Abstract
Macrophages are heterogeneous cells that play a key role in inflammatory and tissue reparative responses. Over the past decade it has become clear that shifts in cellular metabolism are important determinants of macrophage function and phenotype. At the same time, our appreciation of macrophage diversity in vivo has also been increasing. Factors such as cell origin and tissue localization are now recognized as important variables that influence macrophage biology. Whether different macrophage populations also have unique metabolic phenotypes has not been extensively explored. In this article, we will discuss the importance of understanding how macrophage origin can modulate metabolic programming and influence inflammatory responses.
Keywords: Inflammation, metabolism, glycolysis, mitochondria, nitric oxide
1. Introduction: Macrophage heterogeneity and immunometabolism
It is now well accepted that macrophages shift their metabolism in response to environmental cues. In turn, these metabolic adaptations drive specific effector functions in macrophages. Some of the more common events that trigger macrophage metabolic reprogramming include activation of pathogen recognition receptors, such as toll-like receptors (TLRs), or nutrient based signals that engage lipid nuclear receptors (PPARγ, ERRγ) and/or kinases (mTOR or AMP kinase). The regulation of metabolism has traditionally been equated with energetics. From this perspective, metabolic shifts occur primarily to maintain the balance of ATP supply and demand. In tissues like heart and skeletal muscle, which have high ATP demand, energy production is indeed the primary job of metabolic pathways. However, the role of metabolic reprogramming clearly extends beyond ATP production and includes regulation of lipid synthesis, nucleotide biosynthesis, cell signaling, and gene expression. Over the past several years the study of metabolism in immune cells has highlighted the importance of the non-ATP generating functions of cellular metabolism. In particular, the ability of specific metabolites and/or metabolic signaling events to regulate cell differentiation and effector function is now appreciated [1, 2].
Macrophages have diverse functions in tissue homeostasis and inflammation. Evidence is emerging that the metabolic features of these cells regulate their function, including cytokine release and cell surface receptor expression [3]. One of the clearest examples of this concept comes from the comparison of classically activated “inflammatory” macrophages (CAMs) and alternatively activated “reparative” macrophages (AAM). In general, CAMs are highly glycolytic, whereas AAMs utilize fatty acid metabolism and mitochondrial oxidative phosphorylation (OXPHOS) [4, 5]. The distinct metabolic programing of these macrophage subsets is thought to generate unique metabolites that are important for their specific effector functions. At the same time, it has also been recognized that macrophages in vivo are heterogeneous in function based on factors like tissue localization and cell origin [6]. To date, the overlay of immunometabolism with the macrophage diversity has not been explored and represents a critical direction for future research.
In this review we will expand upon established metabolic concepts by exploring how macrophage origin may influence metabolic programming. To accomplish this aim we will take two approaches: 1) review and discuss data comparing the metabolic features of classically activated bone marrow derived macrophages (BMDMs) vs. elicited peritoneal macrophages (pMACs) as a proof of concept that cell origin can influence metabolic behavior; 2) review the data from already existing pools of gene expression profiling to identify metabolic modules that define macrophages from distinct tissues as evidence that these concepts are globally relevant to understanding macrophage biology.
2. Macrophage Phenotype and Immunometabolism
2.1 Macrophage polarization
Macrophages play important roles in inflammation (cytokine release, phagocytosis) and tissue repair (stem cell proliferation, angiogenesis, fibrosis). The concept that macrophages can be directed towards inflammatory or reparative functions, so called “macrophage polarization” by cues from their microenvironments has been a useful construct to describe macrophage behavior. Activation of TLRs on macrophages by pathogen products or alarmins produces a CAM phenotype whereas IL-4 or efferocytosis promotes an AAM phenotype. CAMs produce inflammatory cytokines and reactive oxygen species, which are important for host defense against infection and the early response to tissue damage. In contrast, AAMs release anti-inflammatory cytokines and are thought to promote angiogenesis and fibrosis. AAMs also mediate host responses to parasites. Over the past decade, several studies have demonstrated that the development of CAMs and AAMs is dependent on distinct modes of metabolic reprogramming (see e.g. [5]). These observations have fueled the concept that metabolic modulation could be used to alter macrophage function in disease states. This topic has been discussed in several recent reviews in detail, so we will only briefly review these established concepts [3, 7–9].
2.2 Metabolic phenotype of classically activated macrophages
CAMs are characterized by high rates of aerobic glycolysis, a metabolic feature referred to as Warburg metabolism [10]. In addition to producing ATP, enhanced flux of glucose into glycolysis and the pentose phosphate shunt generates building blocks needed for nucleic acid synthesis, protein synthesis, and lipid synthesis. The other metabolic hallmark of the CAM phenotype observed in bone marrow macrophages is suppression of mitochondrial OXPHOS. This is thought to occur because inflammatory signaling suppresses tricarboxylic acid (TCA) cycle flux at 2 distinct steps [2, 5]. One block comes at the level of isocitrate dehydrogenase (IDH) which leads to the accumulation of citrate and the other at the level of succinate dehydrogenase (SDH). Reduced IDH activity correlates with TLR-induced suppression of IDH gene expression and leads to increased shunting of citrate from the mitochondria to the cytosol where it can be converted by acetyl-CoA carboxylase (ACC) to malonyl-CoA or support itaconate synthesis via Irg1[5]. Malonyl-CoA is used for fatty acid synthesis, a key precursor for membrane remodeling and expansion of organelles such the endoplasmic reticulum [11]. Unlike IDH, Irg1 is induced by TLR4 signaling and drives the production of itaconate [12, 13]. Itaconate has been described as metabolite possessing both anti-inflammatory and anti-microbial properties [2, 14]. While the exact mechanism of action of itaconate is not understood, such duality might stem from its inhibitory properties: being a structural mimetic of succinate, itaconate inhibits both mammalian SDH and microbial Icl enzymes. These two possibilities are not mutually exclusive, however. Strikingly, the amount of itaconate required for microbial killing exceeds the amounts produced by activating macrophages [2, 14, 15], suggesting that anti-microbial effects of itaconate have to be highly localized to achieve sufficient concentration, e.g. inside phagosomes. Thus, mitochondrial or cytosolic itaconate might play regulatory role while phagosomal itaconate participates in anti-microbial action. The fact that itaconate has roles beyond anti-bacterial/antifungal responses stems from the fact that itaconate and Irg1 are highly induced during anti-viral response as well. Metabo-regulatory roles of itaconate are evident from analysis of the activation of Irg1 knockout (KO) macrophages which show complete absence of succinate accumulation during macrophage activation, and accordingly do not demonstrate SDH breakpoint of the TCA cycle [2]. Thus, itaconate modulates activity of SDH, also known as complex II of the electron transport chain. This places itaconate on the critical intersection of cellular bioenergetics and TCA cycle. Functional importance of this intersection stems from previously reported connection between succinate and Hif1α-IL-1β axis [16, 17]. Strikingly, Irg1 KO macrophages are more pro-inflammatory then their wild type counterparts in spite of the fact that succinate does not accumulate. This leads to conclusion that the SDH breakpoint and succinate accumulation per se are not absolutely required for the proinflammatory phenotype of macrophages.
2.3 Macrophage polarization in vivo
The investigation of immunometabolism using ex vivo macrophage systems has led to several key discoveries about the interplay between metabolism and cell function. However, the extent to which these findings translate to the diverse populations of macrophages that exist in vivo remains to be defined. Although the distinction between the CAM and AAM phenotype is black and white in vitro, the in vivo reality is more complicated with macrophages often possessing a mixture of CAM and AAM features [18, 19]. It is well established that resident macrophage gene expression profiles vary dramatically across tissues; however, the influence of these unique profiles on metabolic regulation is not clear. In addition, there is further diversity in the monocyte-derived macrophages that enter into tissues in response to infection and or injury whereupon they encounter nutrient and inflammatory cues that shape their metabolic and functional response. Although it is tempting to speculate that monocyte-derived cells resemble CAMs and resident cells are more like AAMs the question of whether this is also true for their metabolic characteristics remains to be answered.
3. Metabolic comparison of classically activated BMDMs and pMACs
3.1 Characteristics of BMDMs and pMACs
The majority of research on the links between macrophage metabolism and effector phenotype has come from the study of BMDMs. The BMDM system is very powerful because large numbers of homogeneous cells can be generated for functional metabolism assays and metabolomics. However, macrophage heterogeneity is more complex than previously appreciated leading to questions about how findings in BMDMs translate to other macrophage subtypes. pMACs are another commonly used primary cell system to investigate macrophage function ex vivo. These macrophages are monocyte-derived cells that differentiate into macrophages in vivo [20]. After stimulation with LPS pMACs are ~ 90% similar to BMDMs at the gene expression level [21]. Moreover, both subtypes of macrophages release high levels of inflammatory cytokines, become microbicidal, and take a “CAM” like phenotype following TLR4 activation. However, despite the large amount of data in the literature using these distinct macrophage subtypes, no direct comparison of their metabolic phenotypes has been reported. Recently, studies conducted with LPS-activated pMACs indicate that despite sharing many functional attributes with BMDMs, these cells differ dramatically in their mitochondrial phenotype [22, 23]. Understanding how and why these macrophage subtypes have divergent metabolic responses downstream of a common activation signal creates a unique opportunity to gain new insights into the links between metabolic reprogramming and cell function. In the following section we will discuss the metabolic comparison of CAMs generated from BMDMs or pMACs as an example of how the study of metabolic phenotypes from macrophages of different origin can shed light onto the intersection of inflammation and metabolism.
3.2 Metabolic Reprogramming in classically activated BMDMs and pMACs
The immediate metabolic response to TLR4 stimulation in both BMDMs and pMACs is to upregulate glycolysis (Fig. 1 A, C) [17, 24]. The functions of enhanced glycolytic flux include the synthesis of building blocks for lipids, nucleic acids, and proteins in addition to the generation of ATP. At the same time, mitochondrial respiration is suppressed in LPS activated BMDMs and this is thought to be a consequence of the broken TCA cycle (Fig. 1B) [5, 17]. Surprisingly, this feature is not conserved in pMACs where treatment with LPS increased mitochondrial OXPHOS (Fig. 1D) [22, 23]. The fact that BMDMs and pMACs have polar opposite mitochondrial phenotypes following LPS indicates that TCA cycle and OXPHOS reprogramming are uncoupled in these macrophage subtypes. This observation prompts several provocative questions: What is the mechanism that explains the divergent metabolic phenotypes? Is the TCA cycle still broken in pMACS? Since both macrophage subtypes are capable of generating a strong inflammatory response to LPS what is the role of mitochondrial OXPHOS in the inflammatory phenotype? How do the differences in mitochondrial respiration affect macrophage behavior? Although the answer to most of these questions will require additional investigation there are a few important considerations that will be discussed below.
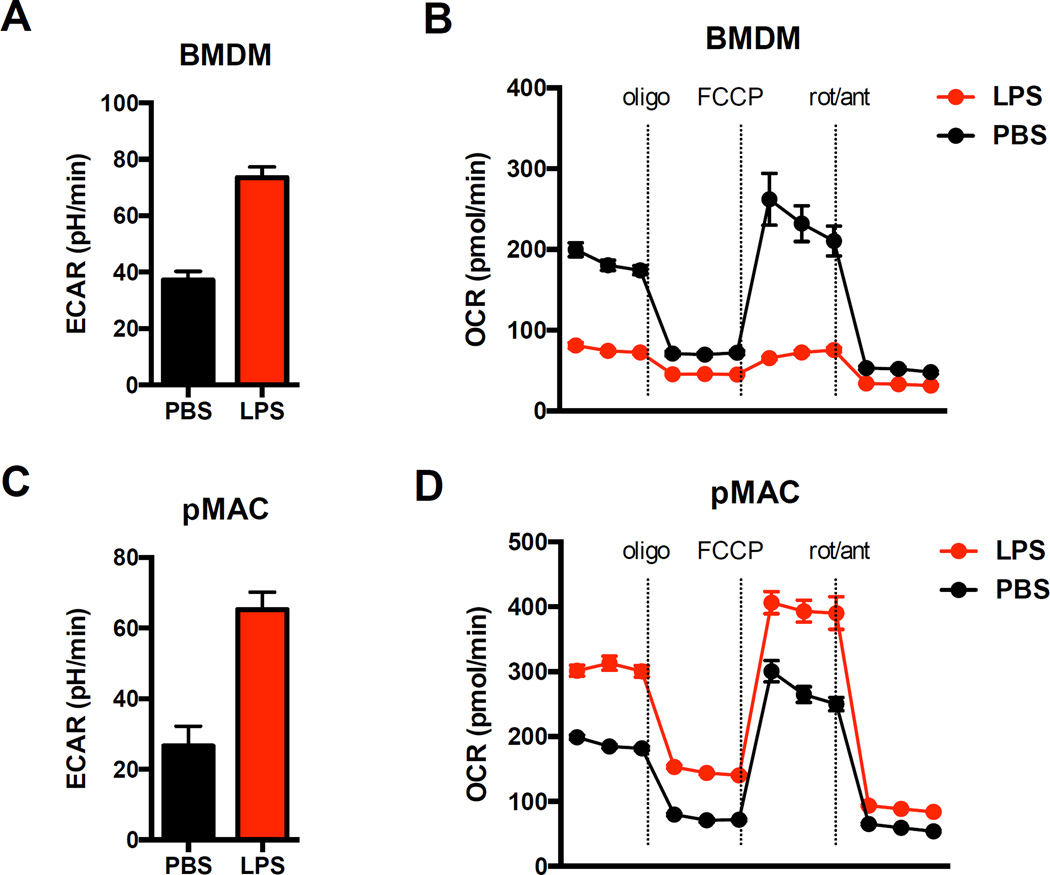
Figure 1. BMDMs and pMACs have divergent mitochondrial responses to LPS activation.
BMDMs or day 4 thioglycollate elicted peritoneal macrophages (pMACs) were plated into 96 well seahorse plates and mitochondrial function was compared using a mitochondrial stress test (oligomycin (10 µM) , FCCP (1.5 µM) , rotenone (100 nM)/antimycin (1µM)) on a seahorse flux analyzer. (A, C) Baseline ECAR was measured 16 h after stimulation with PBS (black) or LPS (red; 100 ng/ml) as an indicator of glycolysis. (B, D) Mitochondrial oxygen consumption (OCR) was quantified 16h after LPS injection at baseline and after the indicated injections. The key observation is that in response to LPS both macrophage types share glycolytic phenotypes but have profoundly different mitochondrial OCR with BMDMs showing suppression and pMACs displaying enhancement of mitochondrial respiration.
3.3 The broken TCA cycle
One possibility to explain the preservation of OXPHOS in pMACs is that unlike BMDMs the activity of IDH and/or SDH is not suppressed allowing for uninterrupted TCA cycle flux. To gain insight into this issue, we analyzed publically available gene expression data comparing LPS treated BMDMs to pMACs which revealed that Irg1 expression is similar at baseline and increases robustly in both macrophage subtypes following LPS treatment (Fig. 2A)[21]. Similarly, IDH expression was suppressed by LPS in pMACs and BMDMs alike, although baseline expression was higher in pMACs (Fig. 2B). Consistent with these gene expression changes succinate levels increased in pMACs after LPS treatment as did the citrate metabolites acetyl-CoA and malonyl-CoA (Fig. 2C). These observations support the concept that mitochondrial respiration remains intact despite evidence of a broken TCA cycle in pMACS. This observation suggests that the TCA cycle is replenished by anapleurotic flux from glutamine and the aspartate-argininosuccinate shunt (Fig. 2C).
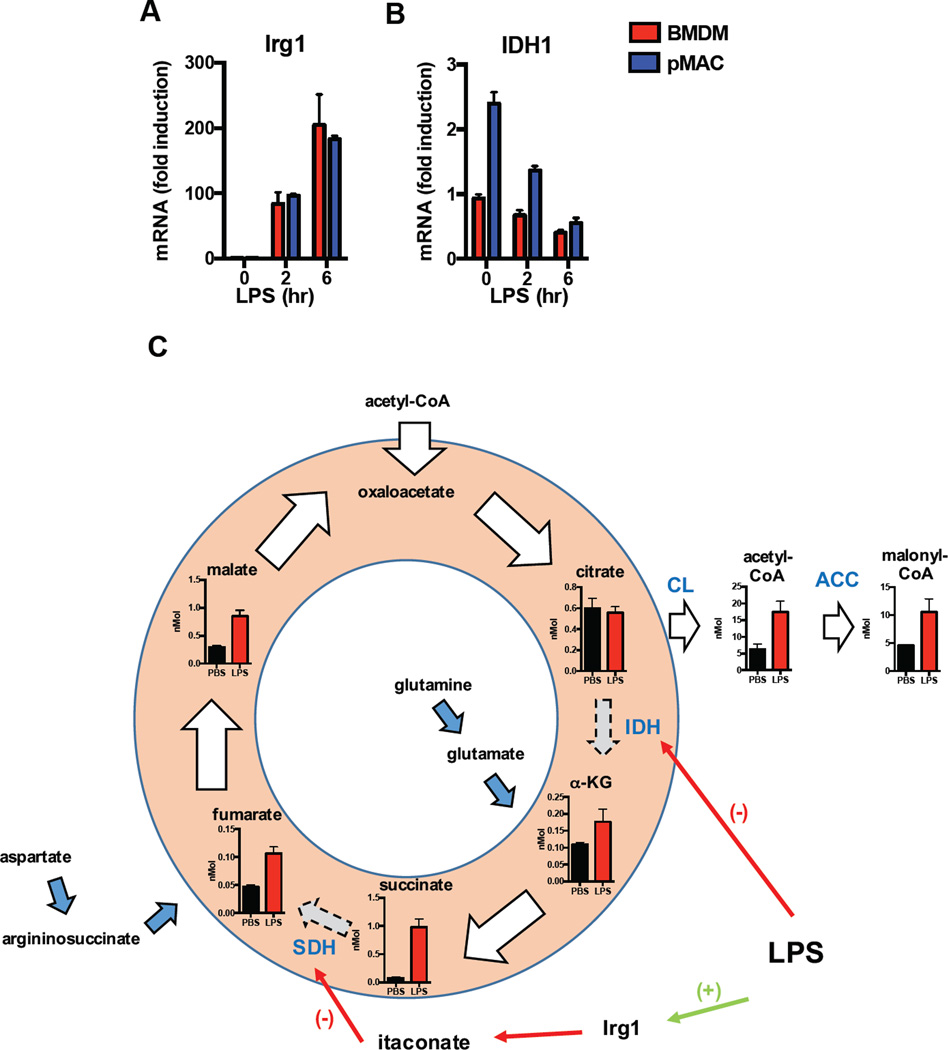
Figure 2. TCA reprogramming in LPS activated macrophages.
(A, B) Kinetic gene expression analysis for BMDMs (red bars) and pMACs (blue bars) treated with LPS were obtained from available previously published data (ref). The relative expression of Irg1 (A) and IDH (B) for these macrophage subtypes are shown. (C) Schematic diagram of the TCA cycle in pMACs with quantified levels of the indicated metabolites shown. Whole cell concentrations of TCA metabolites were generated from 2×106 pMACs treated with PBS (black bars) or LPS (red bars; 100 ng/ml) in triplicate for 16h. After stimulation, the cells were snap frozen in liquid nitrogen and TCA intermediates were quantified by LC-MS/MS at the Sanford Burnham Prebys metabolomics core (Lake Nona, USA). The enzymatic steps disrupted in the broken TCA cycle induced by LPS occur at IDH1 and SDH and these reactions are indicated by blue text. IDH is transcriptionally suppressed whereas SDH is inhibited by itaconate, a product of the enzyme Irg1. When flux though IDH is reduced, a greater proportion of citrate is diverted out of the mitochondria where it contributes to the cytosolic acetyl-CoA pool and the production of malonyl-CoA via citrate lyase (CL) and acetyl-CoA carboxylase (ACC). Anapleurotic flux into the TCA cycle from glutamine to α-ketoglutarate (α-KG) and the argininoosuccinate shunt to fumarate, which can maintain TCA function, are indicated with the blue arrows.
How might the divergent mitochondrial responses of these macrophage subtypes be explained? To understand this one must first consider the interplay between the TCA cycle and the ETC. In addition to its role in the TCA cycle, SDH is also known as complex II of the ETC where it feeds electrons generated by succinate oxidation to complex III via ubiquinone. Therefore, inhibition of SDH not only causes succinate to build up in the cell, but it also reduces electron flux through the ETC. This concept is illustrated by Irg1 knockout BMDMs where itaconate-mediated suppression of SDH is lost and as a consequence mitochondrial OXPHOS is enhanced, rather than suppressed, after LPS treatment [2]. The fact that Irg1 is strongly induced and succinate accumulates in both pMACs and BMDMs argues that SDH inhibition cannot fully explain the differences in mitochondrial phenotype. Together these observations argue that pMACs must continue to feed reducing equivalents to complex I of the ETC whereas BMDMs do not. How this occurs and what affect it has on macrophage function are not known.
3.4 Nitric oxide and the electron transport chain
One of the well described inhibitors of complex I is nitric oxide (NO). NO is produced by the enzyme iNOS which is induced by TLR activation [25]. To evaluate whether the balance of NO could contribute to the mitochondrial differences between BMDM and pMACs we reanalyzed gene expression data from Schroder et al which revealed delayed induction of iNOS transcript in pMACs compared to BMDMs (Fig 3A) [21]. NO production is also influenced by the activity of arginase, an enzyme which competes with iNOS for arginine and generates ornithine [26]. Kinetic analysis of arginase expression after LPS stimulation revealed that following an initial decline, its mRNA abundance actually increases compared to baseline by 24h in both BMDMs and pMACs. However, the more striking finding was that arginase expression at baseline was significantly increased in pMACs compared to BMDMs, a finding that has been seen by others [25]. In line with this, metabolite analysis of pMACs revealed a significant increase in ornithine levels after LPS treatment (Fig. 3C). The delayed induction of iNOS in combination with increased expression of arginase would be expected to suppress NO generation in pMACs and lessen the inhibition of the ETC (Fig. 3D). Therefore, the differences in mitochondrial respiration between pMACs and BMDMs appear to result from differential suppression of the ETC by NO rather than the broken TCA cycle per se. A similar effect of NO has been described with bone marrow derived DCs where inhibition or deletion of iNOS reverses the suppression of mitochondrial OXPHOS by LPS [27] . Of importance, human macrophages also do not produce significant NO and like pMACS they have increased mitochondrial respiration following LPS stimulation [21, 28]. Further investigation is warranted to dissect the role of reactive nitrogen and oxygen species on mitochondrial function in diverse macrophage subtypes across species.
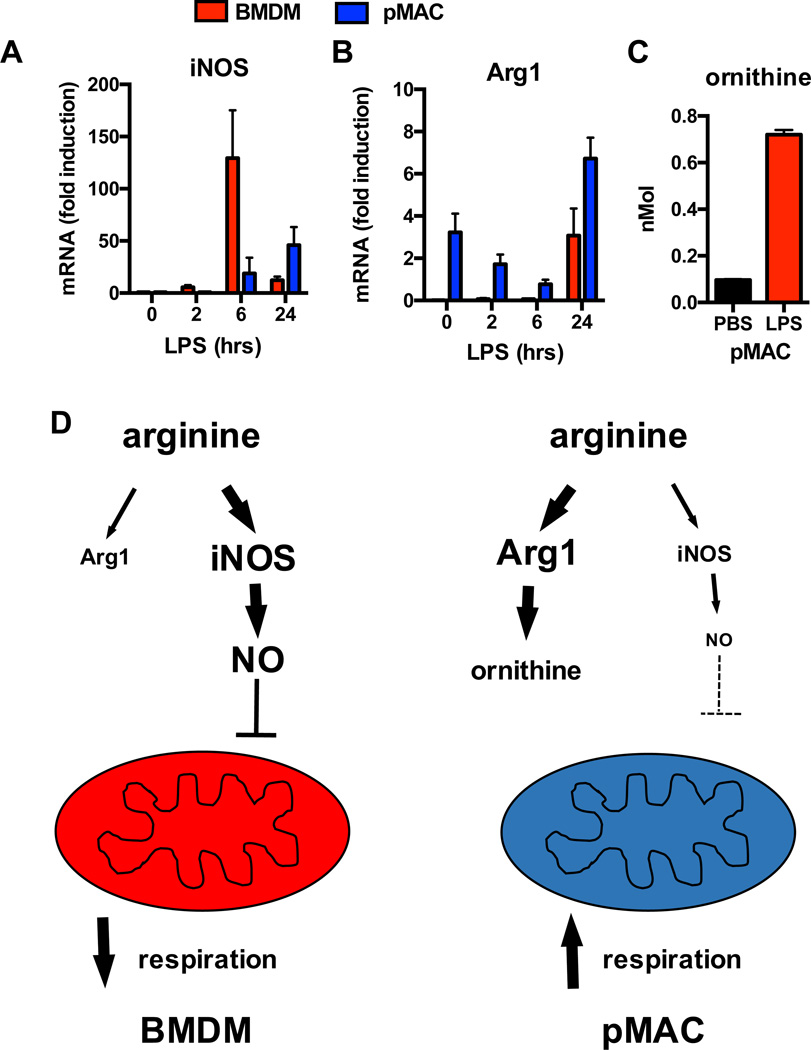
Figure 3. Nitric oxide in the suppression of macrophage mitochondrial respiration.
(A, B) Kinetic gene expression of iNOS and arginase 1 (Arg1) in BMDMs (red bars) and pMACs (blue bars) illustrates delayed induction of iNOS and higher expression of Arg1 in pMACs. (C) Consistent with increased arginase activity, pMACs demonstrate a significant increase in ornithine production upon LPS activation. (D) This profile suggests a model in which arginine metabolism favors iNOS in BMDMs and Arg1 in pMACs. The net effect of this shift is to reduce NO release and increased mitochondrial respiration in LPS stimulated pMACs, whereas high level NO production in BMDMs suppresses mitochondrial function.
The comparison of metabolic phenotypes between pMACs and BMDMs serves as an illustrative example currently in the literature of how macrophages from different origins can have unique metabolic responses to similar stimuli. The divergent mitochondrial phenotypes observed with these macrophage subtypes also informs our understanding of how specific metabolic pathways relate to inflammatory function. For example, enhanced mitochondrial respiration has generally been associated with anti-inflammatory macrophage phenotypes. However, even with an oxidative mitochondrial phenotype pMACS produce inflammatory cytokines like TNFα, IL-6, and IL-1β, often exceeding levels seen with BMDMs [29]. This profile is also exemplified by other immune cell types upon TLR activation [30]. Altogether these observations have several important implications: 1) The broken TCA cycle can be sustained by anapleurotic flux allowing for continued delivery of reducing equivalents to the mitochondrial ETC 2) mitochondrial respiration is not required for the CAM phenotype, but can augment cytokine release 3) mitochondrial OXPHOS is not uniformly anti-inflammatory and therefore other signaling cues or upstream substrate choices must direct macrophage fate 4) baseline differences in gene expression can prime cells towards particular metabolic responses in response to activation. This last point is of particular importance and will be the subject of the remainder of this article. Gene expression data is readily obtainable from macrophages of various origins in vivo and has the potential to provide significant insight into metabolic diversity. Therefore, we will explore baseline metabolic priming first using the example discussed above with pMACs and BMDMs and then in more diverse populations of macrophages from different tissues.
4. Metabolic gene expression and macrophage origin
4.1 Metabolic transcriptional signatures of BMDMs and pMACs
To understand the variation in metabolic gene expression networks between macrophage populations we have reviewed and analyzed Immgen data profiling macrophages in various tissues [31] using network analysis approaches developed in our group [5, 32, 33]. First, we have compared metabolic networks in the non-activated bone-marrow macrophages against non-activated peritoneal macrophages combining all the types of peritoneal macrophages available in Immgen. As Fig. 4 shows, a number of pathways are consistently regulated in a differential fashion between BMDMs and pMACs. Specifically, iron regulation (heme biosynthesis), glycolysic enzyme expression, serine biosynthesis, glutathione and folate metabolism, the latter three forming 1-carbon metabolism [34] show distinctly different patterns of expression between two types of macrophages.
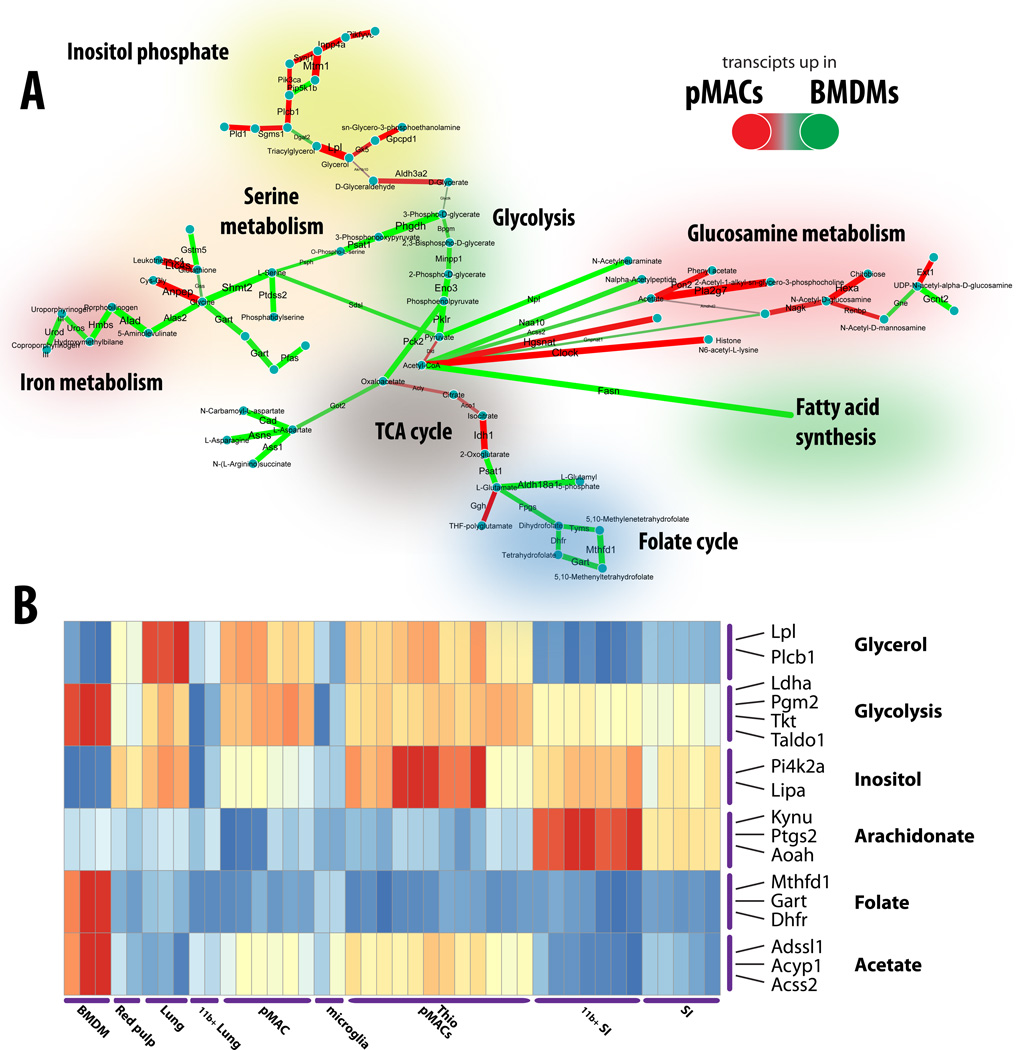
Figure 4. Metabolic-transcriptional profiling of diverse tissue macrophages.
(A) Metabo-transcriptional network representing differences between BMDM and pMACs based on the gene expression data from Immgen Consortium. Edges are colored according to specificity of enzyme expression – green implies upregulation of corresponding enzymes in BMDMs, and red – in pMACs. (B) Metabo-transcriptional clustering of the multiple resident macrophage types profiled in Immgen reveals the metabolic modules differentially regulated in different tissue macrophages.
A number of pathways were specific to pMACs. Strikingly, inositol phosphate metabolism/Phosphatidylinositol signaling system was more active at the transcriptional level in pMACs (Fig. 4A). Furthermore, acetyl-CoA metabolism appeared as one of the transcriptional marks differentiating between two types of macrophages:while BMDMs direct this molecule towards fatty acid synthesis (based on gene expression data), peritoneal macrophages utilize it more actively in TCA cycle (see increased expression of genes of the TCA cycle connecting pyruvate to 2-oxoglutarate (AKG): Dld, Acly, Aco1, Idh1). Additional features that discriminated the metabolism of two macrophage types includedhistone acetylation (through upregulated Clock), and glucosamine metabolism. Extension of these observations using functional metabolomics will be important to validate these findings and to determine the functional impacts of these differences.
4.2 Metabolic transcriptional signatures in diverse tissue macrophages
This approach can be extended to analysis of multiple subsets of macrophages to yield a global picture of the differential metabolism at baseline (Fig. 4B). Such analysis shows a number of distinct features corresponding to specific tissue macrophages. First, microglia and CD11b+ lung macrophages appear to have low metabolic activity at their basal state. At the same time, macrophages purified from small intestine show increased expression of a metabolic associated with cholesterol biosynthesis. Lung resident macrophages (CD11chigh) maintain active glycerophospholipid metabolism relative to other resident macrophages. Finally, inositol phosphate metabolism remains a characteristic feature of the peritoneal macrophages, while bone marrow macrophages are enriched in iron metabolism, glycolysis and 1-carbon metabolism genes.
It is feasible therefore, that basal differences in macrophage metabolism might define the specific trajectory of metabolic remodeling that is required to satisfy the needs of macrophage activation in the context of adaption to environment, production of toxic intermediates (e.g. ROS) and fulfill the immune activation program. Future research into these differences will be necessary to unravel the complex relationship between baseline metabolic programming and macrophage responses.
5. Conclusion
The last decade has seen an explosion of knowledge in the fields of macrophage immunometabolism and macrophage ontogeny/diversity. However, our understanding of how these disciplines intersect is limited. In this article, we begin to explore this interplay by using data from the study of BMDMs and pMACs to provide evidence that metabolic diversity exists in macrophages of disparate origins. This discussion provided proof-of-concept evidence that unique metabolic programs can be induced even within the well-defined system of LPS-induced macrophage activation. Moreover, comparing these macrophage subtypes also revealed that crosstalk between the TCA cycle and mitochondrial respiration in inflammatory macrophages may be more complicated than previously thought. This theme was expanded by gene expression data that demonstrated significant variability in basal metabolic programming observed between macrophages from distinct tissues, further illustrating the ability of tissue environment to influence macrophage metabolic responses. One of the major challenges of the next decade will be to further dissect the complicated relationships that exist between immunometabolic phenotype and macrophage localization and origin, including across species. This should be an area of emphasis for future research in immunometabolism.
Acknowledgments
Funding: This work was support by the National Institute of Health (RO1 DK11003401 to JDS) and the Diabetes Research Center (P30 DK02057937 to JDS).
Abbreviations
- BMDMs
bone marrow derived macrophages
- pMACs
elicted peritoneal macrophages
- TCA cycle
tricarboxylic acid cycle
- CAM
classically activated macrophage
- AAM
alternatively activated macrophage
- OXPHOS
oxidative phosphorylation
- ETC
electron transport chain
- TLR
toll like receptor
- NO
nitric oxide
References
- 1.Loftus RM, Finlay DK. Immunometabolism: Cellular Metabolism Turns Immune Regulator. The Journal of biological chemistry. 2016;291:1–10. doi: 10.1074/jbc.R115.693903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lampropoulou V, et al. Itaconate Links Inhibition of Succinate Dehydrogenase with Macrophage Metabolic Remodeling and Regulation of Inflammation. Cell metabolism. 2016;24:158–166. doi: 10.1016/j.cmet.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Neill LA, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. The Journal of experimental medicine. 2016;213:15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Odegaard JI, Chawla A. Alternative macrophage activation and metabolism. Annual review of pathology. 2011;6:275–297. doi: 10.1146/annurev-pathol-011110-130138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jha AK, et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. 2015;42:419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Epelman S, et al. Origin and functions of tissue macrophages. Immunity. 2014;41:21–35. doi: 10.1016/j.immuni.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corcoran SE, O'Neill LA. HIF1alpha and metabolic reprogramming in inflammation. The Journal of clinical investigation. 2016 doi: 10.1172/JCI84431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mills EL, O'Neill LA. Reprogramming mitochondrial metabolism in macrophages as an anti-inflammatory signal. European journal of immunology. 2016;46:13–21. doi: 10.1002/eji.201445427. [DOI] [PubMed] [Google Scholar]
- 9.Namgaladze D, Brune B. Macrophage fatty acid oxidation and its roles in macrophage polarization and fatty acid-induced inflammation. Biochimica et biophysica acta. 2016;1861:1796–1807. doi: 10.1016/j.bbalip.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Palsson-McDermott EM, O'Neill LA. The Warburg effect then and now: from cancer to inflammatory diseases. BioEssays : news and reviews in molecular, cellular and developmental biology. 2013;35:965–973. doi: 10.1002/bies.201300084. [DOI] [PubMed] [Google Scholar]
- 11.Everts B, et al. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKvarepsilon supports the anabolic demands of dendritic cell activation. Nature immunology. 2014;15:323–332. doi: 10.1038/ni.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nemeth B, et al. Abolition of mitochondrial substrate-level phosphorylation by itaconic acid produced by LPS-induced Irg1 expression in cells of murine macrophage lineage. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2016;30:286–300. doi: 10.1096/fj.15-279398. [DOI] [PubMed] [Google Scholar]
- 13.Strelko CL, et al. Itaconic acid is a mammalian metabolite induced during macrophage activation. Journal of the American Chemical Society. 2011;133:16386–16389. doi: 10.1021/ja2070889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michelucci A, et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7820–7825. doi: 10.1073/pnas.1218599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammerer F, et al. Small Molecule Restores Itaconate Sensitivity in Salmonella enterica: A Potential New Approach to Treating Bacterial Infections. Chembiochem : a European journal of chemical biology. 2016;17:1513–1517. doi: 10.1002/cbic.201600078. [DOI] [PubMed] [Google Scholar]
- 16.Tretter L, et al. Succinate, an intermediate in metabolism, signal transduction, ROS, hypoxia, and tumorigenesis. Biochimica et biophysica acta. 2016;1857:1086–1101. doi: 10.1016/j.bbabio.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 17.Tannahill GM, et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brancato SK, Albina JE. Wound macrophages as key regulators of repair: origin, phenotype, and function. The American journal of pathology. 2011;178:19–25. doi: 10.1016/j.ajpath.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sager HB, et al. Proliferation and Recruitment Contribute to Myocardial Macrophage Expansion in Chronic Heart Failure. Circulation research. 2016;119:853–864. doi: 10.1161/CIRCRESAHA.116.309001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gautier EL, et al. Systemic analysis of PPARgamma in mouse macrophage populations reveals marked diversity in expression with critical roles in resolution of inflammation and airway immunity. Journal of immunology. 2012;189:2614–2624. doi: 10.4049/jimmunol.1200495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schroder K, et al. Conservation and divergence in Toll-like receptor 4-regulated gene expression in primary human versus mouse macrophages. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E944–E953. doi: 10.1073/pnas.1110156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He L, et al. Inhibition of mTOR reduces lipotoxic cell death in primary macrophages through an autophagy-independent mechanism. Journal of leukocyte biology. 2016 doi: 10.1189/jlb.3A1015-463R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He L, et al. Glutamine Modulates Macrophage Lipotoxicity. Nutrients. 2016;8:215. doi: 10.3390/nu8040215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez-Prados JC, et al. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. Journal of immunology. 2010;185:605–614. doi: 10.4049/jimmunol.0901698. [DOI] [PubMed] [Google Scholar]
- 25.Takeda N, et al. Differential activation and antagonistic function of HIF-{alpha} isoforms in macrophages are essential for NO homeostasis. Genes & development. 2010;24:491–501. doi: 10.1101/gad.1881410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rutschman R, et al. Cutting edge: Stat6-dependent substrate depletion regulates nitric oxide production. Journal of immunology. 2001;166:2173–2177. doi: 10.4049/jimmunol.166.4.2173. [DOI] [PubMed] [Google Scholar]
- 27.Krawczyk CM, et al. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood. 2010;115:4742–4749. doi: 10.1182/blood-2009-10-249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu TF, et al. NAD+-dependent sirtuin 1 and 6 proteins coordinate a switch from glucose to fatty acid oxidation during the acute inflammatory response. The Journal of biological chemistry. 2012;287:25758–25769. doi: 10.1074/jbc.M112.362343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber K, Schilling JD. Distinct lysosome phenotypes influence inflammatory function in peritoneal and bone marrow-derived macrophages. International journal of inflammation. 2014;2014:154936. doi: 10.1155/2014/154936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu D, et al. Type 1 Interferons Induce Changes in Core Metabolism that Are Critical for Immune Function. Immunity. 2016;44:1325–1336. doi: 10.1016/j.immuni.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gautier EL, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nature immunology. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sergushichev AA, et al. GAM: a web-service for integrated transcriptional and metabolic network analysis. Nucleic acids research. 2016;44:W194–W200. doi: 10.1093/nar/gkw266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vincent EE, et al. Mitochondrial Phosphoenolpyruvate Carboxykinase Regulates Metabolic Adaptation and Enables Glucose-Independent Tumor Growth. Molecular cell. 2015;60:195–207. doi: 10.1016/j.molcel.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 34.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nature reviews. Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]