Abstract
The unfolded protein response (UPR) is a conserved signalling pathway activated on the accumulation of unfolded proteins within the endoplasmic reticulum (ER), termed ER stress. Upon ER stress, HAC1/XBP1 undergoes exon/intron-specific excision by inositol requiring enzyme 1 (IRE1) to remove an intron and liberate the 5′ and 3′ exons. In yeast, the 5′ and 3′ HAC1 exons are subsequently ligated by tRNA ligase (Rlg1p), whereas XBP1 ligation in mammalian cells is catalysed by a recently identified ligase, RtcB. In the present study, RNA ligase activity of the human RtcB (hRtcB) involved in the unconventional splicing of XBP1/HAC1 mRNA was explored in an rlg1-100 mutant yeast strain. Distinct from Escherichia coli RtcB and Rlg1p, expression of hRtcB alone inefficiently complemented HAC1/XBP1 splicing and the hRtcB cofactor (archease) was required to promote enzymatic activity of hRtcB to catalyse RNA ligation.
Keywords: archease, human RtcB, human IRE1α/XBP1, Rlg1p, rlg1-100
Introduction
In eukaryotic cells, one-third of total proteins are destined to the secretory pathway, intracellular compartments, the plasma membrane or the extracellular space [1]. Perturbation of endoplasmic reticulum (ER) protein-folding homoeostasis results in the accumulation of unfolded proteins in the ER lumen, termed ‘ER stress’, which can be lethal to the cell. To restore protein-folding homoeostasis, the unfolded protein response (UPR) in the ER lumen is activated by the conserved sensor inositol requiring enzyme 1 (IRE1) as well as two additional UPR sensors in metazoan cells, activating transcription factor 6 (ATF6) and protein kinase RNA (PKR)-like ER kinase (PERK) [2,3]. Under ER stress, IRE1 forms oligomers to promote trans-autophosphorylation resulting in the allosteric activation of its RNase activity [4].
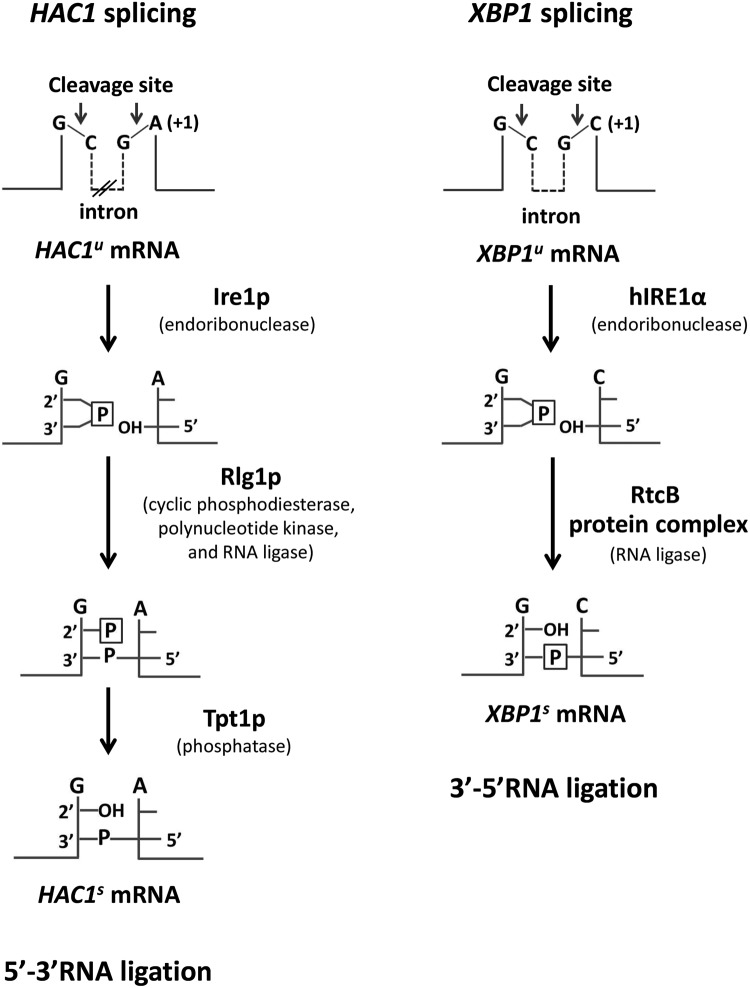
In Saccharomyces cerevisiae, the unspliced HAC1 (HAC1u) mRNA precursor is recognized and cleaved by active Ire1p to remove a 252-nt intron [5,6]. In a similar manner, unspliced XBP1 (XBP1u) undergoes human IRE1α (hIRE1α)-mediated cleavage to remove a 26-nt intron in human cell. As a consequence of Ire1p and hIRE1α cleavage, a 2′,3′-cyclic phosphate at the 3′-end of the 5′ exon and a 5′-OH at the 5′-end of the 3′ exon are generated (Figure 1). Following the cleavage reaction in HAC1 splicing, Rlg1p conducts its enzymatic activities (cyclic phosphodiesterase, polynucleotide kinase and RNA ligase) to catalyse RNA ligation between the two exons via 5′–3′ RNA ligation generating spliced HAC1 (HAC1s) transcript. By this mechanism, the phosphate utilized for phosphodiester bond formation between two exons is obtained from a nucleotide triphosphate cofactor, whereas the unincorporated 2′ phosphate that originates from the 2′,3′-cyclic phosphate precursor at the splice-site junction is ultimately removed by Tpt1 phosphatase [7,8]. In contrast, RNA ligation in XBP1 splicing mediated by RtcB goes through a 3′–5′ RNA ligation in which the phosphodiester bond is directly formed between the phosphate derived from the 2′,3′-cyclic phosphate precursor and the 5′-OH generating spliced XBP1 (XBP1s) [9,10] (Figure 1).
Figure 1. HAC1 and XBP1 splicing.
HAC1/XBP1 are cleaved specifically by Ire1 (Ire1p/hIRE1α) endo-RNase to generate a 2′,3′-cyclic phosphate and 5′-OH end at 3′ and 5′ exons respectively. In yeast, the two ends are modified before ligation by a multifunctional protein Rlg1p with cyclic phosphodiesterase, polynucleotide kinase and RNA ligase activities leaving a 2′-phosphate at the splice junction. The 2′-phosphate is finally removed by Tpt1p phosphatase. This ligation is defined as 5′–3′ RNA ligation. In mammals, the two ends are directly ligated by the RtcB protein complex activity via 3′–5′ RNA ligation. This protein complex is composed of five subunits: ASW, CGI-99, DDX1, FAM98B and archease. The nucleotides at +1 position in the 3′-splice site junction of HAC1 and XBP1 are indicated. P represents the phosphate group derived from 2′,3′-cyclic phosphate.
RtcB, the catalytic subunit of the tRNA ligase complex and its cofactor (archease) have been identified as the RNA ligase that mediates tRNA splicing [9–11] and XBP1 mRNA splicing both in vitro and in vivo in many organisms [12–14], except in fungi and plants as they have Rlg1p and Arabidopsis thaliana tRNA ligase respectively, for mediating tRNA splicing [15,16]. Archease is required for accelerating RtcB RNA ligation with GTP and is Mn2+-dependent [17]. However, some RtcBs are archease-independent in mediating RNA ligation such as Thermus thermophilus RtcB [18].
Interestingly, Escherichia coli RtcB is able to compensate for Rlg1p function in HAC1 splicing in rlg1-defective yeast [10]. Likewise, E. coli RtcB with a possible accessory role of mammalian RtcB in HAC1/XBP1 splicing is not well characterized. In the present study, we used an rlg1-100 yeast strain as a surrogate platform for elucidating whether human RtcB (hRtcB) could mediate RNA ligation to provide insight into the HAC1/XBP1 splicing event. We demonstrate that hRtcB is unable to catalyse RNA ligation by itself unless human archease is simultaneously expressed.
Materials and methods
Yeast strains and bacterial strains
S. cerevisiae Δire1/Δhac1 and the Δire1/Δhac1/rlg1-100 triple deletion yeast strain was developed from CF203 [16]. The HAC1 and URA3 gene loci of CF203 were replaced with Zeocinr and kanMX respectively, by double homologous recombination. E. coli DH5α was used for propagation and construction of all plasmids.
Plasmids construction
pYES-Ire1 and pYES-hIRE1α were used for expressing exogenous yeast Ire1 and human IRE1α in yeast cells respectively. These two expression plasmids were constructed in the pYES2 vector under GAL1 inducible promoter as previously described [19].
YCplac111-mtHAC1 was constructed for expressing HAC1u mRNA that carries a single point mutation (adenine to cytosine) at the +1 position in the 3′-splice site junction. We have recently demonstrated that wild-type HAC1 was unable to be spliced by hIRE1α unless adenine (A) at +1 position in the 3′-splice site junction was substituted with cytosine (C) [20]. To construct this expression plasmid, the 2.5-kb mtHAC1 coding sequence including the ADH1 promoter and CYC1 transcription terminator from pTB-mtHAC1 was subcloned into YCplac111 between the SmaI and XbaI sites. YCplac111-dmXBP1-3′BE was constructed as an dmXBP1u expression plasmid in yeast. The 3′BE refers to the 3′ bipartite element in the 3′-UTR of HAC1 that acts as an ER-membrane targeting signal [21]. We utilized YCplac111-mtHAC1 as a template to replace the mtHAC1 coding sequence with the unspliced Drosophila melanogaster XBP1 (dmXBP1u) coding sequence from pcDNA-dmXBP1u [22].
pTB-hRtcB was created to express hRtcB. The 1645-bp fragment from hRtcB cDNA was amplified from HeLa cells using HSPC117F and HSPC117R primers and cloned into pTB326 between the EcoRI and KpnI restriction sites. pTB-RtcB was created as an expression plasmid for E. coli RtcB in yeast. The bacterial RtcB coding sequence was amplified from DH5α genomic DNA using RtcB primers (Table 1). The 1300-bp PCR product corresponding to E. coli RtcB coding sequence was inserted into pTB326 between the KpnI and EcoRI sites.
Table 1.
Primers used in the present study
| Primers | Sequence (5′-3′) |
|---|---|
| HSPC117 F | TGA ATT CAG ATC TGT GCT CTG AGA AGC CGG ACT AC |
| EcoRl | |
| HSPC117 R | GAG GTA CCC AGT CCA CTT CCA CTT CAG AG |
| Kpnl | |
| RtcB F | GGG TAC CTT ATC ATC GAC AGG CTC AGC |
| Kpnl | |
| RtcB R | GGA ATT CTC TCT AGA CAC CAT GAA TTA CGA ATT ACT G |
| EcoRl | |
| ARCH F | GCG GAT CCT AGA GCG GAA GTA GTA ACT C |
| BamHl | |
| ARCH R | CCA GTC GAC ACA GTT CTT CGT AGG AGT C |
| Sall | |
| HAC1flkF | GCC CAA GAG TAT GCG CGA TTC CG |
| HAC1flkR | ACC CTC TTG CGA TTG TCT TCA TG |
| HAC 1s F | GCC CAA GAG TAT GCG CGA TTC CG |
| HAC 1s R | CAA ACC TGA CTG CGC TGC TGG |
| hlRE1α F | TC TAT CCA TGC CCA ATG CAC ACG |
| hlRE1αR | GTC GCT CAC GTC CTG GAA GAA C |
| lre1 F | ACC GCA TCC CTT TAA TCC TGG TGA |
| lre1 R | GAC TTC CAT CGT TCA CAG CAC CTT C |
| dmXBP1sF | CGA ACT GAA GCA GCA ACA GCA G |
| dmXBP1sF2 | GGC TGG ATC CCA GCC CAA GGC CAA GAA GC |
| dmXBP1sR | GTA TAC CCT GCG GCA GAT CCA AGG |
| actin F | CAT CTA TCG TCG GTA GAC CAA G |
| actin R | GGA GCA ATG ATC TTG ACC TTC ATG |
| qRT KAR2F | TGG GTG GTG GTA CTT TCG ATG TCT |
| qRT KAR2R | AGC TAG GGC CTT GTT GTT GTC AGA |
| qRT actin F | CAC GTC GTT CCA ATT TAC GCT GGT |
| qRT actin R | TCG AAG TCC AAG GCG ACG TAA CAT |
pAG-archease was created as a recombinant plasmid for expression of human archease in yeast cells. The cDNA fragment of human archease was produced by PCR using ARCH primers and inserted into pTEF413 between the BamHI and SalI sites. This vector is an intermediate, for which the human archease including TEF promoter and CYC1 terminator from pTEF413 backbone was further blunt-end ligated with pAG26. All PCR reactions were performed using Phusion DNA polymerase (Thermo Scientific) and sequences were confirmed by DNA sequencing. Primers used for constructing each recombinant plasmid are shown in Table 1.
Exogenous gene expression in yeast
The Δire1/Δhac1 or Δire1/Δhac1/rlg1-100 triple deleted yeast strain was transformed with recombinant plasmid for expressing IRE1 (pYES-Ire1 or pYES-hIRE1α), unconventional RNA substrate (YCplac111-mtHAC1 or YCplac111-dmXBP1-3′BE), RNA ligase (pTB-hRtcB or pTB-RtcB) and pAG-archease using an electroporation protocol [23]. The transformants were selected on a synthetic dropout medium that corresponded to auxotrophic marker of each vector backbone containing 2% (v/v) D-glucose. To determine the RNA ligase activity of candidate proteins, yeast cells were cultured in dropout medium containing 2% (v/v) D-glucose. The IRE1 expression was induced by addition of 2% (v/v) D-galactose. Finally, cells were exposed to ER stress by treatment with dithiothreitol (DTT). For maintaining the pAG expression vector, yeast cells were cultured in medium supplemented with hygromycin B (250 μg/ml). A similar protocol was used for testing the ability for Rlg1p to complement hIRE1α/XBP1 splicing in the Δire1/Δhac1 double deleted yeast strain.
RT-PCR
Yeast cells were pretreated with lyticase (Sigma) in buffer containing 1 M sorbitol, 0.1 M EDTA, pH=8.0 and 0.1% (v/v) β-mercaptoethanol. The reaction was incubated at 30ºC for 30 min and then the yeast-cell pellet was resuspended with 1 ml TRIzol Reagent (MRC) and RNA isolation was performed following the manufacturer’s instructions. First-strand cDNA was synthesized from DNase I-treated RNA using oligo d(T) with Improm-II reverse transcriptase (Promega). The HAC1u and HAC1s transcripts were monitored by RT-PCR using primers that flank the 252-nt intron (HAC1flkF and HAC1flkR). Alternatively, the HAC1s transcript was selectively monitored using HAC1sF and HAC1sR primers. In a similar manner, dmXBP1s transcripts were measured using dmXBP1sF and dmXBP1sR primers. The transcription levels of hIRE1α, Ire1, hRtcB, E. coli RtcB, archease and actin were measured using specific primers (Table 1). The HAC1s and dmXBP1s PCR detections were performed at 69 and 74.7ºC annealing temperatures respectively. The PCR was performed using DNA polymerase (Thermo Scientific).
Quantitative PCR (qPCR)
The same preparations of cDNAs used in RT-PCR were diluted 10-fold before analysis of gene expression by qRT-PCR using the SYBR fluorescent dye (KAPA). The HAC1s, dmXBP1s, KAR2 and actin mRNAs were monitored with specific primers (Table 1). The PCR protocol for analysing HAC1s was performed on following parameters: 95ºC for 3 min followed by 40 cycles of 95ºC for 10 s, 69ºC for 30 s and 72ºC for 20 s. The quantification of dmXBP1s, KAR2 and actin mRNAs was performed using a similar protocol except the annealing temperatures being 74.7ºC, 57ºC and 57ºC respectively. The amplification data were analysed according to 2−ΔΔCT method normalizing with actin mRNA levels.
Western blot of hRtcB
To determine expression level of hRtcB, yeast cells were cultured, pelleted and resuspended in 4× lysis buffer [24]. Cell suspensions were then boiled for 5 min. The yeast cell extract was resolved on 4–15% gradient gels (Bio–Rad Laboratories). The blot was probed with rabbit polyclonal antibody hRtcB protein (Santa Cruz Biotechnology) and then the antigen–antibody complexes were monitored by horseradish-peroxidase-conjugated with anti-rabbit IgG (Sigma–Aldrich) using the West Dura chemiluminescent substrate (Thermo Scientific) detection system.
Results
Rlg1p inefficiently rescues hIRE1α-mediated splicing in yeast cells
Previous studies demonstrated that Rlg1p is essential for splicing of Ire1p/HAC1 mRNA in yeast cells [16]. Interestingly, we found that Rlg1p is also able to complement hIRE1α/mtHAC1 splicing in ∆ire1/∆hac1/RLG1 [20], however, HAC1 splicing efficiency was much less than Ire1p-mediated splicing (Figure 2A, compare lanes 7–8 with 5–6 and Supplementary Figure S1A). Since HAC1 mRNA is not a natural substrate for hIRE1α, it is possible that the HAC1 splicing substrates generated by Ire1p and hIRE1α are different. To test this hypothesis, we analysed XBP1 mRNA as a splicing substrate to investigate hIRE1α cleavage activity. Using the same analysis platform, dmXBP1u was co-expressed with hIRE1α. Under ER stress conditions induced by DTT, a single PCR product of 106 bp corresponding to dmXBP1s was clearly detected exclusively upon hIRE1α co-expression, but not presented upon dmXBP1 expression alone (Figure 2B, compare lanes 5–8 with 3–4, Supplementary Figure S1B). By Western blot analysis, the predicted pXBP1u (~37 kDa) and pXBP1s (~50 kDa) were undetectable from yeast cell lysates (results not shown). This might be due to instability of XBP1 protein in the yeast cells [25]. Notably, overexpression of hIRE1α was capable to mediate dmXBP1 splicing even under non-stress condition as we previously reported [19]. Interestingly, we found that dmXBP1 could be processed by Ire1p similar to hIRE1α mediated splicing. The dmXBP1s was easily detected upon Ire1 co-expression (Figure 2B, lanes 5 and 6). However, dmXBP1 substitution did not increase the hIRE1α splicing efficiency in this wild-type tRNA ligase RLG1 yeast strain. Regardless of the mRNA substrate, our results suggest that Rlg1p is not compatible to complement the hIRE1α-mediated splicing process.
Figure 2. Rlg1p is able to mediate XBP1 splicing in yeast.
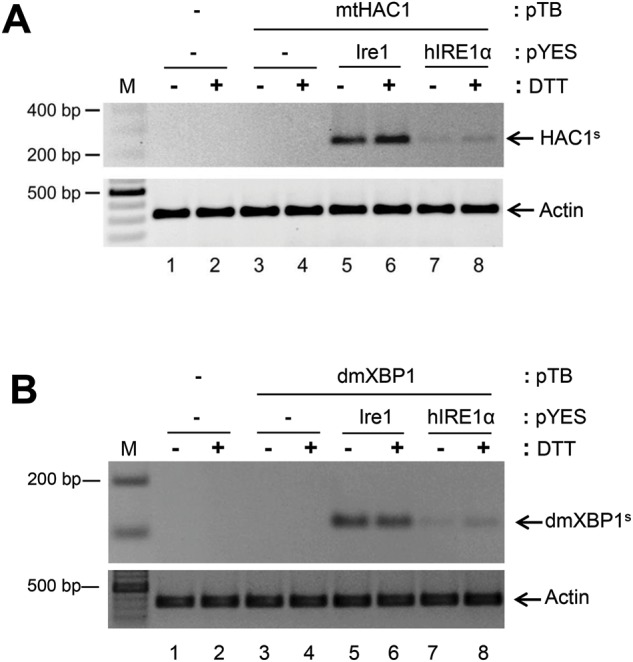
The Δire1/Δhac1/RLG1 yeast strain was transformed with a recombinant plasmid to express mtHAC1/dmXBP1 with hIRE1α or Ire1. Splicing of dmXBP1/mtHAC1 was analysed after induction of IRE1 expression for 17 h with D-galactose and subsequently treating the cells with 5 mM DTT for 2 h. Splicing was quantified by RT-PCR using splice-specific primers. (A) RT-PCR of HAC1s. (B) RT-PCR of spliced dmXBP1 (dmXBP1s). Actin transcription level was included as internal control. M: GeneRuler DNA ladder.
E. coli RtcB, not hRtcB alone can complement HAC1/XBP1 splicing
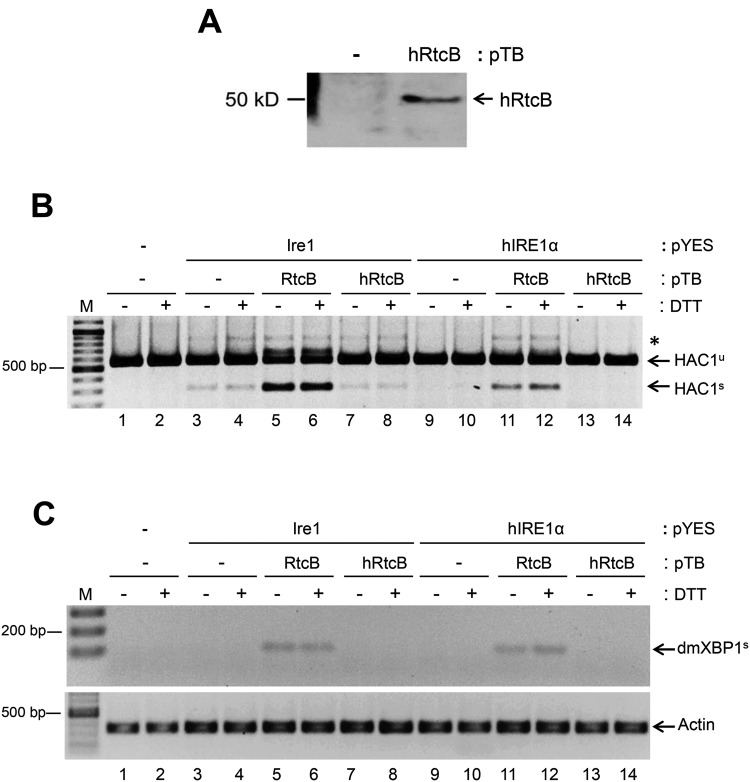
It was reported that rlg1-100 deficiency severely impairs HAC1 splicing but does not affect the pre-tRNA maturation process [16]. Taking this advantage, we asked whether hRtcB can compensate for Rlg1p to complement the hIRE1α requirement in HAC1 mRNA splicing. For this purpose, we transformed Δire1/Δhac1/rlg1-100 yeast strain with three different recombinant plasmids encoding mtHAC1, hIRE1α or Ire1p and hRtcB. From the transformed yeast-cell lysates, the hRtcB protein (~55 kDa) was detected as predicted (Figure 3A). We found that rlg1-100 was not totally defective in RNA ligase activity. Indeed, it is still sufficient to complement HAC1 splicing i.e. there is a faint band of HAC1s appearing in Ire1 co-expressing cells (Figure 3B, lanes 3 and 4). Although, HAC1 splicing was markedly rescued when E. coli RtcB was co-expressed with Ire1 included as positive control (Figure 3B, lanes 5 and 6). Unexpectedly, HAC1 splicing process was defective in yeast with co-expression of hRtcB (Figure 3B, lanes 7 and 8). Even though a faint band intensity of HAC1s was detected upon hRtcB co-expression, it was not significantly different compared with Ire1 expression alone (Supplementary Figure S2A). A similar result was obtained in the presence of hIRE1α co-expression (Figure 3B, lanes 9–14).
Figure 3. E. coli RtcB efficiently mediate HAC1/XBP1 splicing in yeast.
The Δire1/Δhac1/rlg1-100 yeast strain was transformed with three different expression plasmids to express mtHAC1/dmXBP1, hIRE1α/Ire1 and hRtcB/E.coli RtcB (RtcB). The splicing efficiencies of dmXBP1 and mtHAC1 were measured after induction of IRE1 expression with D-galactose for 17 h and ER stress by treatment with 5 mM DTT for 2 h. (A) Western blot analysis of hRtcB. (B) RT-PCR of HAC1 using an intron-flanking primers. Asterisk (*) represents a hybrid between unspliced and spliced forms of mutant HAC1 transcript. (C) RT-PCR of dmXBP1s using specific primers. Actin mRNA level was used as an internal control. M: GeneRuler DNA ladder.
To confirm our finding, the RNA ligase activity of hRtcB was further tested by its ability to ligate the dmXBP1 RNA substrate. Surprisingly, rlg1-100 did not mediate the ligation reaction (Figure 3C, lanes 3–4 and 9–10) in contrast with HAC1 splicing. As previous observed, the dmXBP1 splicing was exclusively complemented only by E. coli RtcB co-expression (Figure 3C, lanes 5–6 and 11–12) but not in hRtcB co-expression (Figure 3C, lanes 7–8 and 13–14, Supplementary Figure S2B). We propose that hRtcB by itself is not able to ligate RNA and the reaction required an additional cofactor(s) for hRtcB RNA ligase-mediated mRNA splicing.
Archease facilitates hRtcB RNA ligase activity
Distinct from Rlg1p, mammalian RtcB is associated with ASW, DDX1, FAM98B, CGI-99 and archease in the tRNA ligase complex [9,26 ]. Of all these associated proteins, archease is known as an activator for RtcB to mediate in vitro RNA ligation of cleaved pre-tRNA transcripts [27]. However, the ability of human archease to directly stimulate hRtcB in the HAC1/XBP1 splicing reaction in vivo has not been reported. Hence, the hRtcB RNA ligase activity was further analysed upon archease co-expression. We found that HAC1 splicing in yeast was still defective upon archease expression alone. This indicates that archease has no RNA ligase activity and the rlg1-100 cannot mediate HAC1 splicing under these conditions. Consistently, HAC1 splicing was complemented with E. coli RtcB (Figure 4A, lanes 1 and 2) but not with hRtcB expression alone (Figure 4A, lanes 3 and 4). Interestingly, HAC1 splicing was restored when archease was simultaneously expressed with hRtcB (Figure 4A, lanes 5 and 6, Supplementary Figure S3A). We observed the expression of KAR2, a downstream target of the Hac1 transcription factor, was increased upon archease co-expression (Figure 4B). The collaboration between hRtcB and archease in mediating RNA ligation was confirmed upon analysis of dmXBP1 splicing (Figure 4C and Supplementary Figure S3B). Taken together, our results indicate that hRtcB and archease are fundamental subunits required for RNA ligase activity in the unconventional splicing reaction.
Figure 4. Archease is required for hRtcB RNA ligase activity.
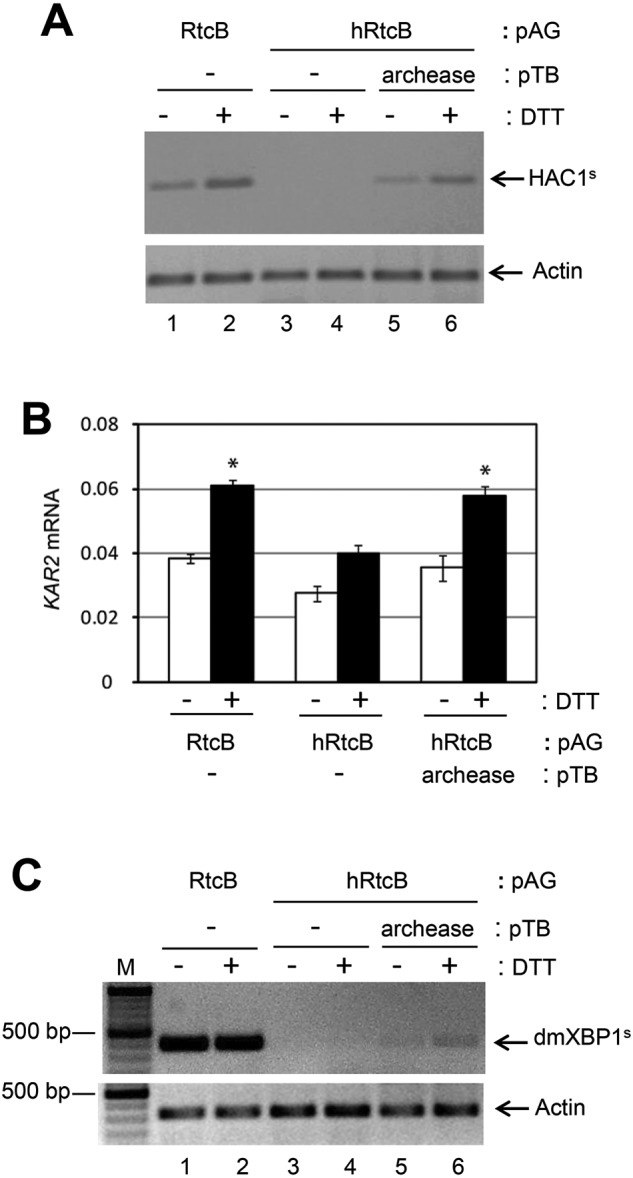
The Δire1/Δhac1/rlg1-100 yeast strain was transformed with four different plasmids to express hIRE1α/Ire1, mtHAC1/dmXBP1, hRtcB/RtcB and archease. The effect of archease in hRtcB function was determined. (A) RT-PCR of HAC1s using HAC1s-specific primers after induction of IRE1 expression for 18 h including 2-h DTT treatment. (B) qPCR of KAR2. The mRNA level was normalized with yeast actin mRNA level and is shown as mean ± S.E.M. and subjected to one-way ANOVA test (*P<0.01, compared with hIRE1α, mtHAC and hRtcB co-expression at the same condition). (C) RT-PCR of dmXBP1s using splice-specific primers (dmXBP1sF2 and dmXBP1sR) (Table 1) after induction of hIRE1α expression for 45 h, then a 3-h DTT treatment. Actin mRNA levels were used as an internal control. M: GeneRuler DNA ladder.
Discussion
In the present study, we reconstituted the entire splicing process of XBP1/HAC1 in the yeast mutant strain rlg1-100. Using this system, we studied functional roles of hRtcB in HAC1/XBP1 splicing. Our results demonstrate that the hRtcB subunit expressed in yeast cell cannot complement HAC1/XBP1 splicing as previous attempts to identify the RNA ligase using a purified fly RtcB and hRtcB from HEK293T cells as candidate protein, both were unable to complement in vitro splicing of XBP1/hIRE1α [28]. In the present study, we demonstrated that hRtcB RNA ligase activity is recruited when its cofactor (archease) is co-expressed in yeast cells. The exact functional role of archease to promote hRtcB RNA ligase activity in XBP1 splicing is unclear. It likely involves a guanylylation process that is an initial step to activate the archaeal RtcB RNA ligase activity [17]. Although, the hRtcB and archease function involving RNA ligation was previously reported by RNAi knockdown where XBP1 splicing was almost completely impaired when hRtcB and archease were simultaneously depleted, whereas knockdown of either hRtcB or archease alone did not effectively diminish splicing [10]. However, by RNAi-mediated RtcB knockdown, the RtcB-associated protein DDX1 and FAM98B were unexpectedly co-depleted with RtcB consistent with RtcB conditional knockout mice [9,12].
Supporting our findings, RtcB is the responsible for RNA ligase in pre-tRNA splicing that occurs in the nucleus, however, some RtcBs are found at the ER membrane where XBP1u mRNA and hIRE1α are located [12,26]. RtcB is thought to interact with XBP1u mRNA specifically at intron-flanking regions [29]. However, our data not only demonstrated that hRtcB can catalyse dmXBP1 ligation but it can also mediate HAC1 ligation, although the putative RtcB binding site in XBP1u is not present in HAC1u mRNA. Likewise, Rlg1p and E. coli RtcB can ligate both dmXBP1 and HAC1 mRNA. Together the findings suggest that RNA ligase and RNA substrate interactions are not sequence specific, for example, it was shown that RtcB associates with hIRE1α at the ER membrane [12]. Thus, the RNA ligase may indirectly interact with its RNA substrate through IRE1. By this scenario, it is implicated that Rlg1p and other RtcBs associate with hIREα as hRtcB do during XBP1 splicing in yeast.
In the present study, we showed that hRtcB RNA ligase activity on XBP1/HAC1 splicing is very low compared with E. coli RtcB activity (Supplementary Figures S3A and S3B). An in vitro-ligation assay suggested that DDX1 helicase is another factor required for RtcB activation to mediate pre-tRNA ligation [27]. Therefore, it is possible that DDX1 is required for full activation of hRtcB in XBP1 splicing in addition to archease. This is in contrast with E. coli RtcB that is able to catalyse RNA ligation by itself without any cofactor [10]. Besides DDX1, it is interesting to investigate the exact function of other RtcB cofactors (ASW, FAM98B and CGI-99) in the RNA ligase complex. This might be useful for discriminating functional roles of each component of the tRNA ligase complex in pre-tRNA maturation process, XBP1 splicing and possibly RIDD (regulated IRE1-dependent decay) [30]. The IRE1α cleavage reaction generates a 5′-OH and a 2′,3′-cyclicphosphate terminus that are potential substrates for RtcB.
Acknowledgments
The authors wish to thank Dr. Peter Walter (University of California, San Francisco) for providing CF203 yeast strain.
Abbreviations
- dmXBP1s
spliced Drosophila melanogaster XBP1
- dmXBP1u
unspliced Drosophila melanogaster XBP1
- ER
endoplasmic reticulum
- HAC1s
spliced HAC1
- HAC1u
unspliced HAC1
- hIRE1α
human IRE1α
- hRtcB
human RtcB
- IRE1
inositol requiring enzyme 1
- qPCR
quantitative PCR
- UPR
unfolded protein response
- XBP1
X-box-binding protein 1
- XBP1s
spliced XBP1
- XBP1u
unspliced XBP1
- 3′BE
3′ bipartite element
Author contribution
J.P. designed and performed the experiments, analysed the data and wrote the manuscript. R.J.K. and W.T. designed the experiments, analysed the data and edited the manuscript.
Funding
This work was supported by the Royal Golden Jubilee Ph.D. Program (PHD/0332/2551) to JP. .and WT. .under Thailand Research Fund and Mahidol University. Portions of this work were supported by NIH/NCI Grants R37DK042394, R01DK103185, R24DK110973 and R01CA198103 (R.J.K.), and Sanford Burnham Prebys NCI Cancer Center Grant P30 CA030199].
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Schroder M. and Kaufman R.J. (2005) The mammalian unfolded protein response. Annu. Rev. Biochem. 74, 739–789 [DOI] [PubMed] [Google Scholar]
- 2.Bernales S., Papa F.R. and Walter P. (2006) Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 22, 487–508 [DOI] [PubMed] [Google Scholar]
- 3.Wang M. and Kaufman R.J. (2016) Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 529, 326–335 [DOI] [PubMed] [Google Scholar]
- 4.Walter P. and Ron D. (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 [DOI] [PubMed] [Google Scholar]
- 5.Cox J.S. and Walter P. (1996) A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87, 391–404 [DOI] [PubMed] [Google Scholar]
- 6.Sidrauski C. and Walter P. (1997) The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90, 1031–1039 [DOI] [PubMed] [Google Scholar]
- 7.Culver G.M., McCraith S.M., Consaul S.A., Stanford D.R. and Phizicky E.M. (1997) A 2′-phosphotransferase implicated in tRNA splicing is essential in Saccharomyces cerevisiae. J. Biol. Chem. 272, 13203–13210 [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez T.N., Sidrauski C., Dörfler S. and Walter P. (1999) Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J. 18, 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Popow J., Englert M., Weitzer S., Schleiffer A., Mierzwa B., Mechtler K. et al. (2011) HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science 331, 760–764 [DOI] [PubMed] [Google Scholar]
- 10.Tanaka N., Meineke B. and Shuman S. (2011) RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J. Biol. Chem. 286, 30253–30257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Englert M., Sheppard K., Aslanian A., Yates J.R. III and Soll D. (2011) Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc. Natl. Acad. Sci. U.S.A. 108, 1290–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jurkin J., Henkel T., Nielsen A.F., Minnich M., Popow J., Kaufmann T. et al. (2014) The mammalian tRNA ligase complex mediates splicing of XBP1 mRNA and controls antibody secretion in plasma cells. EMBO J. 33, 2922–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kosmaczewski S.G., Edwards T.J., Han S.M., Eckwahl M.J., Meyer B.I., Peach S. et al. (2014) The RtcB RNA ligase is an essential component of the metazoan unfolded protein response. EMBO Rep. 15, 1278–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu Y., Liang F.X. and Wang X. (2014) A synthetic biology approach identifies the mammalian UPR RNA ligase RtcB. Mol. Cell 55, 758–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Englert M. and Beier H. (2005) Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res. 33, 388–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sidrauski C., Cox J.S. and Walter P. (1996) tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell 87, 405–413 [DOI] [PubMed] [Google Scholar]
- 17.Desai K.K., Cheng C.L., Bingman C.A., Phillips G.N. Jr and Raines R.T. (2014) A tRNA splicing operon: archease endows RtcB with dual GTP/ATP cofactor specificity and accelerates RNA ligation. Nucleic Acids Res. 42, 3931–3942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai K.K., Beltrame A.L. and Raines R.T. (2015) Coevolution of RtcB and archease created a multiple-turnover RNA ligase. RNA 21, 1866–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poothong J., Sopha P., Kaufman R.J. and Tirasophon W. (2010) Domain compatibility in Ire1 kinase is critical for the unfolded protein response. FEBS Lett. 584, 3203–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poothong J., Sopha P., Kaufman R.J. and Tirasophon W. (2016) IRE1α nucleotide sequence cleavage specificity in the unfolded protein response. FEBS lett., 10.1002/1873-3468.12546 [DOI] [PubMed] [Google Scholar]
- 21.Aragon T., van Anken E., Pincus D., Serafimova I.M., Korennykh A.V., Rubio C.A. et al. (2009) Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature 457, 736–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plongthongkum N., Kullawong N., Panyim S. and Tirasophon W. (2007) Ire1 regulated XBP1 mRNA splicing is essential for the unfolded protein response (UPR) in Drosophila melanogaster. Biochem. Biophys. Res. Commun. 354, 789–794 [DOI] [PubMed] [Google Scholar]
- 23.Wu S. and Letchworth G.J. (2004) High efficiency transformation by electroporation of Pichia pastoris pretreated with lithium acetate and dithiothreitol. Biotechniques 36, 152–154 [DOI] [PubMed] [Google Scholar]
- 24.Kushnirov V.V. (2000) Rapid and reliable protein extraction from yeast. Yeast 16, 857–860 [DOI] [PubMed] [Google Scholar]
- 25.Tirosh B., Iwakoshi N.N., Glimcher L.H. and Ploegh H.L. (2006) Rapid turnover of unspliced Xbp-1 as a factor that modulates the unfolded protein response. J. Biol. Chem. 281, 5852-5860. [DOI] [PubMed]
- 26.Perez-Gonzalez A., Pazo A., Navajas R., Ciordia S., Rodriguez-Frandsen A. and Nieto A. (2014) hCLE/C14orf166 associates with DDX1-HSPC117-FAM98B in a novel transcription-dependent shuttling RNA-transporting complex. PLoS ONE 9, e90957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Popow J., Jurkin J., Schleiffer A. and Martinez J. (2014) Analysis of orthologous groups reveals archease and DDX1 as tRNA splicing factors. Nature 511, 104–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwawaki T. and Tokuda M. (2011) Function of yeast and amphioxus tRNA ligase in IRE1alpha-dependent XBP1 mRNA splicing. Biochem. Biophys. Res. Commun. 413, 527–531 [DOI] [PubMed] [Google Scholar]
- 29.Baltz A.G., Munschauer M., Schwanhausser B., Vasile A., Murakawa Y., Schueler M. et al. (2012) The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690 [DOI] [PubMed] [Google Scholar]
- 30.Hollien J., Lin J.H., Li H., Stevens N., Walter P. and Weissman J.S. (2009) Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 186, 323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]