (CLANs) are present in lamina cribrosa cells. Brit J Ophthalmol. 2010;94:1388–1392. doi: 10.1136/bjo.2009.176032. [DOI] [PubMed] [Google Scholar]
- Kim JY, Oh MH, Bernard LP, Macara IG, Zhang H. The RhoG/ELMO1/Dock180 signaling module is required for spine morphogenesis in hippocampal neurons. J Biol Chem. 2011;286:37615–37624. doi: 10.1074/jbc.M111.268029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurin M, Cote JF. Insights into the biological functions of Dock family guanine nucleotide exchange factors. Genes Dev. 2014;28:533–547. doi: 10.1101/gad.236349.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Collins C, Kiosses WB, Murray AM, Joshi M, Shepherd TR, Fuentes EJ, Tzima E. A novel pathway spatiotemporally activates Rac1 and redox signaling in response to fluid shear stress. J Cell Biol. 2013;201:863–873. doi: 10.1083/jcb.201207115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montalvo-Ortiz BL, Castillo-Pichardo L, Hernandez E, Humphries-Bickley T, De La Mota-Peynado A, Cubano LA, Vlaar CP, Dharmawardhane S. Characterization od EHop-16, novel small molecule inhibitor of Rac GTPase. J Biol Chem. 2012;287:13228–13238. doi: 10.1074/jbc.M111.334524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon MS, Gomez TM. Balanced Vav2 GEF activity regulates neurite outgrowth and branching in vitro and in vivo. Mol Cell Neurosci. 2010;44:118–128. doi: 10.1016/j.mcn.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores SL, Selfors LM, Fredericks J, Breit T, Fujikawa K, Alt FW, Brugge JS, Swat W. Vav family proteins couple to diverse cell surface receptors. Mol Cell Biol. 2000;20:6364–6373. doi: 10.1128/mcb.20.17.6364-6373.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nature Review Molecular Cell Biology. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- Sauzeau V, Sevilla MA, MJM, Bustelo XR. The Rho/Rac exchange factor Vav2 controls nitric oxide-dependent responses in mouse vascular smooth muscle cells. J Clin Invest. 2010;120:315–330. doi: 10.1172/JCI38356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayedyahossein A, Dagnino L. Integrins and small GTPases as modulators of phagocytosis. Int Rev Cell Mol Biol. 2013;302:321–354. doi: 10.1016/B978-0-12-407699-0.00006-6. [DOI] [PubMed] [Google Scholar]
- Sayedyahossein A, Nini L, Irvine TS, Dagino L. Essential role of integrin-linked kinase in regulation of phagocytosis in keratinocytes. FASEB Journal. 2012;26:4218–4229. doi: 10.1096/fj.12-207852. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- Shutes A, Onesto C, Picard V, Leblond B, Schweighoffer F, Der CJ. Specificity and Mechanism of Action of EHT 1864, a Novel Small Molecule Inhibitor of Rac Family Small GTPases. J Biol Chem. 2007;282:35666–35678. doi: 10.1074/jbc.M703571200. [DOI] [PubMed] [Google Scholar]
- Springelkamp H, Iglesias AI, Cuellar-Partida G, Amin N, Burdon KP, van Leeuwen EM, Gharahkhani P, Mishra A, van der Lee SJ, Hewitt AW, Rivadeneira F, Viswanathan AC, Wolfs RC, Martin NG, Ramdas WD, van Koolwijk LM, Pennell CE, Vingerling JR, Mountain JE, Uitterlinden AG, Hofman A, Mitchell P, Lemij HG, Wang JJ, Klaver CC, Mackey DA, Craig JE, van Duijn CM, MacGregor S. ARHGEF12 influences the risk of glaucoma by increasing intraocular pressure. Hum Mol Gen. 2015;24:2689–2699. doi: 10.1093/hmg/ddv027. [DOI] [PubMed] [Google Scholar]
- Stam JC, Sander EE, Michiels F, van Leeuwen FN, Kain HET, van der Kammen RA, Collard JG. Targeting of Tiam1 to the plasma membrane requires the cooperative function of the N-terminal pleckstrin homology domain and an adjacent protein interaction domain. J Biol Chem. 1997;272:28447–28454. doi: 10.1074/jbc.272.45.28447. [DOI] [PubMed] [Google Scholar]
- Subauste MC, Von Herrath M, Benard V, Chamberlain CE, Chuang TH, Chu K, Bokoch GM, Hahn KM. Rho family proteins modulate rapid apoptosis induced by cytotoxic T lymphocytes and Fas. J Biol Chem. 2000;275:9725–9733. doi: 10.1074/jbc.275.13.9725. [DOI] [PubMed] [Google Scholar]
- Tzircotis G, Braga VN, Caron R. RhoG is required for both FcγR- and CR3-mediated phagocytosis. J Cell Sci. 2011;124:2897–2902. doi: 10.1242/jcs.084269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.