Abstract
Here, we identify the LIM protein lipoma-preferred partner (LPP) as a binding partner of a specific protein phosphatase 2A (PP2A) heterotrimer that is characterised by the regulatory PR130/B″α1 subunit (encoded by PPP2R3A). The PR130 subunit interacts with the LIM domains of LPP through a conserved Zn2+-finger-like motif in the differentially spliced N-terminus of PR130. Isolated LPP-associated PP2A complexes are catalytically active. PR130 colocalises with LPP at multiple locations within cells, including focal contacts, but is specifically excluded from mature focal adhesions, where LPP is still present. An LPP–PR130 fusion protein only localises to focal adhesions upon deletion of the domain of PR130 that binds to the PP2A catalytic subunit (PP2A/C), suggesting that PR130–LPP complex formation is dynamic and that permanent recruitment of PP2A activity might be unfavourable for focal adhesion maturation. Accordingly, siRNA-mediated knockdown of PR130 increases adhesion of HT1080 fibrosarcoma cells onto collagen I and decreases their migration in scratch wound and Transwell assays. Complex formation with LPP is mandatory for these PR130-PP2A functions, as neither phenotype can be rescued by re-expression of a PR130 mutant that no longer binds to LPP. Our data highlight the importance of specific, locally recruited PP2A complexes in cell adhesion and migration dynamics.
Keywords: Cell migration, Cell adhesion, LIM domain, Lipoma-preferred partner, Protein phosphatase 2A, Zn2+-finger
Introduction
Cell migration is a dynamic process requiring integration and coordination of many cellular events, including the establishment of cell polarity, the regulation of dynamic actin and microtubule polymerisation, and the dynamic regulation of cell–cell and cell–substratum interactions (Ridley et al., 2003; Ridley, 2011). In all of these spatially and temporally controlled signalling processes, protein phosphorylation – on tyrosine as well as on serine or threonine (Ser/Thr) residues – plays a vital regulatory role. Although the involvement and manipulation of specific, locally recruited Ser/Thr protein kinases has been well documented (Huang et al., 2004; Boeckeler et al., 2010; Rossé et al., 2010; Howe, 2011; McDonald, 2014), it is much less understood whether the localised action of counteracting Ser/Thr phosphatases is important in these events.
Protein phosphatase 2A (PP2A) is a ubiquitously expressed phosphatase with a complex structure and regulation that is increasingly coming into focus as a tumour suppressor (Janssens et al., 2005; Westermarck and Hahn, 2008) and novel anti-cancer target (Perrotti and Neviani, 2013). Currently, relatively little is known about the role of specific PP2A complexes in control of cytoskeletal dynamics, (cancer) cell adhesion, migration or invasion (Sontag and Sontag, 2006; Basu, 2011). Through the use of okadaic acid, a phosphatase inhibitor affecting all PP2A holoenzymes (Lambrecht et al., 2013), it has been shown that PP2A can restrict migration of various cell types, in line with its tumour suppressor function (Maier et al., 1995; Young et al., 2002; Pullar et al., 2003). By contrast, other studies have reported impaired cell motility upon pharmacologic inhibition of PP2A, rather suggesting a pro-migratory role for PP2A (Basu et al., 2007; Wu et al., 2014). Although these inhibitor studies are informative, it is difficult to draw specific conclusions about the in vivo roles of PP2A because the global suppression of phosphatase activity affects many cellular processes, and can result in indirect and even opposing effects. Dissecting the in vivo roles of PP2A requires detailed molecular analyses of the contexts into which the phosphatase is drawn through the particular targeting and regulatory devices that comprise the plethora of PP2A complexes.
PP2A enzymes typically exist as trimers comprising a catalytic C subunit, a structural A subunit and a variable regulatory B-type subunit (Fig. 1A). Regulation occurs through the interaction of the catalytic subunit – through the A subunit – with these regulatory subunits, which act as targeting and/or substrate-specifying entities (Janssens and Goris, 2001; Lambrecht et al., 2013). PR72 (B″α2) and PR130 (B″α1) belong to the B″-family of PP2A regulatory subunits (Fig. 1A), whose physiological roles are still poorly understood. These particular B″ subunits are derived from the same gene (PPP2R3A) through differential splicing (Fig. 1A); they harbour specific N-terminal domains of 45 and 664 amino acids that are encoded by alternatively spliced exons, and have identical C-termini of 486 amino acids (Hendrix et al., 1993; Stevens et al., 2003; Zwaenepoel et al., 2008). The common C-terminus contains two A-subunit-interaction domains (Li and Virshup, 2002), a conserved hydrophobic motif (Davis et al., 2008) and two Ca2+-binding EF-hands, which mediate Ca2+-dependent changes in phosphatase activity and provide an important interaction interface with the A subunit (Janssens et al., 2003; Ahn et al., 2007; Park et al., 2013). Recent structural studies on the related PR70 (B″β1) subunit (encoded by PPP2R3B) have largely confirmed these biochemical data (Wlodarchak et al., 2013; Dovega et al., 2014). Functionally, it has been shown that although PR72 and PR130 both interact with the Wnt antagonist Naked cuticle (Nkd), PR72 represses canonical Wnt signalling through this interaction (Creyghton et al., 2005), whereas PR130 rather restricts the ability of Nkd to function as a Wnt inhibitor and thus promotes canonical Wnt signalling (Creyghton et al., 2006). It has therefore been suggested that these opposing cellular functions are likely to be mediated by their specific N-terminal domains (Creyghton et al., 2006). In contrast, the interaction of PR130 with the inositol polyphosphate 5-phosphatase SHIP2 occurs through the 486-amino-acid common domain, and positively contributes to EGF-mediated signalling by preventing EGF-receptor degradation (Zwaenepoel et al., 2010). Thus, the few molecular data on PR130 cellular function, so far, have merely suggested a proto-oncogenic role for the PR130-PP2A holoenzyme.
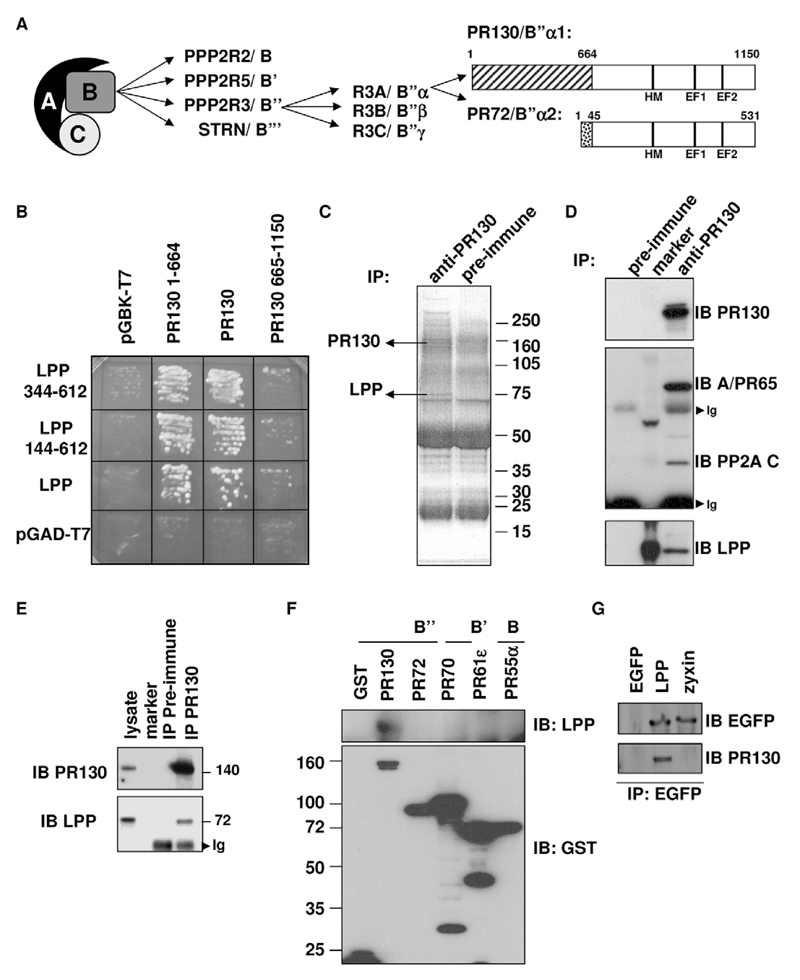
Fig. 1. PP2A PR130/B″α1 forms an endogenous complex with LPP.
(A) Scheme of PP2A holoenzyme structure, and the domain structure of PR130/B″α1 and PR72/B″α2, two splice variants encoded by PPP2R3A. Together with PR70/B″β1 and PR70/B″β2 (both encoded by PPP2R3B) and G5PR/B″γ (encoded by PPP2R3C), these subunits constitute the human B″ subunits of PP2A (Zwaenepoel et al., 2008). Interaction with the A (and C) subunit is mediated by the shared 486-amino-acid C-terminal domain that contains a hydrophobic motif (HM) and two Ca2+-binding EF-hands (EF1 and EF2). (B) Validation of the interactions identified by yeast two-hybrid screening of a human testis library with PR130 (residues 1–664) as bait. The AH109 yeast strain was retransformed with the indicated plasmids and plated on growth medium without Leu and Trp for selection of double transformants. One colony of each condition was replated on medium lacking Leu, Trp, His and adenine to assay for expression of the HIS and ADE reporter genes. (C) Mass spectroscopic fingerprinting from co-immunoprecipitating proteins in anti-PR130 immunoprecipitates (IPs) from NIH3T3 cell lysates. Immunoprecipitation with the pre-immune serum served as negative control. SDS-PAGE-separated proteins were stained with Coomassie Brilliant Blue. (D) Co-immunoprecipitation of endogenous PR130, LPP, the PP2A A subunit PR65 (encoded by PPP2R1A) and C subunit from NIH3T3 cell lysates. PR130 was immunoprecipitated with anti-PR130 antibodies, and the presence of LPP, the PP2A A subunit PR65 and the C subunit in the immunoprecipitates was evaluated by immunoblotting (IB). Ig, immunoglobulins. (E) Co-immunoprecipitation of endogenous PR130 and LPP from HT1080 cell lysates. PR130 was immunoprecipitated with anti-PR130 antibodies, and the presence of LPP in the immunoprecipitates was revealed by immunoblotting (IB). Ig, immunoglobulins. (F) Specific interaction between LPP and the PR130/B″α1 (PR130) subunit of PP2A. GST, GST-tagged PP2A subunit representatives of the PR55/B (α isoform, PR55α), PR61/B′(ε isoform, PR61ε) and PR72/B″ families (PR70/B″β1 isoform, PR70), GST-tagged PR130/B″α1 (PR130) and the closely related PPP2R3A splice variant PR72/B″α2 (PR72) were ectopically expressed in COS7 cells. Following GST pull down, co-precipitating LPP was visualised by immunoblotting (IB). (G) No interaction of PR130 with zyxin, a LIM-domain protein that is closely related to LPP. EGFP, EGFP-tagged LPP and EGFP-tagged zyxin were ectopically expressed in COS7 cells and immunoprecipitated with anti-EGFP antibodies. The presence of co-immunoprecipitating PR130 was visualised by immunoblotting (IB).
By exploiting the specific PR130 N-terminus as bait in a yeast two-hybrid screen, we now describe a new cellular complex comprising PR130-PP2A and the focal adhesion protein lipoma-preferred partner (LPP) that appears to be functionally important in the control of (cancer) cell adhesion and migration. Our data highlight the importance of specific, locally recruited trimeric PP2A complexes in cell adhesion and migration dynamics.
Results
Identification of LPP as a cellular PR130-binding partner
To obtain insight into the poorly established physiological functions and substrates of the PR130-PP2A holoenzyme, we performed a yeast two-hybrid screen exploiting the unique PR130-specific N-terminus (PR130 amino acids 1–664) as bait. We identified five independent N-terminally-truncated clones of LPP (Petit et al., 1996) starting at amino acid residues 144, 146, 309, 314 and 344. We re-tested both the shortest (LPP 344–612) and the longest of these clones (LPP 144–612), together with full-length LPP (1–612) and confirmed the interaction with LPP, both for full-length PR130 and its specific N-terminal domain (PR130 1–664) (Fig. 1B). To validate this observation on endogenous proteins, we used a PR130-specific antibody (Zwaenepoel et al., 2008) and identified the co-immunoprecipitating proteins using mass spectroscopy. Three different LPP peptides (Materials and Methods) were unambiguously identified from a specific co-precipitating protein with an apparent molecular mass of 75 kDa (Fig. 1C). To confirm these data, we counter-stained immunoprecipitates that had been isolated with an antibody against PR130 from NIH3T3 cells with a specific LPP antibody, revealing LPP immunoreactivity (Fig. 1D). Higher stringency washes of these immunoprecipitates (increasing NaCl concentrations up to 600 mM) could not completely disrupt the complex, suggesting that binding is strong (results not shown). The complex could also be identified in HT1080 (Fig. 1E) and COS cells (results not shown), indicating that complex formation is not cell type-specific. By contrast, LPP failed to interact with other PP2A B-type subunits from the same subclass (PR72/B″α2 and PR70/B″β1) or other subclasses (PR55/Bα and PR61/B′ε, encoded by PPP2R2A and PPP2R5E, respectively) (Fig. 1F), indicating that the interaction of LPP with PP2A is specific for PR130. Similarly, PR130 did not interact with zyxin, a closely related LPP LIM-protein-family member (Fig. 1G).
PR130 interacts with the LIM domains of LPP through a cysteine-rich Zn2+-finger-like domain
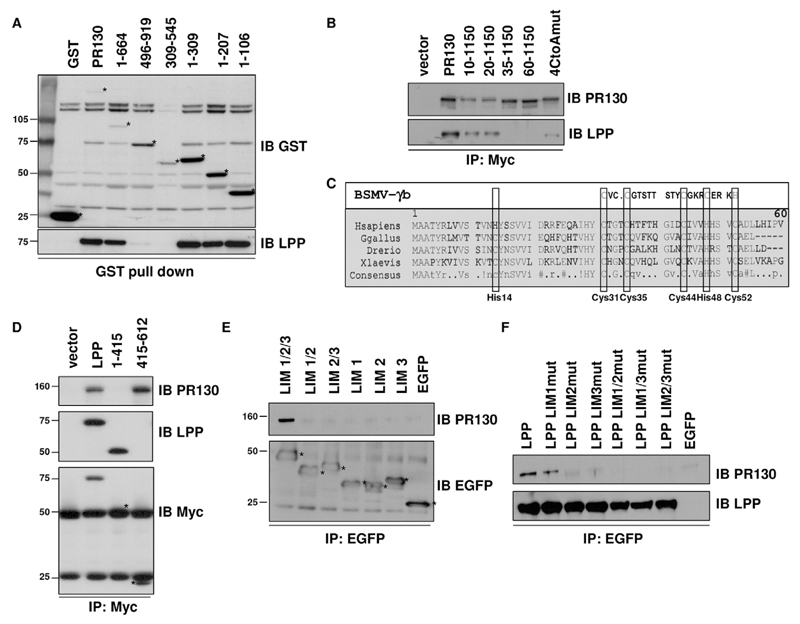
The LPP-interaction domain within PR130 was mapped to the N-terminus (1–106) (Fig. 2A). Deletion of the first 34 or 59 amino acids of PR130 abolished the interaction with LPP, whereas deletion of the first 9 or 19 amino acids decreased the LPP interaction only in relation to the decrease in recovery of the truncated PR130 protein itself (Fig. 2B). An alignment of the PR130 N-terminal region from different vertebrates revealed a high overall degree of conservation, especially within the first 55 amino acids, where several conserved cysteine and histidine residues are present. This cysteine-rich domain shows homology with a known C1-like Zn2+-finger domain of the barley stripe mosaic viral γb protein (Bragg et al., 2004) (Fig. 2C). Alanine substitutions of all cysteine residues within the C1-like domain (Cys31, Cys35, Cys44 and Cys52) severely diminished the interaction with LPP (Fig. 2B), consistent with a functional role of this domain in mediating LPP binding.
Fig. 2. PR130 interacts with the LIM domains of LPP through a conserved cysteine-rich Zn2+-finger-like domain.
(A) Mapping the LPP-interacting domain within PR130, through GST pull down from COS7 cells overexpressing GST fusion proteins of different PR130 deletions. The presence of endogenous LPP in the pull downs was detected with LPP antibodies. Asterisks indicate the bands corresponding to the GST fusion proteins. (B) Sequential deletions and site-directed mutagenesis of the LPP-binding domain of PR130. C-terminally Myc-tagged PR130, PR130(10–1150), PR130(20–1150), PR130(35–1150), PR130(60–1150) and a PR130 4CtoA mutant in which Cys31, Cys35, Cys44 and Cys52 had been mutated into alanine residues, were expressed in COS7 cells. The presence of endogenous LPP in the anti-Myc immunoprecipitates (IP) was evaluated with anti-LPP antibodies. IB, immunoblot. (C) Alignment of the LPP interaction domain of PR130 proteins from different species, illustrating the evolutionary conservation within the first 60 amino acids, and the similarity with a C1-like domain of the BSMV γb protein. The conserved cysteine and histidine residues are boxed. (D) Mapping the PR130-binding domain within LPP, through anti-Myc immunoprecipitation from COS7 cells overexpressing C-terminally Myc-tagged LPP, the LPP non-LIM region (residues 1–415) or the LIM domains (residues 415–612). The presence of endogenous PR130 in the anti-Myc immunoprecipitates (IPs) was detected with anti-PR130 antibodies. LPP counter-staining was performed with LPP- and Myc-specific antibodies, because the LPP-specific antibody does not recognise the LPP LIM region. (E) Requirement of all three LIM domains for the interaction with PR130. N-terminally EGFP-tagged LPP(415–612) comprising the LIM1, LIM2 and LIM3 domains (LIM1/2/3); LPP(415–535) comprising the LIM1 and LIM2 domains (LIM 1/2); LPP(475–612) comprising the LIM2 and LIM3 domains (LIM 2/3), LPP(415–475) comprising the LIM1 domain (LIM1), LPP(475–535) comprising the LIM2 domain (LIM2); and LPP(535–612) comprising the LIM3 domain (LIM3) were overexpressed in COS7 cells. The presence of endogenous PR130 in the recovered anti-EGFP immunoprecipitates was detected with PR130-specific antibodies. (F) Differential effects of mutation of a single LPP LIM domain on the interaction with PR130. The structure of any single LPP LIM domain was disrupted by mutating four Zn2+-binding cysteine and histidine residues into alanine residues (Petit et al., 2003). N-terminally EGFP-tagged LPP, LPP-LIM1mut, LPP-LIM2mut, LPP-LIM3mut, LPP-LIM1/2mut, LPP-LIM1/3mut and LPP-LIM2/3mut were overexpressed in COS7cells, and the presence of endogenous PR130 was detected in the recovered EGFP immunoprecipitates using PR130-specific antibodies.
As supported by the yeast two-hybrid data, PR130 specifically interacted with the LPP LIM domains (residues 415–612), but not with the non-LIM region (residues 1–415) (Fig. 2D). LPP encompasses three LIM domains (Petit et al., 2003), and deletion of any LIM domain in the context of the isolated three LIM domains inhibited binding to PR130, suggesting that all LIM domains contribute to the interaction (Fig. 2E). We also generated LIM-domain mutants in the context of full-length LPP by changing four structurally important cysteine or histidine residues per LIM domain into alanine residues (Petit et al., 2003). Although the double LIM-domain mutants and two of the single-domain LIM mutants (LPP-LIM2mut and LPP-LIM3mut) failed to bind to PR130, the single LPP LIM1-domain mutant still retained some binding capacity (Fig. 2F). Thus, in the context of the complete LPP protein, the integrity of both the LIM2 and LIM3 domains is more important for the interaction with PR130 than the integrity of the LIM1 domain, although, in isolation, the LIM2 and LIM3 domains alone are insufficient for PR130 binding.
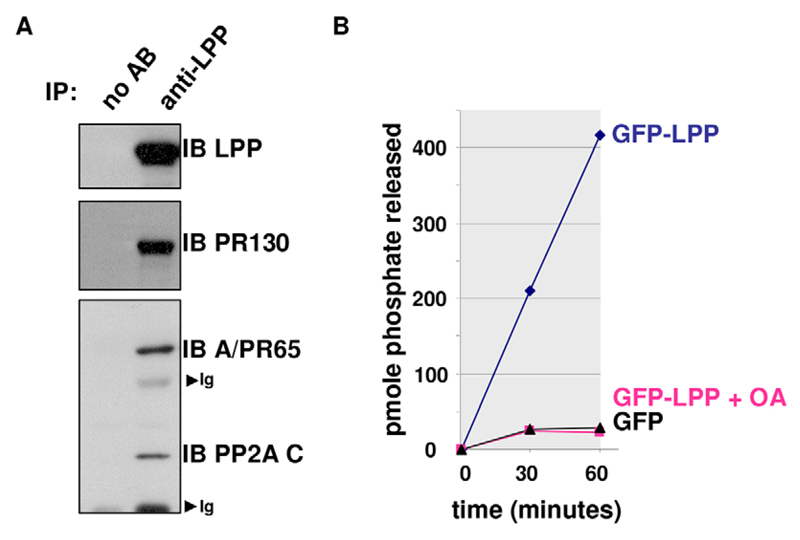
LPP binds to a catalytically competent PR130-containing PP2A trimer
To further characterise the endogenous LPP–PR130 complex, we performed the reciprocal immunoprecipitation with an antibody against LPP and revealed that, besides PR130, the PP2A A and C subunits (the catalytic subunit is referred to as PP2A/C) are also present in the complex (Fig. 3A). Therefore, a PP2A heterotrimer is bound to LPP. This LPP-associated PR130-PP2A holoenzyme proved catalytically competent, as okadaic-acid-sensitive phosphatase activity was measured in GFP-trapped complexes retrieved from GFP–LPP-expressing HEK293T cells, but not from cells expressing GFP alone (Fig. 3B).
Fig. 3. LPP associates with a catalytically competent PR130-PP2A holoenzyme.
(A) Co-immunoprecipitation of endogenous LPP, PR130, the PP2A A subunit PR65 (A/PR65) and C subunit (PP2A C) from NIH3T3 cell lysates. LPP was immunoprecipitated (IP) with anti-LPP antibodies and the presence of PR130, the PP2A A subunit PR65 and PP2A/C in the immunoprecipitates was evaluated by immunoblotting (IB). Ig, immunoglobulins; AB, antibody. (B) PP2A activity assay on GFP-trapped complexes from HEK293T cells expressing EGFP-tagged LPP or EGFP alone. The pmole number of phosphate released from the phosphopeptide (RRApTVA) was determined by using malachite green. Okadaic acid (10−8M; OA) was added as a specificity control. A representative assay is shown.
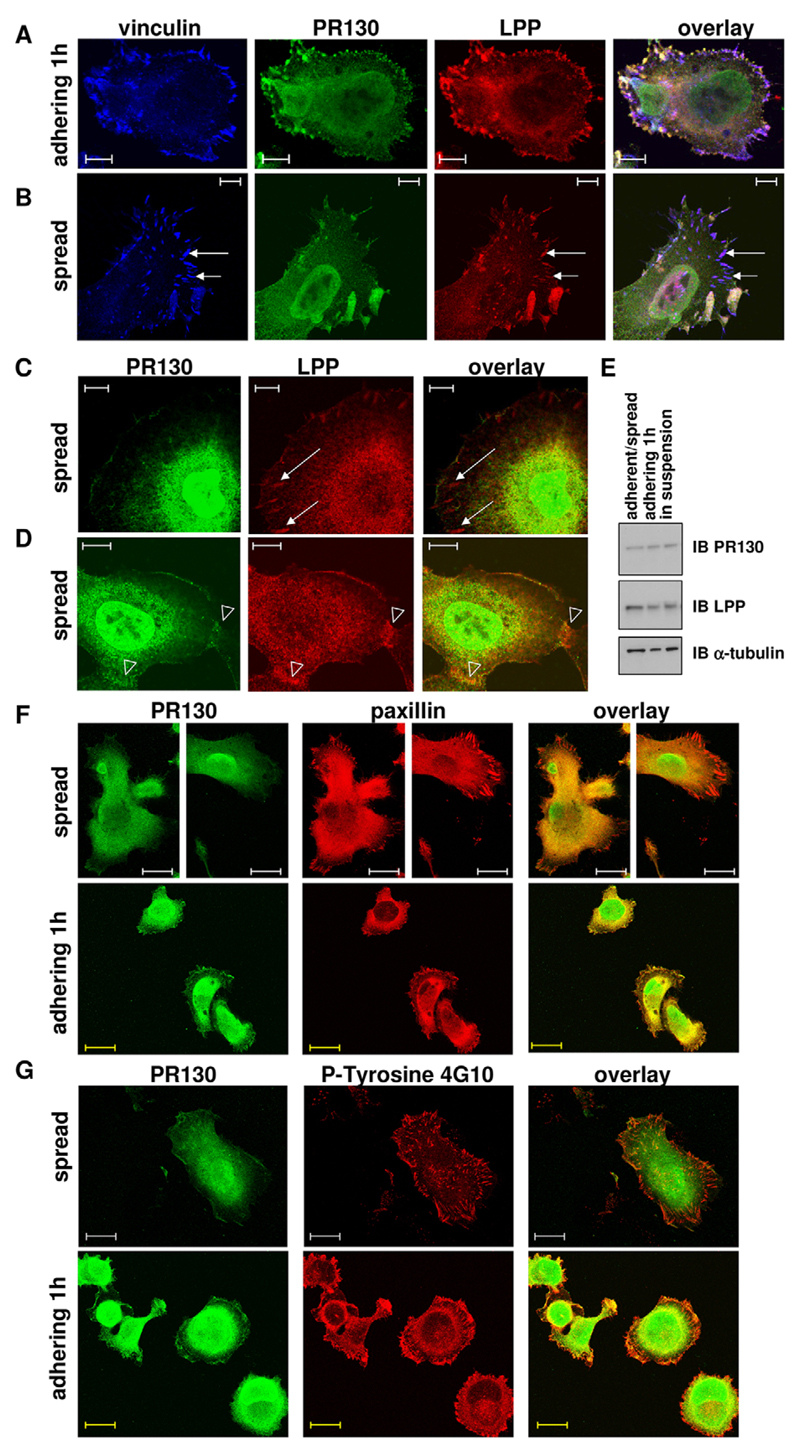
In contrast with LPP, PR130 is excluded from mature focal adhesions
We next determined the localisation of the endogenous LPP–PR130 complex by direct immunofluorescence (Fig. 4). It is well established that all three LIM domains of LPP cooperate to provide targeting to cell–cell and cell–matrix contacts, particularly to focal adhesions where LPP colocalises, for example, with the focal adhesion marker vinculin (Petit et al., 2003). Shortly after plating, when stable focal adhesions have not yet completely formed, PR130 and LPP colocalised at the cell periphery of HT1080 fibrosarcoma cells, presumably at focal contacts (Fig. 4A). In contrast, PR130 was strikingly absent from mature focal adhesions of spread HT1080 cells, where LPP (and vinculin) were clearly present (Fig. 4B,C). Further, there was a good colocalisation of both proteins at cell–cell contacts (Fig. 4D), in the (perinuclear) cytoplasm (Fig. 4C,D) and in the nucleus, including the nuclear membrane or lamina (Fig. 4D). Upon nuclear accumulation of LPP, by blocking its nuclear export with leptomycin B (Petit et al., 2000), this nuclear colocalisation was further intensified (results not shown). The difference in LPP and PR130 colocalisation between adhering and spread cells was not due to potential changes in PR130 expression levels in these conditions (Fig. 4E).
Fig. 4. Cellular distribution of the endogenous LPP–PR130 complex in fibrosarcoma cells.
(A) In adhering HT1080 cells. Cells were fixed and stained 1 h after seeding. The localisation of PR130 (Alexa-Fluor-488), LPP (Cy3) and vinculin (Cy5) was visualised by performing direct immunofluorescence analysis. Scale bars: 10 µm. (B) The same experiment as described in A in spread HT1080 cells. Scale bars: 10 µm. Note the absence of PR130 in elongated mature focal adhesions where vinculin and LPP are clearly present (arrows). (C,D) In spread HT1080 cells. The localisation of PR130 (Alexa-Fluor-488) and LPP (Cy3) was visualised by performing direct immunofluorescence analysis. Arrows indicate focal adhesions (C). Open arrowheads indicate cell–cell contacts (D). Scale bars: 10 µm. (E) Immunoblots of LPP and PR130 in protein lysates of trypsinised (‘in suspension’), adhering (1 h after plating) and adherent and/or spread (8 h after plating, adherent/spread) HT1080 cells. (F) Colocalisation of PR130 and paxillin in spread and adhering HT1080 cells. The localisation of PR130 (Alexa-Fluor-488) and paxillin (Alexa-Fluor-594) was visualised by performing direct immunofluorescence 6 h (spread) and 1 h (adhering) after plating. White scale bars: 20 µm; yellow scale bars: 14 µm. (G) Colocalisation of PR130 and phospho-tyrosine in spread and adhering HT1080 cells. The localisation of PR130 (Alexa-Fluor-488) and phospho-tyrosine residues (P-tyrosine 4G10, Alexa-Fluor-594) was visualised by performing direct immunofluorescence 6 h (spread) and 1 h (adhering) after plating. White scale bars: 20 µm; yellow scale bars: 14 µm.
To provide further support for our interpretation of the immunofluorescence data that LPP and PR130 might colocalise at focal contacts, but no longer do so in more mature focal adhesions, we stained for phosphorylated (phospho-)tyrosine residues (clone 4G10 antibody), a marker of focal adhesions, and for paxillin (clone 5H11 antibody), a marker of both focal contacts and focal adhesions (Madsen et al., 2015). Again, in spread cells, there was no colocalisation of PR130 with either paxillin (Fig. 4F) or phosphotyrosine (Fig. 4G) at focal adhesions, whereas in adhering cells, the peripherally accumulated ‘PR130 patches’ did colocalise substantially with paxillin (Fig. 4F), but much less with phosphotyrosine (Fig. 4G). Thus, the endogenous PR130–LPP complex can be visualised at multiple locations within cells, including the focal contacts, but is strikingly absent from mature focal adhesions.
Exclusion of PR130 from focal adhesions is a function of PP2A activity
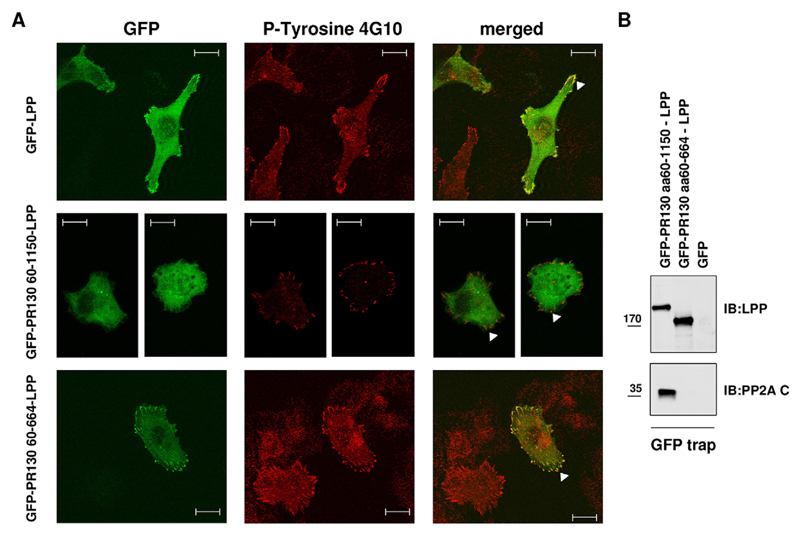
The absence of PR130 from mature focal adhesions suggests that the PR130–LPP complex is not constitutive and might dissociate during focal adhesion maturation. It has been reported that β-galactosidase can be targeted to focal adhesions by covalent binding to GFP–LPP or GFP-tagged LIM domains of LPP (Petit et al., 2003). To further explore why PR130 is excluded from focal adhesions, we generated a GFP–PR130(60–1150)–LPP fusion protein, in an attempt to force the complex into focal adhesions. We deliberately excluded the LPP–binding domain of PR130 (residues 1–59) from this fusion protein to eliminate the potential influence of endogenous LPP binding during the subcellular targeting of the fusion protein. In contrast to GFP–LPP, expression of the truncated fusion protein did not lead to an accumulation in the focal adhesions (Fig. 5A), suggesting that the forced presence of PR130(60–1150) in the LPP complex somehow inhibits the focal-adhesion-targeting capacity of LPP. However, deletion of the PP2A/C-binding domain of PR130 (amino acids 665–1150) did lead to an accumulation of the resulting GFP–PR130(60–664)–LPP fusion protein in 4G10-positive focal adhesions (Fig. 5A), demonstrating a requirement for phosphatase association with PR130 to block focal adhesion accumulation. GFP-trapping of the fusion proteins confirmed their binding behaviour towards PP2A/C (Fig. 5B). Thus, an active dissociation mechanism might operate for the LPP–PR130-PP2A complex in order to permit entry into or maturation of focal adhesions.
Fig. 5. Exclusion of PR130 from focal adhesions is a function of PP2A activity.
(A) Colocalisation of GFP–LPP–PR130 fusion proteins and phospho-tyrosine in spread HT1080 cells. GFP–LPP (positive control), GFP–PR130(60–1150)–LPP and GFP–PR130(60–664)–LPP fusion proteins were expressed in HT1080 cells. At 48 h post transfection, cells were stained with the 4G10 clone anti-phospho-tyrosine monoclonal antibody (P-Tyrosine 4G10, Alexa-Fluor-594) to visualise the focal adhesions (arrowheads). Scale bars: 20 µm. (B) Binding of GFP–LPP–PR130 fusion proteins to PP2A/C. GFP–PR130(60–1150)–LPP, GFP–PR130(60–664)–LPP or GFP alone were isolated on GFP-trapping beads, and the presence of PP2A/C (PP2A C) in the trapped complexes was evaluated by immunoblotting (IB). Expression and trapping of the fusion proteins was verified by anti-LPP immunoblotting. aa, amino acids.
PR130 inhibits HT1080 cell adhesion onto collagen I in an LPP-dependent way
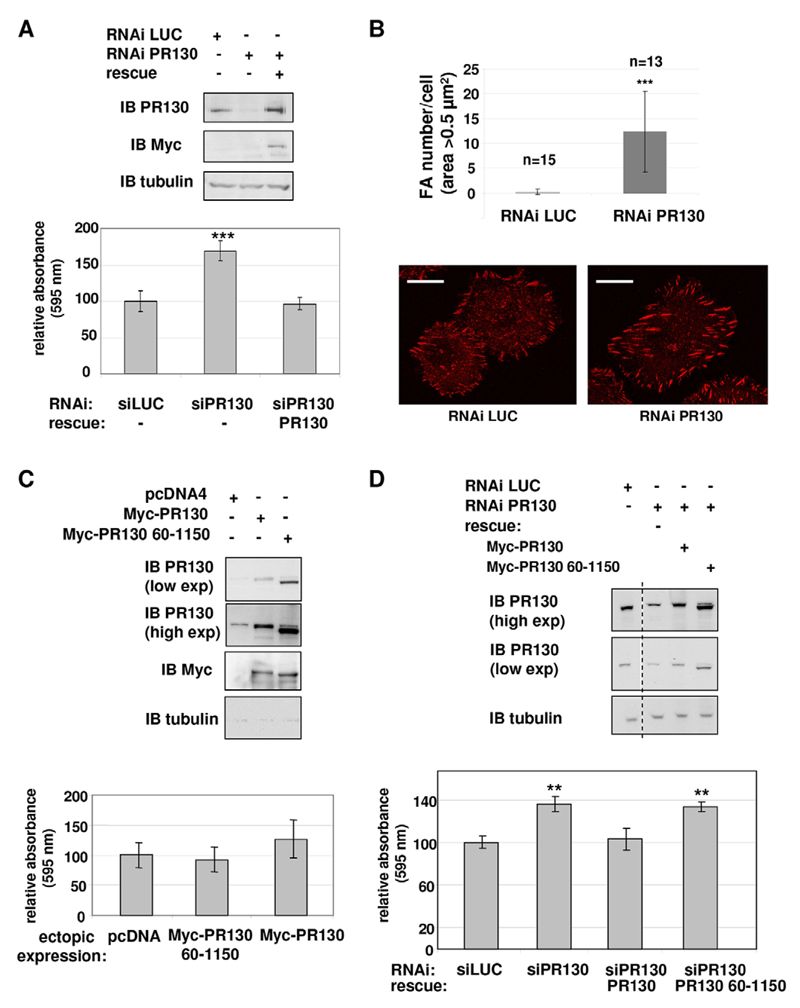
The former observations (Fig. 5) suggest that LPP-dependent recruitment of PR130-associated PP2A activity (Fig. 3) is unfavourable for focal adhesion maturation and predict that PR130 exerts a negative effect on cell–matrix adhesion. To test this hypothesis, PR130 expression was suppressed by performing RNA interference (RNAi) in HT1080 cells, and cell adhesion was measured on collagen-I-coated plates 48–72 h after transfection. When compared to cells that had been transfected with a small interfering (si)RNA against luciferase, PR130-depleted cells indeed showed increased adhesion (Fig. 6A). Morphometric analysis of 4G10-stained focal adhesions demonstrated a significant increase in the number of focal adhesions per cell with an area >0.5 µm2 (measured 45 min after seeding) in PR130-depleted cells (Fig. 6B). Overexpression of PR130 or the LPP-binding deficient PR130(60–1150) mutant did not significantly affect cell–matrix adhesion (Fig. 6C). Increased adhesion in PR130-depleted cells could be rescued by reintroduction of a Myc-tagged RNAi-resistant form of PR130 (Fig. 6A,D), but not by the LPP-binding deficient PR130 (60–1150) mutant (Fig. 6D), not only demonstrating specificity of the siRNA, but also showing a requirement for LPP binding. Immunoblotting confirmed the knockdown, as well as the reconstitution, of PR130 and PR130(60–1150) (Fig. 6A,D).
Fig. 6. PR130 inhibits HT1080 cell adhesion on collagen I in an LPP-dependent manner.
(A) Representative immunoblot (IB) of total lysates of HT1080 cells used in adhesion assays, 72 h after transfection with siRNA against PR130 (RNAi PR130), with or without co-expression of RNAi-resistant Myc-tagged PR130 (rescue). The average of triplicate measurements is shown for each condition tested in the adhesion assays, calculated relative to that of the sample transfected with an siRNA against luciferase (RNAi LUC). Significance was determined with Student’s t-test (***P=0.0005). Adhesion assays were repeated three times, and the observed increase in adhesion in PR130-depleted cells was always in proportion with the degree of PR130 knockdown (which slightly differed between experiments). (B) Focal adhesion (FA) morphometry in PR130-depleted cells. HT1080 cells that had been transfected with siRNA against luciferase or PR130 (72 h post-transfection) were plated on collagen-I-coated cover slips, fixed 45 min later and stained with the phospho-tyrosine-specific 4G10 antibody. Morphometric analysis was performed on individual cells (n=13–15) using ImageJ. Only focal adhesions with an area above 0.5 µm2 were taken into account for analysis. Representative images of staining with the 4G10 antibody in both conditions are shown. Scale bars: 20 µm. (C) Same adhesion experiment as shown in panel A, but in HT1080 cells ectopically expressing empty vector (pcDNA4), Myc–PR130 or Myc–PR130(60–1150). Three independent assays were performed, with triplicate measurements for each condition in each assay. exp, exposure. (D) The same adhesion experiment as in panel C, but in HT1080 cells that had been additionally depleted for endogenous PR130 with siRNA. The dotted line indicates the represented lanes were originally not adjacent. Data are from a single representative assay, with averages of triplicate measurements displayed for each condition, calculated relative to results of the RNAi LUC condition. Significance was determined with Student’s t-test (**P<0.005). Data are mean±s.d. si, siRNA against the indicated protein.
PR130 is required for efficient cell migration, through an LPP- and PP2A/C-dependent mechanism
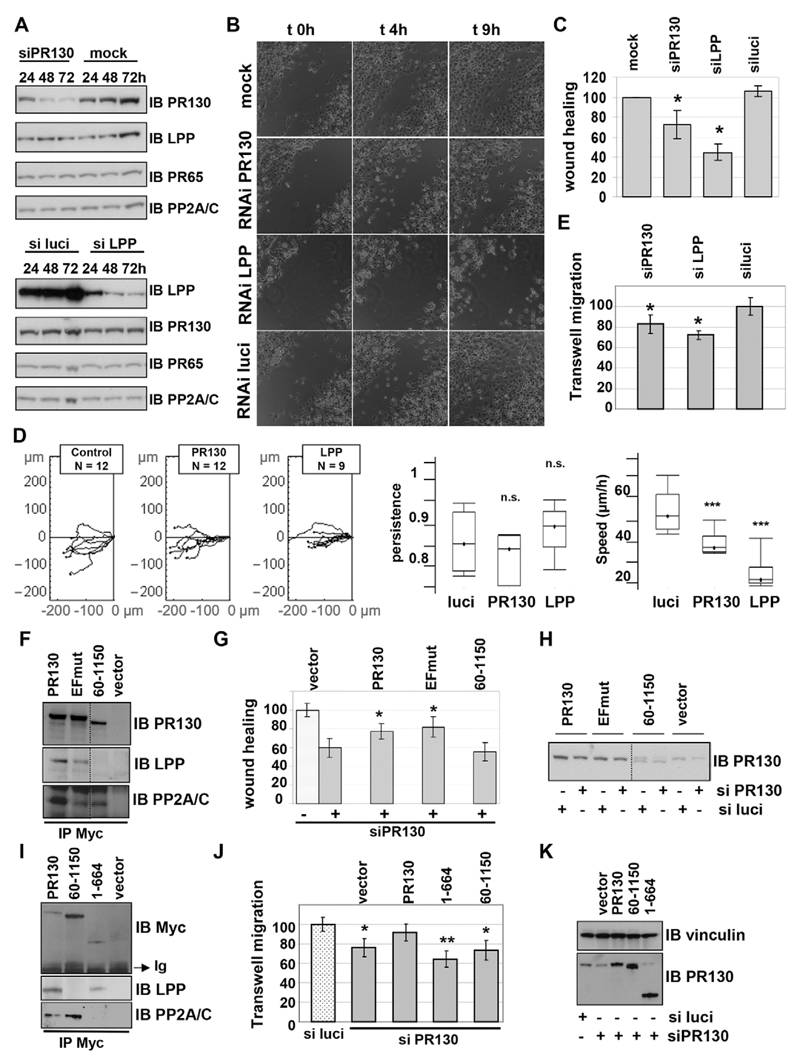
Our adhesion data inferred a role for the LPP–PR130-PP2A/C complex in promoting dynamic focal contacts, which might also play a role in cell migration. Previously, a positive role for LPP has been reported in the migration of smooth muscle cells (Gorenne et al., 2006), mouse embryonic fibroblasts (Vervenne et al., 2009) and breast cancer cells (Ngan et al., 2013; Van Itallie et al., 2014). Following transfection of HT1080 cells with siRNA against LPP or siRNA against PR130, time-dependent knockdown was observed for both LPP and PR130, without any compensatory changes in the expression levels of their binding partners (PR130, LPP, PP2A/C and the PP2A A subunit) (Fig. 7A). At 72 h post transfection, a significant delay in wound closure was observed under conditions where either LPP or PR130 was depleted, but not in the cultures that had been transfected with buffer only (mock) or siRNA against luciferase (luciferase siRNA) (Fig. 7B,C; Movies 1–3). Single-cell tracking revealed that, upon knockdown of LPP or PR130, the observed motility defects are attributable to decreased migration speed, whereas the persistence of movement did not significantly differ from that of the luciferase siRNA control (Fig. 7D). The LPP-knockdown migration phenotype was found to be significantly stronger than the PR130-knockdown phenotype (Fig. 7C), possibly because the LPP knockdown was consistently more efficient than the PR130 knockdown, and LPP-depleted cells showed difficulties in reaching complete confluence at 72 h post transfection. The latter is consistent with the previously reported role for LPP in E-cadherin-dependent cell–cell adhesion (Hansen and Beckerle, 2006; Van Itallie et al., 2014). To circumvent the need for a confluent cell monolayer and, additionally, to exclude potential matrix denudation influences in the wound healing assay, we repeated the migration assays in Boyden chambers, which recapitulate single-cell migration, rather than two-dimensional sheet migration, and do not involve any potential denudation of extracellular matrix. Similar results were found – both LPP- and PR130-depleted cells showed decreased migration (Fig. 7E). Notably, in some experiments, the LPP-knockdown cells (but not the PR130-knockdown cells) showed signs of decreased viability, compromising an unambiguous interpretation of the LPP-depleted cell phenotype. However, in light of previous results (Gorenne et al., 2006; Vervenne et al., 2009; Ngan et al., 2013; Van Itallie et al., 2014), at least part of the LPP-knockdown phenotype could be ascribed to a cell migration defect. Thus, we confirmed the positive involvement of LPP in cell migration and revealed the same now for PR130.
Fig. 7. PR130 promotes efficient HT1080 cell migration in an LPP-dependent manner.
(A) RNAi-mediated knockdown of PR130 and LPP. HT1080 cells were transfected with buffer (mock) or an siRNA duplex (10 nM) directed against PR130, LPP or luciferase (siPR130, siluci and siLPP, respectively). Extracts were prepared in SDS-sample buffer at the indicated times post transfection and subjected to immunoblotting. The same blot was developed, stripped and re-probed with the indicated antibodies. (B) Wound healing assays. A wound was made in a confluent cell layer of HT1080 cells 72 h post transfection with the indicated siRNA duplexes (mock=buffer). Wound closure was followed as a function of time by taking a picture every 10 min. After 9 h, the wounds of the control cells (mock and RNAi luciferase) had virtually closed, whereas this was not the case in cells with suppressed PR130 or LPP expression. These images are of a representative experiment. Time-lapse videos of the migrating cells can be found in Movies 1–3. (C) Quantification of migration speed in the different conditions. The reduction of wound area was measured as a function of time using MetaMorph software. All data were related to the mock-transfected cells (control), for which the migration speed was set to 100%. The results represent the mean±s.d. of three independent experiments. A Student’s t-test was applied to assess statistical significance of the observed differences with the control (mock) condition (P=0.022 for siPR130; P=0.007 for siLPP; *P<0.05). There is also a statistical difference between siPR130 and siLPP (P=0.011). (D) Individual cell tracking. This analysis was performed with the tracking software for the indicated number of cells transfected with siRNAs against the indicated proteins (luciferase, control). Statistical analysis of the tracks was performed with Mathematica software, and is represented in the two right-most box and whisker diagrams (the box represents the first and third quartile of the measured values of the given parameter, the line represents the median of the measured values of the given parameter and the whiskers represent the minimal and the maximal measured values of the given parameter). (E) Transwell migration assays. Single-cell migration through an 8.0-µm-pore membrane along a gradient of 0–10% FCS was determined at 72 h post-transfection of HT1080 cells with siRNAs against luciferase (luci), LPP and PR130. Migrated cells were labelled with Calcein-AM and quantified by measuring fluorescence. Results represent the mean of two independent experiments, in which each condition was measured in triplicate. Asterisks indicate a statistically significant difference with cells transfected with an siRNA against luciferase (*P<0.05). (F–H) Rescue of the migration defect in the wound healing assays by wild-type PR130 and PR130 EFmut, but not by the non-LPP-binding PR130(60–1150) mutant. Polyclonal HT1080 cells stably expressing a putative rescue plasmid encoding C-terminally Myc-tagged RNAi-resistant PR130, PR130 EFmut or PR130(60–1150), or Myc alone were selected with zeocin. (F) Anti-Myc immunoprecipitations (IP) confirmed the expression of the rescue proteins (anti-PR130 blot), and their binding behaviour to endogenous LPP and PP2A/C. (G) At 72 h after transfection with the siRNA duplex against PR130 or luciferase, the stable cell lines were subjected to a wound healing assay. (H) 12 h later, when the wound of the control condition was completely closed, extracts were prepared in SDS sample buffer to evaluate knockdown of endogenous PR130 and RNAi-resistance of the rescue proteins. The wound healing data are expressed as the mean±range of two independent experiments in which each condition was performed in duplicate. Asterisks indicate a statistically significant difference with the control (vector-transfected HT1080 cells treated with siPR130) – for wild-type PR130 rescue, P=0.034; for PR130 EFmut rescue, P=0.016. The western blot (IB) in panel H shows a single representative experiment. (I–K) Rescue of the migration defect in the Transwell assays by wild-type PR130, but not by the non-PP2A/C-binding PR130(1–664) mutant or the non-LPP-binding PR130(60–1150) mutant. (I) Polyclonal HT1080 cells stably expressing a putative rescue plasmid encoding C-terminally Myc-tagged RNAi-resistant PR130, PR130(60–1150), PR130(1–664) or Myc alone were selected with zeocin. Anti-Myc immunoprecipitations (IPs) confirmed expression of the rescue proteins (anti-Myc blot) and their binding behaviour to endogenous LPP and PP2A/C. (J) At 72 h after transfection with the siRNA duplexes against PR130 or luciferase, the stable cell lines were subjected to a Transwell migration assay. (K) A small aliquot of each cell suspension was kept to prepare protein extracts and to evaluate the knockdown of endogenous PR130 and RNAi-resistance of the rescue proteins (IB with PR130- and Myc-specific antibodies). Each condition was performed in triplicate, and means±s.d. are displayed of a single representative experiment. Asterisks indicate a statistically significant difference with the control condition (si-luci-treated cells). *P<0.05; **P<0.005 (Student’s t-test).
The fact that knockdown of either protein in the LPP–PR130 complex gives rise to similar phenotypes is consistent with a functional role for this complex in cell migration. To further confirm this notion, we performed rescue experiments with RNAi-resistant wild-type and mutant PR130 forms, which were Myc-tagged at the C-terminus and used to generate stable polyclonal cell lines. In the wound healing assay (Fig. 7F–H), two different mutants were used – PR130 EFmut, predicted to lack the ability to bind Ca2+ and to bind less PP2A/C (Janssens et al., 2003); and PR130(60–1150), which lacks the LPP-binding domain (Fig. 2). In the Boyden chamber assay (Fig. 7I–K), PR130(1–664), predicted to lack the ability to bind PP2A/C, and the LPP-binding deficient PR130(60–1150) mutant were used. Immunoprecipitations of Myc from lysates of the stable cell lines showed the binding behaviour of the PR130 variants towards LPP and PP2A/C (Fig. 7F,I). In the wound healing assays, we observed that wild-type PR130 and PR130 EFmut partially rescued the migratory defect caused by PR130 knockdown, whereas the non-LPP-binding PR130(60–1150) mutant could not (Fig. 7G). In the Boyden chamber assays, neither PR130(1–664) nor PR130 (60–1150) rescued the migration defect, as opposed to expression of wild-type PR130, which resulted in a near-complete rescue (Fig. 7J). These data indicate the specificity of the siRNA effects on PR130 and further show a requirement for an interaction between LPP and PP2A/C in the functional role of PR130 in cell migration. Western blotting confirmed the knockdown and RNAi-resistance of the rescue plasmids (Fig. 7H,K).
Discussion
One of the major questions regarding Ser/Thr phosphatase regulation is how target specificity is achieved. For PP2A, local recruitment or targeting to defined locales is largely determined by the regulatory B-type subunits (Slupe et al., 2011). The study here provides a basis on which to understand a positive role for a specific PP2A complex in cell migration and a negative role in cell–substratum adhesion. This complex contains PR130/B″α1 as the ‘third’ distinguishing regulatory subunit, a specific splice variant with a long 664-amino-acid N-terminal domain of unknown function. We have used this domain as a bait to screen for interacting partners and have identified the LIM protein LPP as an interesting candidate. In parallel, LPP was identified, by performing mass spectroscopy, as a specific co-immunoprecipitating protein in PR130 immunocomplexes. Reciprocal immunoprecipitations confirmed the presence of an endogenous PR130–LPP complex, in which both the PP2A A and C subunits were also present. The isolated LPP-associated PP2A complexes proved catalytically active. PR130 specifically interacted with the LIM domains of LPP – at least two (LIM2 and LIM3), and probably all three, LIM domains of LPP are required for the interaction, possibly because they fold into a ‘superstructure’, as previously suggested (Petit et al., 2003). Although LIM domains are postulated to be protein interaction interfaces (Dawid et al., 1998; Bach, 2000), no discrete consensus binding sequence or structural element has emerged in their binding partners (Kadrmas and Beckerle, 2004). Here, we show that PR130 interacts with the LPP LIM domains through a well-conserved cysteine-rich, Zn2+-finger-like domain, implying that the PR130–LPP interaction occurs through a Zn2+-finger–Zn2+-finger interface. Despite the high degree of similarity within the LIM domains of LPP and zyxin (Petit et al., 1996), PR130 did not interact with zyxin in our binding assays. Such selectivity has been demonstrated before for other LPP interacting proteins, including Scrib (Petit et al., 2005a) and shelterin (Sheppard et al., 2011). Thus, we have newly identified PR130 as a cellular ligand for the LPP LIM domains and LPP as an adaptor for a specific catalytically active PP2A complex, harbouring PR130 as the regulatory B-type subunit.
An intriguing observation deriving from our localisation studies of endogenous LPP and PR130 is that the two proteins colocalise in dynamic focal contacts but that there is a striking absence of PR130 from mature (non-dynamic) focal adhesions, where LPP clearly accumulates (Petit et al., 2003). It is well established that all three LPP LIM domains cooperate to provide consistent targeting to focal adhesions (Petit et al., 2003). Because PR130 interacts with these LIM domains, focal adhesion targeting of LPP should therefore co-occur with loss of PR130 binding, implying that LPP–PR130 complex formation is not constitutive but dynamic. Loss of PR130 from the focal-adhesion-associated LPP complex might perhaps occur through competition with other LIM-domain-interacting proteins that do colocalise with LPP in focal adhesions, such as, for example, palladin (Jin et al., 2007) or supervillin (Takizawa et al., 2006). An alternative disassembly mechanism might involve selective proteolysis of PR130 by calpains, which are known to regulate focal adhesion dynamics in cancer cells (Storr et al., 2011). PR130 is indeed subject to limited proteolysis of its entire specific N-terminal domain and part of its common C-terminal domain by these proteases (Janssens et al., 2009). Regardless of the precise dissociation mechanism, our localisation data also imply that the sites of cell–substratum contact, where the LPP–PR130 complex proteins colocalised, might be particularly dynamic in nature because of the presence of PR130-PP2A. Such a hypothesis is well supported by the failure of focal adhesion localisation of the GFP–PR130(60–1150)–LPP fusion protein, whereas deletion of the PP2A/C-binding domain reconstituted targeting of the fusion protein to focal adhesions. Our functional cell adhesion and migration assays provide additional support for this view. Loss of PR130 expression indeed increased HT1080 cell adhesion onto collagen I, whereas it inhibited cell migration in wound healing and Transwell assays. Both PR130 functions require a functional interaction with LPP because a non-LPP-binding PR130 mutant failed to rescue the increase in cell adhesion and the decrease in cell migration of PR130-depleted cells. The pro-migratory role of PR130 also requires a functional interaction with PP2A/C because a non-PP2A/C-binding PR130 mutant did not rescue the migration defect in cells with suppressed PR130 expression in a Transwell assay. Although in the wound healing assay potential matrix denudation might specifically have influenced the PR130-depleted cells, this possibility was excluded in the Transwell assay, favouring the conclusion that the LPP–PR130-PP2A interaction contributes to the dynamic nature of cell–substratum contacts in moving cells. Although the requirement for LPP and PP2A/C binding can be well documented, it is less clear whether Ca2+ binding is also important for the function of PR130 in cell migration. Both re-expression of wild-type PR130 or PR130 EFmut could indeed rescue the migration defect of PR130-depleted cells, suggesting that Ca2+-binding or potential Ca2+-induced changes in PR130-PP2A phosphatase activity is not very important in the pro-migratory role of PR130. Although the PR72EFmut construct interacts substantially less with PP2A/C as compared to wild-type PR72 (Janssens et al., 2003; Ahn et al., 2007), the interaction of PR130 EFmut with PP2A/C was diminished to a much lesser extent as compared to wild-type PR130 (Fig. 7F), indicative of additional contributing interaction interfaces in PR130, such as the reported hydrophobic motif (Davis et al., 2008; Wlodarchak et al., 2013). Hence, the ability of PR130 EFmut to rescue knockdown remains consistent with a functional role for the phosphatase complex in these events.
In line with our observations describing a positive role for PR130 in cell migration, it has been reported that PR130 is involved in planar cell polarity signalling in Xenopus embryogenesis (Creyghton et al., 2006). More recently, a similar role has been demonstrated for LPP in the regulation of convergence-extension movement in zebrafish (Vervenne et al., 2008). Consistently, LPP−/− mouse embryonic fibroblasts exhibit reduced migration capacity in a wound healing assay (Vervenne et al., 2009), and depletion of LPP reduces the migration of smooth muscle cells (Gorenne et al., 2006) and breast cancer cells (Ngan et al., 2013; Van Itallie et al., 2014). These reports thus confirm a positive role for PR130 and LPP in cell motility. We speculate that a major function of LPP in determining this cell behaviour is to act as a scaffold that brings a specific PP2A heterotrimer into close contact with potential substrates, the dynamic (de)phosphorylation of which might efficiently steer cell migration or prevent focal adhesion maturation. Such candidate substrates might be Scrib, vasodilator-stimulated phosphoprotein (VASP), LIM and SH3 protein 1 (LASP-1) or palladin – which are all established LPP interaction partners (Petit et al., 2005b, 2000; Keicher et al., 2004; Jin et al., 2007), phosphoproteins on Ser/Thr residues (Yoshihara et al., 2011; Metodieva et al., 2013; Döppler and Storz, 2013; Butt et al., 2003; Keicher et al., 2004; Asano et al., 2011) and known actin cytoskeleton modulators regulating cell adhesion, migration or polarity (Qin et al., 2005; Döppler and Storz, 2013; Orth et al., 2015; Najm and El-Sibai, 2014). Future research efforts should further clarify whether PR130-PP2A does indeed regulate dephosphorylation of these proteins and how this relates to the pro-migratory role of the LPP–PR130-PP2A complex discovered here. Earlier work has already demonstrated a role for a specific PP2A-B′γ1 complex in regulating paxillin dephosphorylation at focal adhesions (Ito et al., 2000) – further underscoring the importance of localised regulation of protein dephosphorylation at sites of cell–substratum contacts – as well as the major determining role of specific PP2A regulatory B-type subunits in these processes.
The demonstration of a direct, specific and strong interaction between PR130 and LPP might suggest yet other cellular functions of this complex, besides the ones demonstrated here. LPP is indeed also involved in the regulation of (epithelial) cell–cell contacts (Hansen and Beckerle, 2006; Van Itallie et al., 2014) and has been described as a transcriptional co-activator (Guo et al., 2006) and telomere-binding protein (Sheppard and Loayza, 2010) in the nucleus. Given the apparent colocalisation of PR130 and LPP at these specific subcellular locales, it is tempting to speculate that PP2A-PR130 also regulates LPP function in these particular processes.
Although we have identified a role for the LPP–PR130 complex in adhesion and migration control in HT1080 fibrosarcoma cells, the presence of the complex in several independent cell lines, both normal and transformed, suggests a general mechanism. Alongside earlier work highlighting a positive role for PR130 in canonical Wnt signalling (Creyghton et al., 2006) and EGF-dependent signalling (Zwaenepoel et al., 2010), our current findings highlight a positive role for PR130 in (cancer) cell migration and a negative role in (cancer) cell–substratum adhesion through the dynamic interaction with LPP. Thus, alongside its tumour suppressor properties in one complex (Westermarck and Hahn, 2008), PP2A might also be involved in growth stimulation, tumour progression and metastasis in another. Specifically, the latter complexes could constitute interesting therapeutic targets for pharmacological intervention.
Materials and Methods
Generation of plasmids and site-directed mutagenesis
Classic molecular biology techniques were used to subclone PR130, LPP or fragments thereof, into different plasmids. Restriction enzymes, Antarctic phosphatase and T4 DNA ligase were from New England Biolabs. GFP-PR130(60–1150)–LPP and GFP–PR130(60–664)–LPP fusion proteins were generated into pEGFP-C1 using In-Fusion® technology (Clontech). EGFP-tagged zyxin, LPP LIM domains and LIM-domain point mutants have been described previously (Petit et al., 2003). GST fusions were expressed from pGMEX-T3 (Amersham), EGFP-tagged proteins from pEGFP-C1 or pEGFP-N1 (Clontech), Myc-tagged proteins from pcDNA4 Tet-on (Invitrogen). The Myc-tag was included in one of the oligonucleotides used to generate the appropriate PCR products. All PCR reactions were performed with proofreading Pwo DNA polymerase (Roche) and oligonucleotides from Sigma-Genosys. For site-directed mutagenesis, the QuickChange protocol (Stratagene) was used. All inserts of the recombinant plasmids were verified by sequencing.
Antibodies
We used the following commercial antibodies: goat anti-LPP (clone N-20, catalogue number sc-27312) and rabbit anti-GST (clone Z-5, catalogue number sc-459) antibodies (Santa Cruz Biotechnologies); mouse anti-α-tubulin (clone B-5-1-2, catalogue number T5168), anti-vinculin (clone hVIN-1, catalogue number V9264) and anti-FLAG (clone M2, catalogue number F1804) antibodies (Sigma); anti-phospho-tyrosine (clone 4G10, catalogue number 05-321) and anti-paxillin (clone 5H11, catalogue number 05-417) antibodies (Millipore). Mouse anti-Myc (clone 9E10) and anti-EGFP (clone GFP4E12/8) antibodies were from Cancer Research UK monoclonal antibody services; monoclonal anti-PP2A/C (clone F2.6AID) and anti-PP2A subunit A (clone C5.3D10) antibodies were a generous gift from Dr S. Dilworth (University of Middlesex, London, UK). Rabbit anti-PR130 and anti-LPP antibodies were as described previously (Zwaenepoel et al., 2008; Petit et al., 2000). For immunoblotting, dilutions were 1/1000; for immunofluorescence 1/50. Horseradish peroxidase (HRP)-conjugated secondary antibodies for western blotting were donkey anti-rabbit and sheep anti-mouse (Amersham), and swine anti-goat (Dako). Secondary antibodies used for immunofluorescence (dilutions: 1/800) were: Alexa-Fluor-488-conjugated donkey anti-rabbit or anti-mouse, and Alexa-Fluor-594-conjugated donkey anti-mouse antibodies (Molecular Probes), and Cy3-conjugated donkey anti-goat and Cy5-conjugated donkey anti-mouse antibodies (Jackson).
Yeast two-hybrid screening
The cDNA encoding PR130(1–664) was PCR amplified and cloned into SmaI-digested pGBKT7. This plasmid was the bait to screen 2.5×106 clones of a Matchmaker testis pACT library in the AH109 S. cerevisiae strain using standard procedures (Clontech). The initial screening was performed on X-α-Gal-containing medium lacking adenine and histidine. The protocols used are as in the ‘Matchmaker GAL4 Two-Hybrid System 3 user manual’ and ‘Yeast protocol handbook’ at www.clontech.com/clontech/techinfo/manuals. After a second screening round to eliminate false positives, the remaining library plasmids were isolated and the inserts sequenced.
Protein fingerprinting and mass spectroscopy
For identification of PR130 co-immunoprecipitating proteins, lysates of 5×15-cm dishes of confluent NIH3T3 cells were used per condition. Anti-PR130 immunocomplexes were resolved on 4–12% NuPage gels (Invitrogen) and stained with Coomassie Brilliant Blue. Bands of interest were excised, trypsinised and the peptide mixtures analysed on a MALDI-TOF instrument (Micromass). The proteins giving rise to the resulting peptide sequences were identified by using a Mascott MS/MS ion search. Three tryptic peptides of murine LPP were retrieved from the 75-kDa protein band present in anti-PR130 immunoprecipitates: residues 297–311, YYEPYYAAGPSYGGR; residues 330–353, EAAYAPPASGNQNHPGMYPVSGPK; residues 401–416, MLYDMENPPADDYFGR.
RNA interference
All siRNA duplexes were purchased from Qiagen: PR130 sense r(ACCAUGCAGCUUACAGAAAUU), PR130 antisense r(UUUCUGUAAGCUGCAUGGUUU), LPP sense r(GUCCCAAGAAGACCUAUAU)dTdT, LPP antisense r(AUAUAGGUCUUCUUGGGAC)dCdA, luciferase sense r(UACGCGGAAUACUUCGAUU)dTdT, luciferase antisense r(AAUCGAAGUAUUCCGCGUA)dCdG (‘r’ denotes ‘RNA’ and ‘d’ denotes ‘DNA’ nucleotides). They were transfected into HT1080 cells (100,000 cells/well in a 24-well plate) at a final concentration of 10 nM with 3–4 µl of Hiperfect reagent (Qiagen). To make RNAi-resistant PR130, three point mutations were introduced into the PR130 target sequence (ACCATGCAGCTTACAGAAA was mutated into ACCATGCAGCATATCGAAA).
Immunoprecipitations, GST pull downs and GFP-trapping
Total cell extracts were prepared in NET buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 15 mM EDTA, 1% NP-40) supplemented with complete protease and phosphatase inhibitor cocktail (Roche). For immunoprecipitations, the lysates were incubated for 2–3 h with the appropriate antibodies on a rotating wheel at 4°C. Protein G Sepharose beads (Sigma) were added for 15 min, and the beads were washed twice in NET buffer, twice in TBS+0.1% NP-40 and once in TBS. For GST pull downs, the lysates were incubated with glutathione–Sepharose (Amersham) in NENT100 buffer (20 mM Tris-HCl pH 7.4, 1 mM EDTA, 0.1% NP-40, 25% glycerol, 100 mM NaCl), supplemented with 2 mg/ml BSA on a rotating wheel for 1 h at 4°C. The beads were washed twice in NENT300 (NENT100 with 300 mM NaCl instead of 100 mM) plus 2 mg/ml BSA, and three times in NENT300 without BSA. For GFP-trapping, cell lysates were incubated at 4°C for 1 h with wash buffer (10 mM Tris-HCl pH 7.5, 0.5 mM EDTA and 150 mM NaCl) and 15 µl of GFP-trap-A beads (Chromotek). Beads were washed four times in wash buffer. In most cases, bound proteins were eluted in 2× SDS sample buffer at 98°C and further analysed by immunoblotting.
Measurement of PP2A activity
GFP-trapped complexes were washed with 20 mM Tris-HCl pH 7.4+1 mM DTT and resuspended in 80 µl of enzyme dilution buffer (Millipore, catalogue number 20-169). One half (40 µl) was pre-incubated (10 min on ice) with okadaic acid (Calbiochem) at a final concentration of 10−8 M; the other half was left untreated. The phosphatase assay was performed on 300 µM of RRApTVA phospho-peptide (lowercase p denoting phosphorylation of the following threonine residue) for different time points at 30°C, all in the linear range of the assay. The released free phosphate was determined by addition of malachite-green solution, composed of a 10:1 mix of solutions A and B (Millipore, catalogue numbers 20-105 and 20-104, respectively). Absorbance at 630 nm was measured in a multi-channel spectrophotometer. The pmole number of released phosphate was calculated by comparison with a standard curve of KH2PO4.
Cell-based assays
HT1080, COS7 and NIH3T3 cells (all from American Type Culture Collection and used at early passage number after being revived) were grown in DMEM +10% foetal calf serum (Sigma) or 10% donor calf serum (Sigma) for NIH3T3 cells. Plasmid transfections were performed with Fugene6 Reagent (Roche) in Optimem (Life Technologies) according to the manufacturer’s instructions. For selection of stable cell lines, HT1080 cells were split 1 in 4 into growth medium containing 400 µg/ml zeocin (Invitrogen) 36 h after transfection with the pcDNA4T/O-derived plasmids. Resistant clones were pooled and frozen, and the early passages were used for rescue experiments.
For immunofluorescence analysis, cells were grown on acid-washed collagen-I-coated coverslips in 24-well plates. Cells were washed twice in PBS supplemented with 1 mM CaCl2 and 0.5 mM MgCl2 and fixed in 4% paraformaldehyde (PBS) for 15 min at room temperature. Free aldehydes were quenched in 50 mM NH4Cl for 10 min. Cells were permeabilised in 0.2% Triton X-100 (PBS) for 15 min and blocked in 1.5% BSA (PBS) for 30 min. Incubation with primary antibodies was performed for 45 min at room temperature. After three washes in PBS, incubation with the secondary antibodies was for 30–45 min. After three washes in PBS and one wash in water, the cells were mounted in DAPI-containing mounting medium (Molecular Probes). All images were captured on a confocal laser scanning microscope (LSM510, Carl Zeiss) equipped with a 63×/1.4 Plan-Apochromat oil immersion objective.
For adhesion assays, 20,000 HT1080 cells/well were plated onto 24-well plates that had been coated with collagen I (1 mg/ml) (Sigma). Each condition was performed in triplicate. After 20 min of adhesion, non-adhering cells were gently washed off, and adhering cells were incubated for an additional hour at 37°C. Next, cells were washed with PBS, fixed with 4% paraformaldehyde and coloured with 0.5% Crystal Violet (Sigma). After three washing steps in water, cells were lysed in 0.5% Triton X-100, and the released Crystal Violet measured at 595 nm in a spectrophotometer. Morphometric analysis of focal adhesions was executed in cells 45 min after plating on collagen-I-coated cover slips. Focal adhesions were stained using an antibody recognising phospho-tyrosine residues (clone 4G10). Images of several random fields were acquired on a Leica LSM510 inverted confocal microscope using a ×63 1.2 Plan-Apochromat oil objective (Leica) with a pinhole of 0.8 to minimise the z-section. Then, 8-bit images were captured and pixel size was 0.1395×0.1395 µm2. Processing and analysis of the focal adhesions of individual cells (n=13–15) was executed with ImageJ using the tools ‘Background subtraction’, ‘adjustment of brightness and contrast’, ‘edge finding’, ‘adjustment of threshold’ and ‘particle analysis’ with size=0.5 and circularity=0–1 as set parameters. Using these settings, only focal adhesions >0.5 µm2 were included in the final analysis.
To monitor migration, a wound was made in a confluent layer of HT1080 cells seeded on a 24-well plate. The cells were washed with medium to remove all non-adherent cells, and the wound was allowed to close in complete medium supplied with 20 mM Hepes pH 7.4 at 37°C. Wound healing was followed on a light microscope (Carl Zeiss) equipped with a moveable stage, a 10× objective and an Orca ER CCD camera (Hamamatsu). Every 10 min, a picture was taken, and the resulting data were analysed with MetaMorph software. Cell tracking was performed with Tracking software and analysed with Mathematica software. Single-cell migration was monitored in Boyden chambers (ThinCert™ Transwell inserts for 24-well plates, 8.0 µm pore, Greiner Bio-one), largely according to the accompanying ‘Application Note’ protocol for HT1080 cells (www.gbo.com/bioscience). In short, 100,000 cells were seeded in serum-free medium+0.2% BSA (Sigma), and the lower reservoir additionally contained 10% FCS. After incubation at 37°C for 12 h, the inserts were incubated for 45 min at 37°C in serum-free medium containing 0.2% BSA and 8 µM Calcein-AM (Sigma). Cells at the outside bottom of the inserts were trypsinised for 10 min (37°C) in a new well containing 450 µl trypsin (0.05%) with EDTA (0.02%) (Sigma). 200 µl of cell suspension was transferred to a black F-bottom 96-well plate (Greiner Bio-one) and fluorescence (excitation: 485 nm; emission: 520 nm) was determined on a Flex Station 3 instrument (Molecular Probes).
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.175778/-/DC1
To monitor migration, a wound was made in a confluent layer of HT1080 cells seeded in a 24-well plate. The cells were washed with medium to remove all non-adherent cells and the wound was allowed to close in complete medium supplied with 20mM Hepes pH 7.4 at 37°C. Wound healing was followed on a light microscope (Carl Zeiss Inc.) equipped with a moveable stage, a 10x objective and Orca ER CCD camera (Hamamatsu). Every 10’ a picture was taken and time-lapse videos were made with MetaMorph software. In all cases, the wound was made 72h post-transfection with the siRNAs, and the wound healing was followed for at least 9 hours.
Acknowledgements
The authors wish to acknowledge Dr D. Aubyn, Dr C. Gray, Dr P. Jordan and Dr D. Zicha [Microscopy Laboratory, London Research Institute – Cancer Research UK (LRI-CRUK)] for help and advice regarding imaging, and Dr N. Totty (Protein Analysis Laboratory, LRI-CRUK) for mass spectroscopic analyses. We are grateful to Prof. P. Vandenberghe and the KU Leuven Cell Imaging Core for help and advice regarding focal adhesion morphometry, and to Prof. G. Bultynck (KU Leuven) for use of the Flex Station.
Funding
V.J. received a 15-month Mobility grant from the Research Foundation - Flanders to perform this study. Research was supported by a KU Leuven GOA grant [grant number GOA/08/016].
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
V.J., K.Z. and C.R. performed experiments; M.M.R.P. provided plasmids and analytical tools; V.J., K.Z., C.R., M.M.R.P., J.G. and P.J.P. analysed the data; V.J. and P.J.P. wrote the manuscript.
References
- Ahn J-H, Sung JY, McAvoy T, Nishi A, Janssens V, Goris J, Greengard P, Nairn AC. The B″/PR72 subunit mediates Ca2+-dependent dephosphorylation of DARPP-32 by protein phosphatase 2A. Proc Natl Acad Sci USA. 2007;104:9876–9881. doi: 10.1073/pnas.0703589104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano E, Maeda M, Hasegawa H, Ito S, Hyodo T, Yuan H, Takahashi M, Hamaguchi M, Senga T. Role of palladin phosphorylation by extracellular signal-regulated kinase in cell migration. PLoS ONE. 2011;6:e29338. doi: 10.1371/journal.pone.0029338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach I. The LIM domain: regulation by association. Mech Dev. 2000;91:5–17. doi: 10.1016/s0925-4773(99)00314-7. [DOI] [PubMed] [Google Scholar]
- Basu S. PP2A in the regulation of cell motility and invasion. Curr Protein Pept Sci. 2011;12:3–11. doi: 10.2174/138920311795659443. [DOI] [PubMed] [Google Scholar]
- Basu S, Ray NT, Atkinson SJ, Broxmeyer HE. Protein phosphatase 2A plays an important role in stromal cell-derived factor-1/CXC chemokine ligand 12-mediated migration and adhesion of CD34+ cells. J Immunol. 2007;179:3075–3085. doi: 10.4049/jimmunol.179.5.3075. [DOI] [PubMed] [Google Scholar]
- Boeckeler K, Rosse C, Howell M, Parker PJ. Manipulating signal delivery - plasma-membrane ERK activation in aPKC-dependent migration. J Cell Sci. 2010;123:2725–2732. doi: 10.1242/jcs.062299. [DOI] [PubMed] [Google Scholar]
- Bragg JN, Lawrence DM, Jackson AO. The N-terminal 85 amino acids of the barley stripe mosaic virus gamma-b pathogenesis protein contain three zinc-binding motifs. J Virol. 2004;78:7379–7391. doi: 10.1128/JVI.78.14.7379-7391.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt E, Gambaryan S, Göttfert N, Galler A, Marcus K, Meyer HE. Actin binding of human LIM and SH3 protein is regulated by cGMP- and cAMP-dependent protein kinase phosphorylation on serine 146. J Biol Chem. 2003;278:15601–15607. doi: 10.1074/jbc.M209009200. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Roël G, Eichhorn PJA, Hijmans EM, Maurer I, Destrée O, Bernards R. PR72, a novel regulator of Wnt signaling required for Naked cuticle function. Genes Dev. 2005;19:376–386. doi: 10.1101/gad.328905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Roël G, Eichhorn PJA, Vredeveld LC, Destrée O, Bernards R. PR130 is a modulator of the Wnt-signaling cascade that counters repression of the antagonist Naked cuticle. Proc Natl Acad Sci USA. 2006;103:5397–5402. doi: 10.1073/pnas.0507237103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AJ, Yan Z, Martinez B, Mumby MC. Protein phosphatase 2A is targeted to cell division control protein 6 by a calcium-binding regulatory subunit. J Biol Chem. 2008;283:16104–16114. doi: 10.1074/jbc.M710313200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid IB, Breen JJ, Toyama R. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 1998;14:156–162. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- Döppler H, Storz P. Regulation of VASP by phosphorylation: consequences for cell migration. Cell Adh Migr. 2013;7:492–496. doi: 10.4161/cam.27351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovega R, Tsutakawa S, Quistgaard EM, Anandapadamanaban M, Löw C, Nordlund P. Structural and biochemical characterization of human PR70 in isolation and in complex with the scaffolding subunit of protein phosphatase 2A. PLoS ONE. 2014;9:e101846. doi: 10.1371/journal.pone.0101846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorenne I, Jin L, Yoshida T, Sanders JM, Sarembock IJ, Owens GK, Somlyo AP, Somlyo AV. LPP expression during in vitro smooth muscle differentiation and stent-induced vascular injury. Circ Res. 2006;98:378–385. doi: 10.1161/01.RES.0000202802.34727.fd. [DOI] [PubMed] [Google Scholar]
- Guo B, Sallis RE, Greenall A, Petit MMR, Jansen E, Young L, Van de Ven WJ, Sharrocks AD. The LIM domain protein LPP is a coactivator for the ETS domain transcription factor PEA3. Mol Cell Biol. 2006;26:4529–4538. doi: 10.1128/MCB.01667-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen MDH, Beckerle MC. Opposing roles of zyxin/LPP ACTA repeats and the LIM domain region in cell-cell adhesion. J Biol Chem. 2006;281:16178–16188. doi: 10.1074/jbc.M512771200. [DOI] [PubMed] [Google Scholar]
- Hendrix P, Mayer-Jackel RE, Cron P, Goris J, Hofsteenge J, Merlevede W, Hemmings BA. Structure and expression of a 72-kDa regulatory subunit of protein phosphatase 2A. Evidence for different size forms produced by alternative splicing. J Biol Chem. 1993;268:15267–15276. [PubMed] [Google Scholar]
- Howe AK. Cross-talk between calcium and protein kinase A in the regulation of cell migration. Curr Opin Cell Biol. 2011;23:554–561. doi: 10.1016/j.ceb.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Jacobson K, Schaller MD. MAP kinases and cell migration. J Cell Sci. 2004;117:4619–4628. doi: 10.1242/jcs.01481. [DOI] [PubMed] [Google Scholar]
- Ito A, Kataoka TR, Watanabe M, Nishiyama K, Mazaki Y, Sabe H, Kitamura Y, Nojima H. A truncated isoform of the PP2A B56 subunit promotes cell motility through paxillin phosphorylation. EMBO J. 2000;19:562–571. doi: 10.1093/emboj/19.4.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V, Jordens J, Stevens I, Van Hoof C, Martens E, De Smedt H, Engelborghs Y, Waelkens E, Goris J. Identification and functional analysis of two Ca2+-binding EF-hand motifs in the B″/PR72 subunit of protein phosphatase 2A. J Biol Chem. 2003;278:10697–10706. doi: 10.1074/jbc.M211717200. [DOI] [PubMed] [Google Scholar]
- Janssens V, Goris J, Van Hoof C. PP2A: the expected tumor suppressor. Curr Opin Genet Dev. 2005;579:3392–3396. doi: 10.1016/j.gde.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Janssens V, Derua R, Zwaenepoel K, Waelkens E, Goris J. Specific regulation of protein phosphatase 2A PR72/B″ subunits by calpain. Biochem Biophys Res Commun. 2009;386:676–681. doi: 10.1016/j.bbrc.2009.06.096. [DOI] [PubMed] [Google Scholar]
- Jin L, Kern MJ, Otey CA, Wamhoff BR, Somlyo AV. Angiotensin II, focal adhesion kinase, and PRX1 enhance smooth muscle expression of lipoma preferred partner and its newly identified binding partner palladin to promote cell migration. Circ Res. 2007;100:817–825. doi: 10.1161/01.RES.0000261351.54147.de. [DOI] [PubMed] [Google Scholar]
- Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- Keicher C, Gambaryan S, Schulze E, Marcus K, Meyer HE, Butt E. Phosphorylation of mouse LASP-1 on threonine 156 by cAMP- and cGMP-dependent protein kinase. Biochem Biophys Res Commun. 2004;324:308–316. doi: 10.1016/j.bbrc.2004.08.235. [DOI] [PubMed] [Google Scholar]
- Lambrecht C, Haesen D, Sents W, Ivanova E, Janssens V. Structure, regulation, and pharmacological modulation of PP2A phosphatases. Methods Mol Biol. 2013;1053:283–305. doi: 10.1007/978-1-62703-562-0_17. [DOI] [PubMed] [Google Scholar]
- Li X, Virshup DM. Two conserved domains in regulatory B subunits mediate binding to the A subunit of protein phosphatase 2A. Eur J Biochem. 2002;269:546–552. doi: 10.1046/j.0014-2956.2001.02680.x. [DOI] [PubMed] [Google Scholar]
- Madsen CD, Hooper S, Tozluoglu M, Bruckbauer A, Fletcher G, Erler JT, Bates PA, Thompson B, Sahai E. STRIPAK components determine mode of cancer cell migration and metastasis. Nat Cell Biol. 2015;17:68–80. doi: 10.1038/ncb3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier GD, Wright MA, Lozano Y, Djordjevic A, Matthews JP, Young MR. Regulation of cytoskeletal organization in tumor cells by protein phosphatases-1 and -2A. Int J Cancer. 1995;61:54–61. doi: 10.1002/ijc.2910610110. [DOI] [PubMed] [Google Scholar]
- McDonald JA. Canonical and noncanonical roles of Par-1/MARK kinases in cell migration. Int Rev Cell Mol Biol. 2014;312:169–199. doi: 10.1016/B978-0-12-800178-3.00006-3. [DOI] [PubMed] [Google Scholar]
- Metodieva G, Nogueira-de-Souza NC, Greenwood C, Al-Janabi K, Leng L, Bucala R, Metodiev MV. CD74-dependent deregulation of the tumor suppressor scribble in human epithelial and breast cancer cells. Neoplasia. 2013;15:660–668. doi: 10.1593/neo.13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najm P, El-Sibai M. Palladin regulation of the actin structures needed for cancer invasion. Cell Adh Migr. 2014;8:29–35. doi: 10.4161/cam.28024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngan E, Northey JJ, Brown CM, Ursini-Siegel J, Siegel PM. A complex containing LPP and α-actinin mediates TGFβ-induced migration and invasion of ErbB2-expressing breast cancer cells. J Cell Sci. 2013;126:1981–1991. doi: 10.1242/jcs.118315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth MF, Cazes A, Butt E, Grunewald TGP. An update on the LIM and SH3 domain protein 1 (LASP1): a versatile structural, signaling, and biomarker protein. Oncotarget. 2015;6:26–42. doi: 10.18632/oncotarget.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Scheffler TL, Rossie SS, Gerrard DE. AMPK activity is regulated by calcium-mediated protein phosphatase 2A activity. Cell Calcium. 2013;53:217–223. doi: 10.1016/j.ceca.2012.12.001. [DOI] [PubMed] [Google Scholar]
- Perrotti D, Neviani P. Protein phosphatase 2A: a target for anticancer therapy. Lancet Oncol. 2013;14:e229–e238. doi: 10.1016/S1470-2045(12)70558-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit MMR, Mols R, Schoenmakers EFPM, Mandahl N, Van de Ven WJM. LPP, the preferred fusion partner gene of HMGIC in lipomas, is a novel member of the LIM protein gene family. Genomics. 1996;36:118–129. doi: 10.1006/geno.1996.0432. [DOI] [PubMed] [Google Scholar]
- Petit MMR, Fradelizi J, Golsteyn RM, Ayoubi TAY, Menichi B, Louvard D, Van de Ven WJM, Friederich E. LPP, an actin cytoskeleton protein related to zyxin, harbors a nuclear export signal and transcriptional activation capacity. Mol Biol Cell. 2000;11:117–129. doi: 10.1091/mbc.11.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit MMR, Meulemans SMP, Van de Ven WJM. The focal adhesion and nuclear targeting capacity of the LIM-containing lipoma-preferred partner (LPP) protein. J Biol Chem. 2003;278:2157–2168. doi: 10.1074/jbc.M206106200. [DOI] [PubMed] [Google Scholar]
- Petit MMR, Crombez KRMO, Vervenne HBVK, Weyns N, Van de Ven WJM. The tumor suppressor Scrib selectively interacts with specific members of the zyxin family of proteins. FEBS Lett. 2005a;579:5061–5068. doi: 10.1016/j.febslet.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Petit MMR, Meulemans SMP, Alen P, Ayoubi TAY, Jansen E, Van de Ven WJM. The tumor suppressor Scrib interacts with the zyxin-related protein LPP, which shuttles between cell adhesion sites and the nucleus. BMC Cell Biol. 2005b;6:1. doi: 10.1186/1471-2121-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullar CE, Chen J, Isseroff RR. PP2A activation by beta2-adrenergic receptor agonists: novel regulatory mechanism of keratinocyte migration. J Biol Chem. 2003;278:22555–22562. doi: 10.1074/jbc.M300205200. [DOI] [PubMed] [Google Scholar]
- Qin Y, Capaldo C, Gumbiner BM, Macara IG. The mammalian Scribble polarity protein regulates epithelial cell adhesion and migration through E-cadherin. J Cell Biol. 2005;171:1061–1071. doi: 10.1083/jcb.200506094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ. Life at the leading edge. Cell. 2011;145:1012–1022. doi: 10.1016/j.cell.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Rossé C, Linch M, Kermorgant S, Cameron AJM, Boeckeler K, Parker PJ. PKC and the control of localized signal dynamics. Nat Rev Mol Cell Biol. 2010;11:103–112. doi: 10.1038/nrm2847. [DOI] [PubMed] [Google Scholar]
- Sheppard SA, Loayza D. LIM-domain proteins TRIP6 and LPP associate with shelterin to mediate telomere protection. Aging. 2010;2:432–444. doi: 10.18632/aging.100170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SA, Savinova T, Loayza D. TRIP6 and LPP, but not Zyxin, are present at a subset of telomeres in human cells. Cell Cycle. 2011;10:1726–1730. doi: 10.4161/cc.10.11.15676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupe AM, Merrill RA, Strack S. Determinants for substrate specificity of protein phosphatase 2A. Enzyme Res. 2011;2011:398751. doi: 10.4061/2011/398751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag J-M, Sontag E. Regulation of cell adhesion by PP2A and SV40 small tumor antigen: an important link to cell transformation. Cell Mol Life Sci. 2006;63:2979–2991. doi: 10.1007/s00018-006-6300-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens I, Janssens V, Martens E, Dilworth S, Goris J, Van Hoof C. Identification and characterization of B″-subunits of protein phosphatase 2A in Xenopus laevis oocytes and adult tissues. Evidence for an independent N-terminal splice variant of PR130 and an extended human PR48 protein. Eur J Biochem. 2003;270:376–387. doi: 10.1046/j.1432-1033.2003.03398.x. [DOI] [PubMed] [Google Scholar]
- Storr SJ, Carragher NO, Frame MC, Parr T, Martin SG. The calpain system and cancer. Nat Rev Cancer. 2011;11:364–374. doi: 10.1038/nrc3050. [DOI] [PubMed] [Google Scholar]
- Takizawa N, Smith TC, Nebl T, Crowley JL, Palmieri SJ, Lifshitz LM, Ehrhardt AG, Hoffman LM, Beckerle MC, Luna EJ. Supervillin modulation of focal adhesions involving TRIP6/ZRP-1. J Cell Biol. 2006;174:447–458. doi: 10.1083/jcb.200512051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Itallie CM, Tietgens AJ, Aponte A, Fredriksson K, Fanning AS, Gucek M, Anderson JM. Biotin ligase tagging identifies proteins proximal to E-cadherin, including lipoma preferred partner, a regulator of epithelial cell-cell and cell-substrate adhesion. J Cell Sci. 2014;127:885–895. doi: 10.1242/jcs.140475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervenne HBVK, Crombez KRMO, Lambaerts K, Carvalho L, Köppen M, Heisenberg C-P, Van de Ven WJM, Petit MMR. Lpp is involved in Wnt/PCP signaling and acts together with Scrib to mediate convergence and extension movements during zebrafish gastrulation. Dev Biol. 2008;320:267–277. doi: 10.1016/j.ydbio.2008.05.529. [DOI] [PubMed] [Google Scholar]
- Vervenne HBVK, Crombez KRMO, Delvaux EL, Janssens V, Van de Ven WJM, Petit MMR. Targeted disruption of the mouse Lipoma Preferred Partner gene. Biochem Biophys Res Commun. 2009;379:368–373. doi: 10.1016/j.bbrc.2008.12.074. [DOI] [PubMed] [Google Scholar]
- Westermarck J, Hahn WC. Multiple pathways regulated by the tumor suppressor PP2A in transformation. Trends Mol Med. 2008;14:152–160. doi: 10.1016/j.molmed.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Wlodarchak N, Guo F, Satyshur KA, Jiang L, Jeffrey PD, Sun T, Stanevich V, Mumby MC, Xing Y. Structure of the Ca2+-dependent PP2A heterotrimer and insights into Cdc6 dephosphorylation. Cell Res. 2013;23:931–946. doi: 10.1038/cr.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M-Y, Xie X, Xu Z-K, Xie L, Chen Z, Shou L-M, Gong F-R, Xie Y-F, Li W, Tao M. PP2A inhibitors suppress migration and growth of PANC-1 pancreatic cancer cells through inhibition on the Wnt/β-catenin pathway by phosphorylation and degradation of β-catenin. Oncol Rep. 2014;32:513–522. doi: 10.3892/or.2014.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara K, Ikenouchi J, Izumi Y, Akashi M, Tsukita S, Furuse M. Phosphorylation state regulates the localization of Scribble at adherens junctions and its association with E-cadherin–catenin complexes. Exp Cell Res. 2011;317:413–422. doi: 10.1016/j.yexcr.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Young MRI, Kolesiak K, Meisinger J. Protein phosphatase-2A regulates endothelial cell motility and both the phosphorylation and the stability of focal adhesion complexes. Int J Cancer. 2002;100:276–282. doi: 10.1002/ijc.10491. [DOI] [PubMed] [Google Scholar]
- Zwaenepoel K, Louis JV, Goris J, Janssens V. Diversity in genomic organisation, developmental regulation and distribution of the murine PR72/B″ subunits of protein phosphatase 2A. BMC Genomics. 2008;9:393. doi: 10.1186/1471-2164-9-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaenepoel K, Goris J, Erneux C, Parker PJ, Janssens V. Protein phosphatase 2A PR130/B″α 1 subunit binds to the SH2 domain-containing inositol polyphosphate 5-phosphatase 2 and prevents epidermal growth factor (EGF)-induced EGF receptor degradation sustaining EGF-mediated signaling. FASEB J. 2010;24:538–547. doi: 10.1096/fj.09-140228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.