Abstract
Motivation
As the development of the high throughput sequencing technologies, transcriptome can be sequenced with a low price and high efficiency. Sequence assembly approaches have been renewed to meet the new requirements from new sequencing technologies. Assembly strategies are important for biologists who need to assemble the transcriptome generated in their experiments. However, some modern de novo assembly strategies generate a large section of redundant contigs due to sequence variations, which greatly affect downstream analysis and experiments. This work proposed TransPS, a post transcriptome scaffolding method to generate high quality transcriptomes.
Results
TransPS shows promising results on the test transcriptome data sets where the redundancy is greatly reduced by at least 50%, while the coverage is improved considerately.
Availability
The web server and source code are available at https://bioinformatics.cs.vt.edu/zhanglab/transps/
1. Introduction
The rapid development of the next generation sequencing technologies is bringing new development of the genome assembly tools that are able to handle the features of the new technologies as well as the downstream applications based on the quality of the sequencing. The demand of low cost of sequencing has driven high-through and high efficiency modern sequencing technologies, such as 454, Illumina, etc. Despite of the advantages of the next generation sequencing technologies, the length of the sequence generated by these modern instruments is considerably short (~100bp), which brings challenge to sequence assembly algorithms. Similar to short read genome assembly, transcriptome assembly needs to connect short, low quality reads. However, transcriptome assembly is even more difficulty than genome assembly with regards to considering sequencing depth, strand specific or transcript variants (Martin et al., 2011).
There are three typical transcriptome assembly strategies, reference based strategy, de novo strategy or combined strategy that joins the two. Widely used transcriptome assembly tools include Cufflinks (Adam et al., 2011) for reference based assembly, and Trinity (Grabherr et al., 2011) and Oases (Marcel et al., 2012) for de novo assembly. De novo assembler generates large number of contigs due to repetitive elements. They are also very sensitive to sequencing errors, which result in big redundancy in the output contigs. Paired-read sequencing technology could help to reduce the amount of contigs as the known intermediate distance between read pairs can be used to place contigs in their likely order and orientation. Assembly tools like Trinity does not include a scaffolding step, while the majority provide a scaffolding option only as a built-in function which cannot be independently controlled or effectively reduce the amount of contigs.
Very few previous studies about independently scaffolding preassembled transcriptome. SSPACE (Boetzer et al., 2011) is a tool to scaffold pre-assembled genome contigs using paired end data. In this work, we developed TransPS (Transcriptome Post Scaffolding) programming to scaffold pre-assembled transcriptome from any desired assemblers by using a reference species. The feature of TransPS is easy to implement and efficiently remove redundancy in the original config set. TransPS reduced the size of original contigs by at least %50 for our test datasets, while the quality of the scaffolding contigs are greatly enhanced in terms of the coverages.
2. MATERIALS AND METHODS
2.1. Datasets Description
Six test datasets were used to validate the TransPS pipeline, four of which were taken from NCBI Transcriptome Shotgun Assembly (TSA) projects, including Dendroctonus frontalis (TSA record: GAFI01), Dendroctonus ponderosae (TSA record: GAFX01), Diaphorina citric (TSA record: GACJ01) and Ixodes ricinus (TSA record: GACI01). The other two datasets are Aedes albopictus and Aedes hensilli transcriptome, which are newly sequenced via 454 and Illumina RNA-seq by Virginia Bioinformatics Institute. Please see supplementary table S1 for the corresponding reference sequences and pre-assembled methods used for each organism and other details.
2.2 Post Scaffolding
The input of the TransPS are pre-assembled contigs and the search results from NCBI BLASTX programming (Altschul et al., 1997) by aligning the contigs to the reference amino acid sequences. The post scaffolding procedure follows an align-layout-consensus structure, consisting of three major stages. First, original contigs search best alignment from the reference amino acid sequences database using BLASTX programming. Second, contigs are placed in a right order and orientation in terms of the coordinates aligned to the reference. Third, non-redundant sequences matched to the same sequence of the reference are scaffolded into one contig. Supplementary Figure S1 shows an overview of the procedure.
In the alignment stage, BLASTX was used in order to use protein sequence to guide the alignment of DNA sequence by searching a protein database using a nucleotide query. The best target sequence (lowest evalue and highest bitscore) is selected as the best match with the query sequence. It is possible that multiple cintigs match with the same protein sequence due to incomplete assembly of sequences, which is referred as a one-to-many matching, otherwise a one-to-one matching. In the case of one-to-many matching, all the contigs matching with the same protein are connected and put into a set of sequences Ti, where i represents protein i with which all contigs in Ti match.
To produce a high quality set of contigs, it is important to remove the redundant sequences generated by the assemblers. The contigs in Ti are divided into three subsets, contigs that are accepted as a final contig (Ai) that needs no further processing, contigs that are redundant (Ri), and sequences that are used to scaffold into one contig (Si). The one to-one matching contigs goes to set A. For oneto-many matching, sequences in Ti are assigned to Ri, Ai or Si. If two contigs match with the same protein at the same region or they are overlapped with each other for a certain percentage (user defined argument), then one of the two contigs is a redundant and removed to Ri. After moving the redundant and accepted sequences, all the remaining sequences in Ti go to subset Si for scaffolding. In the case of only one config left for scaffolding, it goes to Ai subset. This procedure is summarized in the supplementary Figure S1 (above the dashed line). In the following two stages, we focus on the contigs for scaffolding in Si.
In the layout stage, the purpose is to order the contigs in Si based on the reference coordinates provided by the alignment. Intuitively, to ensure the consistency with the reference genome, the contigs are sorted in terms of the matching start positions on the reference protein in a ascending order.
In the consensus stage, the purpose is to scaffold the original contigs in Si from s1 to sn into one high quality contig based on the order obtained from the previous stage (si ⎿ Si (1≤i≤n)). The most important part for scaffolding is to estimate the “scaffolding parts”, which are determined based on two cases, overlapped contigs or separated configs (see supplementary method for details).
The output of TransPS are contigs in three different groups, accepted contigs (A), scaffolded contigs (S) and unused contigs (R). The contigs in the redundant set (R) could be real redundancy due to genetic variants or different transcripts due to alternative splicing. A matching map between the original contigs and the corresponding reference protein sequences used in the scaffolding algorithm is also provided.
3. RESULTS
Table 3 shows the number of scaffolds from the original contigs that match with at least one reference sequence (No. matched original). The average number of configs per scaffold is 2 to 3. The contigs in the redundant set (R) account for most of the original contigs (% Shrinked). Please see supplementary Table S2 for detailed distribution of original contigs in three different groups.
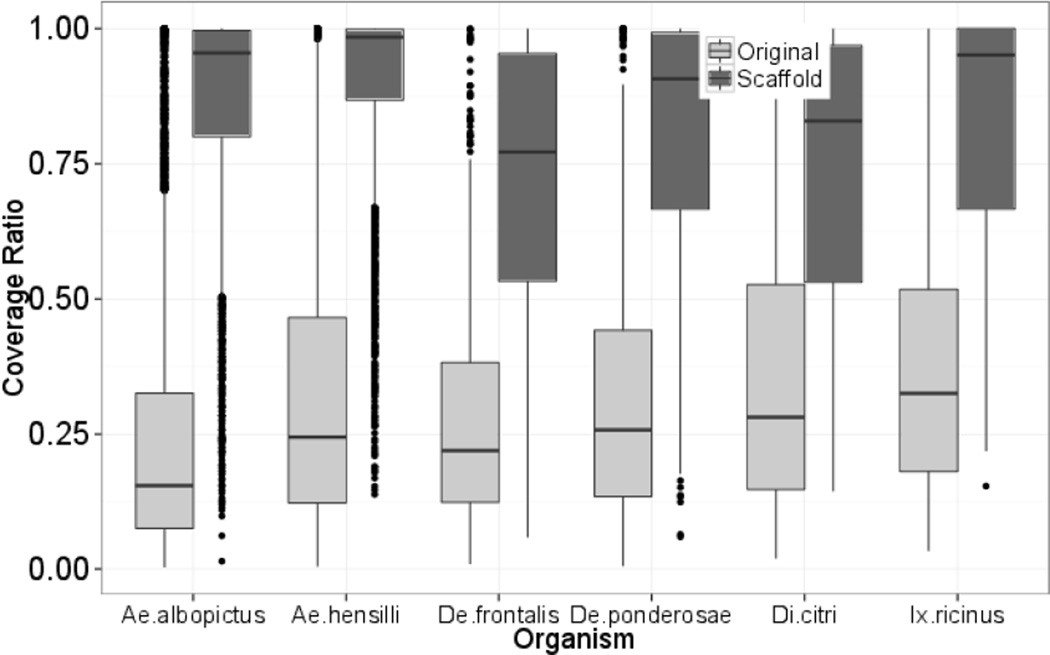
The new scaffolded contigs are compared with the original transcriptome contigs by measuring the coverage ratio against the matched reference protein sequence. Coverage ratio was calculated as the matched percentage between the scaffolded (or original) contig and the matched reference protein sequence in a global sequence alignment. The global sequence alignment was implemented by NAP (Huang et al., 1996), a programming doing global sequence alignment between a amino acid sequence and a nucleotide sequence. The redundant and accepted sequences in the original transcriptome set were also removed for comparison. As shown in Figure 1, the scaffolded contigs have much higher coverage ratio compared with the original ones and the median of scaffolded group is significant higher than the original group (p < 2.2e–16 ) in all the tested datasets.
Fig. 1.
Coverage comparison between scaffolds and original contigs for different data sets
4. CONCLUSION
This paper proposed a referenced based post transcriptome scaffolding method that considerately reduced the size of the original de novo assembled contigs while the qualities are greatly improved with regarding to coverage ratio. As more and more genomes become available in the public databases, it is more likely than not that a species with a complete genome closely related to a genome we are interested in. In this case, a post assembling by taking the information in the existed genome provides advantages to better guide the assembling of the studied genome.
Supplementary Material
Table 1.
Number of scaffolds generated by TransPS from different datasets
| NO. matched original |
NO. scaffolds | NO.contig /scaffold |
%Shrinked | |
|---|---|---|---|---|
| De. Frontal | 15,095 | 496 | 2.17 | 70 |
| De. Ponderosae | 16,457 | 433 | 2.15 | 74 |
| Di. citri | 16,732 | 165 | 2.21 | 90 |
| Ix. ricinus | 7,787 | 71 | 2.04 | 57 |
| Ae. albopictus | 47,445 | 3,863 | 3.36 | 74 |
| Ae. hensilli | 62,088 | 2,651 | 2.73 | 80 |
NO.contig/scaffold: average number of original contigs per scaffold
% Shrinked: percentage of “redundant” contigs
Acknowledgments
The authors would like to acknowledge Dr. Lenwood S Heath for help with the code and discussions.
Funding: National Institute of Health (grant XXX)
REFERENCES
- Martin JA, Wang Z. Article title. Nature Reviews Genetics. 2011;12:671–682. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 2011;7:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam R, et al. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- Marcel HS, et al. Oases: Robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;8:1082–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M, et al. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Zhang J. Methods for comparing a DNA sequence with a protein sequence. Bioinformatics. 1996;12:497–506. doi: 10.1093/bioinformatics/12.6.497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.