Abstract
Endometriosis is a highly prevalent, chronic, heterogeneous, fibro-inflammatory disease that remains recalcitrant to conventional therapy. We previously showed that loss of KLF11, a transcription factor implicated in uterine disease, results in progression of endometriosis. Despite extensive homology, co-expression, and human disease association, loss of the paralog Klf10 causes a unique inflammatory, cystic endometriosis phenotype in contrast to fibrotic progression seen with loss of Klf11. We identify here for the first time a novel role for KLF10 in endometriosis. In an animal endometriosis model, unlike wild-type controls, Klf10−/− animals developed cystic lesions with massive immune infiltrate and minimal peri-lesional fibrosis. The Klf10−/− disease progression phenotype also contrasted with prolific fibrosis and minimal immune cell infiltration seen in Klf11−/− animals. We further found that lesion genotype rather than that of the host determined each unique disease progression phenotype. Mechanistically, KLF10 regulated CD40/CD154-mediated immune pathways. Both inflammatory as well as fibrotic phenotypes are the commonest clinical manifestations in chronic fibro-inflammatory diseases such as endometriosis. The complementary, paralogous Klf10 and Klf11 models therefore offer novel insights into the mechanisms of inflammation and fibrosis in a disease-relevant context. Our data suggests that divergence in underlying gene dysregulation critically determines disease-phenotype predominance rather than the conventional paradigm of inflammation being precedent to fibrotic scarring. Heterogeneity in clinical progression and treatment response are thus likely from disparate gene regulation profiles. Characterization of disease phenotype-associated gene dysregulation offers novel approaches for developing targeted, individualized therapy for recurrent and recalcitrant chronic disease.
Keywords: CD154, CD40, CD40 ligand, endometriosis, epigenetics, inflammation, KLF10
INTRODUCTION
Endometriosis is a chronic disease characterized by peritoneal dissemination of uterine endometrial cells that affects 10% of reproductive aged women [1]. The disease causes chronic pain, infertility, and sexual dysfunction as well as affects abdominal and pelvic organ physiology. Endometriosis is characterized pathologically by progressive inflammation and prolific fibrosis, which results in profound anatomic and physiologic compromise of the urogenital and gastrointestinal systems. Current understanding is that chronic inflammation results in scarring and loss of function.
The ubiquitously expressed, highly homologous transcription factors KLF10 and KLF11 are implicated in several chronic human diseases [2–9]. KLF10 and KLF11 are members of the Sp/KLF family of 24 transcription factors that are widely expressed, and amongst themselves can potentially regulate most expressed genes [10, 11]. Family members exhibit greater than 80% homology in their zinc finger C-terminal DNA-binding region with more variable N-termini [12, 13]. Members are further classified into subgroups that exhibit greater than familial homology; accordingly KLF10 and KLF11 belong to the TIEG (TGFβ induced early genes) subgroup and can compensate for each other [14, 15]. Although KLF10 and KLF11 are both TGFβ- and sex steroid-regulated tumor suppressors, they have unique immunomodulatory and antifibrotic roles, respectively [3–5, 15–19]. Whereas KLF11 has a distinct antifibrotic role in endometriosis by repressing Collagen 1 expression; we describe here for the first time a role of KLF10 in endometriosis [3, 20]. KLF10 has anti-inflammatory roles through activation of T-regulatory cells via modulation and expression of FOXP3 [16]. Accordingly, loss of KLF10 results in unchecked inflammation and progression of disease in an inflammatory bowel disease model [21, 22]. Because endometriosis is predominantly characterized by inflammation and scarring, we evaluated the role of KLF10, the anti-inflammatory paralog of KLF11, in endometriotic progression.
Immune system dysregulation in endometriosis is multifaceted and heterogeneous. Several studies have characterized mechanisms underlying the attachment and establishment of ectopic endometriotic lesions within the peritoneal cavity by analyzing the immune composition of both peritoneal fluid and disease lesions [23]. The disease is associated with infiltration of activated macrophages, immature dendritic cells, and natural killer cells in the peritoneal cavity as well as in the core of lesions [24, 25]. The reasons for their specific recruitment and migration remain incompletely understood. Recent evidence has demonstrated that in addition to immune cells, epithelial cells can also act as immune effectors via cytokine and immune modulator expression and elaboration [26–28]. Endometriosis is characterized by varied inflammatory responses to ectopically implanted epithelial and stromal cells and thus offers a valuable model to investigate these mechanisms that are nevertheless fundamental in a diversity of chronic diseases. This is also the first study that evaluates the role of KLF10, a key sex-steroid responsive TGFβ-regulated disease relevant transcription factor in the reproductive tract and disease [7].
MATERIALS AND METHODS
Ethics Statement
All animal experiments were performed per the recommendations outlined in the Guide for Care and Use of Laboratory Animals from the National Institutes of Health as required by the Mayo Clinic (Rochester, MN). Our study protocol (A54112) was submitted, reviewed, and approved by the Institutional Animal Care and Use Committee at the Mayo Clinic. Animals were treated with ketamine and xylazine anesthesia for surgeries in order to ensure appropriate analgesia. Archived human endometrial and endometriosis samples for tissue microarray (TMA) assays were selected per the Mayo Clinic Institutional Review Board protocol 11-003074.
Tissue and Cell Lines
The Ishikawa cell line was utilized to characterize the immune processes involved in our in vivo model of endometriosis. Ishikawa is an established endometrial adenocarcinoma cell line that is well characterized and was obtained from Dr. P. Goodfellow (Washington University, St. Louis, MO). Ishikawa cells were maintained in Dulbecco-modified Eagle media (DMEM) supplemented with 10% fetal bovine serum (FBS). Primary immortalized endometrial stromal cells are an established cell line and are commercially available from American Type Culture Collection (CRL-4003) [29]. Primary immortalized endometrial stromal cells were grown in DMEM/F-12 and supplemented with 10% FBS. The 12Z endometriotic epithelial cell line was a gift from Dr. Romana Nowak (University of Illinois, Champaign-Urbana, IL). The cell line was created and established by Dr. Anna Starzinski-Powitz, who generously permitted us the use of these cells [30]. The cells were maintained in DMEM/F12 and supplemented with 10% FBS.
Expression Plasmid Constructs and Transfection Assays
Human wild-type (wt) KLF10 complementary DNA was amplified by PCR and cloned into the pcDNA4/His empty expression vector (EV) (Qiagen) to generate pcDNA4/His-KLF10. All constructs were sequenced to verify the correct sequence. Ishikawa cells (∼80% confluent in 10 cm plate) were transfected with 10 μg of pcDNA4/His-KLF10 or EV for 48 h. For KLF10 small interfering RNA (siRNA) knockdown assay, Ishikawa cells in 6-well plates were transfected with KLF10 siRNA (Dharmacon) or a scrambled control (Scr) using Trans-IT-TKO reagent (Invitrogen) per the manufacturer's protocol. Each well received 50 nM of KLF10 or Scr siRNA and 10 μl Trans-IT-TKO for 48 h.
TMA Design
Patients aged 15–45 yr old with a prior histologic diagnosis of endometriosis who had not been on hormonal therapy for at least 3 mo prior to hysterectomy were selected from Mayo Clinic's Department of Laboratory Medicine and Pathology institutional archives for the creation of TMAs. TMA development was approved per the Mayo Clinic Institutional Review Board protocol 11-003074. Following diagnostic confirmation and adequacy of tissue, formalin-fixed paraffin-embedded tissue blocks were selected for construction of the TMA. A matched pair of eutopic (endometrial) and ectopic (endometriosis) tissue was obtained from 28 patients. Normal endometrium from patients without endometriosis was also obtained to evaluate the menstrual cycle. A total of 142 human proliferative and 164 secretory endometrial samples were obtained and evaluated.
RNA Isolation and Quantitative PCR
Total RNA from one 10-cm dish of 80% confluent Ishikawa cells was extracted utilizing the RNeasy kit (Qiagen) per the manufacturer's protocol. Two micrograms of total RNA were used for complementary DNA synthesis using Oligo-dT primer in a SuperScriptTMIII first-strand synthesis system (Invitrogen) per the manufacturer's protocol. For real-time PCR, commercially available gene-specific primers (Qiagen) were used. The reactions were performed using the IQ-SYBR Green Supermix (Bio-Rad) per the manufacturer's protocol in a StepOne Plus Real-Time PCR System (Applied Biosystems). All quantitative PCR measurements were carried out within the linear amplification range. Each experiment was done in triplicate.
Human Innate and Adaptive RT2 Profile PCR Array
The human innate and adaptive RT2 profile PCR array (PAHS-052ZC-12; Qiagen) contains 84 innate and adaptive genes and five endogenous control genes. The protocol for real-time PCR was described above. Supplemental Table S1 (Supplemental data are available online at www.biolreprod.org) lists the genes measured in this assay. Five endogenous genes (B2M, HPRT1, RPL13A, GAPDH, and beta-actin) were used for normalization. Each cycle threshold (Ct) was normalized to the average Ct of the five controls on a per plate basis. The comparative Ct method was used to calculate relative quantification of gene expression. The data was analyzed using the PCR Array Data Analysis Software available from SABiosciences (http://www.sabiosciences.com/pcr/arrayanalysis.php).
Immunohistochemistry
Tissue sections were deparaffinized and rehydrated in xylene and a series of ethanol solutions. Antigen retrieval was performed by heating slides in a steamer for 20 min in 10 mM citrate buffer (pH 6.0). Endogenous peroxide was quenched using a solution of hydrogen peroxide/methanol solution, followed by avidin/biotin blocking (Vector Labs), and then by a blocking solution (CAS Block; Invitrogen). This was followed by incubation with anti-KLF10 992 (1:100 dilution; gift from Dr. John R. Hawse, Mayo Clinic) or CD154 (1:100 dilution; Abcam) overnight at 4°C. For the peptide-binding control, an epitope-corresponding peptide was synthesized at the Mayo Clinic Proteomic Core. The peptide (20 μg peptide and 2 μg antibody in 200 μl) was incubated overnight with anti-KLF10 in 1× PBS at 4°C prior to overnight incubation of tissue sections with the neutralized antibody as above. The sections were then incubated with secondary biotinylated horse anti-rabbit antibody (1:500 dilution; Vector Labs) for 30 min at room temperature followed by incubation with streptavidin (Invitrogen), NovaRED (Vector Labs), and Mayer hematoxylin counterstaining.
Chromatin Immunoprecipitation Assays
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (3, 41). Briefly, Ishikawa cells were grown to confluence and 10 × 106 cells were lysed for analysis. DNA shearing was performed to produce fragments of 200–600 base pairs in size by sonication. Anti-CD154 (1:250 dilution; Abcam), anti-acetyl histone H3 antibody (1:250 dilution; Abcam), or a species-specific control IgG was used for ChIP at 4°C for 16 h with rotation. The immunoprecipitate was analyzed per EZ-ChIP protocol (Millipore). PCR products representing CD154 promoter regions (−600 to −400) containing putative KLF10-binding GC elements were examined on a 2% agarose gel. Primer sequences for ChIP were as follows: forward/reverse, CTTGCATTATCTTTCCAGC/GAAACACATGTCAACAAAAG; quantitative PCR was performed as described above.
Western Blot
Total cell lysate was obtained from one 10-cm cell culture of Ishikawa cells and stromal cells for Western blot analysis. Standard Western blot techniques were used to determine protein expression; 10 μg protein was separated by 12% SDS-PAGE, transferred to a polyvinylidene fluoride membrane, and probed with anti-KLF10 (1:250 dilution; Abcam) or anti-β-tubulin (1:500 dilution; Sigma) at 4°C overnight followed by incubation with secondary antibody (1:5000 dilution; Santa Cruz Biotechnology, Inc.) for 1 h. The enhanced ECL system (GE/Amersham) was used for detection of signals.
Mouse Colony and Animal Endometriosis Model
Whole body C57BL/6 (B6) Klf10 knockout (Klf10−/−) mice were gifts from John Hawse and Malayannan Subramaniam, Department of Biochemistry, Mayo Clinic [18]. Mouse genotypes were confirmed by PCR; 8- to 12-wk-old mice were used for all experiments. Endometriosis was surgically induced in these animals by autologous transplantation of two 5-mm everted segments of uterine horn sutured to the parietal peritoneum (n = 7 animals in each group) [3]. This was performed using a well-characterized surgical approach by excision of two uterine horns followed by autologous transplantation of two 5-mm segments sutured onto the parietal peritoneum. In all experiments, both Klf10−/− and Klf11−/− animals were compared to wt C57BL6 controls.
In Vitro Mutagenesis
Plasmid KLF10EAPP mutant was generated by mutation of E40P and A41P using the QuikChange mutagenesis kit (Stratagene) as previously described for KLF11 and KLF16, which contain homologous N-terminal SIN3A/HDAC-binding domains [31, 32]. The mutant construct was verified by sequencing. Primers for in vitro mutagenesis were generated in the designated software program (Stratagene). Corresponding primer sequences are: forward, cagagaaaagtgattttccgcccgtagaagcacttatgtc, and reverse, gacataagtgcttctacaaaatcacttttctctg, wherein the mutagenized element is italicized.
Luciferase Reporter Assay
The pGL4-basic EV was purchased from Promega. The pGL4 basic promoter-reporter construct containing 609-base pair GC-rich elements in the CD154 promoter were generated by PCR: forward primer, CCCGGTACCTTGCATTATCTTTCCAGC, and reverse primer, CAAGAGCTCGGCAGCATGAGAAGACTGTC. Ishikawa and 12Z cells at 80% confluence were cotransfected with either 2.5 μg of pcDNA3/HIS EV (Invitrogen), pcDNA3/HIS-KLF10, or pcDNA3/HIS-KLF10EAPP construct and 3 μg of a pGL4-CD154-promoter-reporter construct (−600 to +9), corresponding to the region evaluated by ChIP. Forty-eight hours after transfection, cells were lysed and reporter activity was read using the Luciferase assay system (Promega) and a 20/20 luminometer (Turner Designs) per the manufacturer's protocol. Data in relative light units was normalized to lysate protein concentrations as characterized previously. Experiments were performed in triplicate, three independent times.
Statistical Analysis
All the results are expressed as means with a standard error of means (SEM). Each experiment was repeated in triplicate at least three times. The Bonferroni method of multiple comparisons (t-tests) or chi square tests were used as indicated by the data type. All statistical tests were two-sided. For consistency, all P values are reported as less than 0.05 throughout, where significant.
RESULTS
KLF10 Was Expressed in Human Endometrial Cells and Was Selectively Diminished in Ectopic Endometriosis Lesions Compared to Eutopic Uterine Endometrium
Uterine endometrial mucosa consists of sex steroid-responsive stromal and epithelial cells supported by vasculature. We initially determined KLF10 mRNA expression in the Ishikawa endometrial epithelial adenocarcinoma cell line and stromal cells and found it to be expressed in both cell types (Fig. 1A).
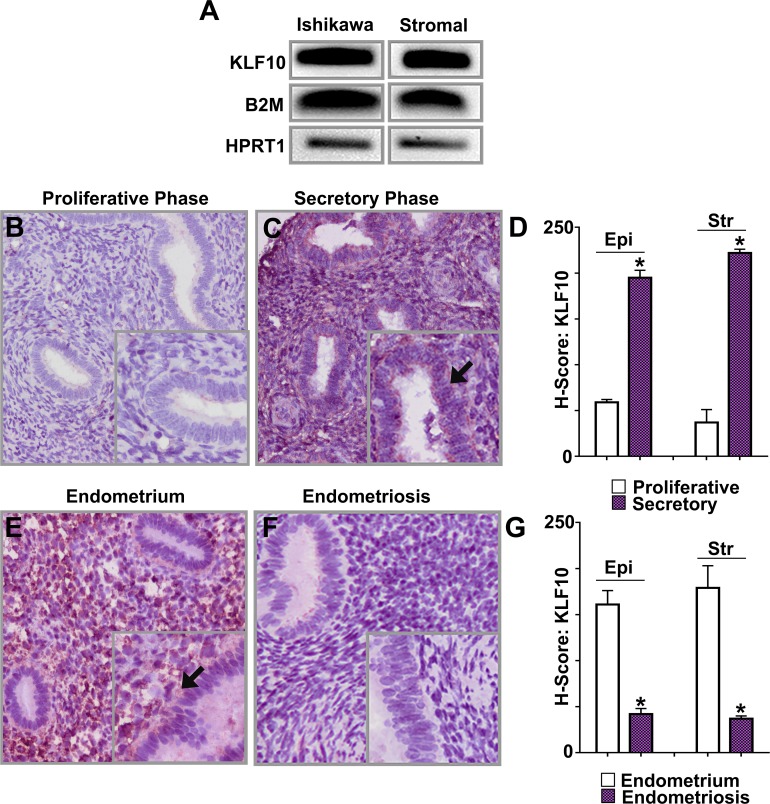
FIG. 1.
KLF10 expression in epithelial, stromal cells, uterine eutopic endometrium, and endometriosis. A) Expression of KLF10 mRNA was assessed in two cell lines: Ishikawa, a well-differentiated endometrial adenocarcinoma cell line, and human endometrial stromal cells (Stromal). KLF10 was expressed in both cell lines utilizing B2M and HPRT1 as reference controls. To demonstrate KLF10 expression in each of these endometrial cell lines, lysate for RNA or protein extraction was obtained from one 10 cm cell culture dish of confluent cells. B, C) Using a tissue microarray (TMA) consisting of 306 samples of benign uterine endometrium during both the proliferative and secretory phases of the menstrual cycle, immunohistochemistry was performed for KLF10. KLF10 expression was significantly increased in the secretory phase of the menstrual cycle as compared to the proliferative phase (magnification ×200; inset ×400). Inset picture with arrow indicates increased staining. D) H scores were 59 ± 3 and 195 ± 8 in epithelial cells (Epi) and 37 ± 14 and 222 ± 4 in stromal cells (Str) during the proliferative and secretory phases (*P < 0.05 as shown). E, F) KLF10 expression was also evaluated in 28 paired samples of eutopic endometrium and ectopic endometrial implants by immunohistochemistry. Representative samples are shown. KLF10 was expressed in nuclei and cytoplasm of epithelial and stromal cells in both eutopic endometrium as well as in endometriotic implants. KLF10 expression was decreased in endometriotic implants compared to eutopic endometrium (magnification ×200; inset ×400). Inset picture with arrow indicates increased staining. G) Corresponding H scores were: 161 ± 15 and 42 ± 6 in epithelial cells and 179 ± 24 and 37 ± 3 in stromal cells in eutopic endometrium and endometriosis, respectively (*P < 0.05 as shown).
Most active genes within the endometrium display cyclic differences in expression suggestive of distinct physiological roles. To determine if in addition to its epithelial and stromal expression, KLF10 also displayed a dynamic endometrial expression pattern suggestive of a role in endometrial physiology, we evaluated its expression in a TMA consisting of 142 human proliferative and 164 secretory endometrial samples (Fig. 1, B–D). KLF10 expression was significantly up-regulated in both endometrial epithelial and stromal cells in the secretory phase compared to levels in the proliferative phase. H scores were 59 ± 3 and 195 ± 8 in epithelial cells (P <0.05) and 37 ± 14 and 222 ± 4 in stromal cells (P < 0.05) during the proliferative and secretory phase, respectively (Fig. 1D). Endometrial KLF10 expression was evaluated using anti-KLF10; antibody-binding specificity was evaluated by pre-incubation with an epitope-specific peptide prior to tissue application. In addition, representative secretory phase peptide pretreated control is shown (Supplemental Fig. S1). KLF10 therefore likely has a role in endometrial differentiation associated with the secretory phase.
To examine KLF10 expression in human endometriosis, we evaluated 28 paired samples of uterine endometrium (eutopic) and endometriosis lesions (ectopic) from the same patients. Like KLF11, KLF10 was also specifically diminished in endometriosis lesions compared to eutopic endometrium (Fig. 1, E and F) [3]. Corresponding H scores were 161 ± 15 and 42 ± 6 in epithelial cells (P < 0.05) and 179 ± 24 and 37 ± 3 in stromal cells (P < 0.05) in eutopic endometrium and endometriosis respectively (Fig. 1G).
KLF10 Modulated the Immune Response to Implantation of Endometriosis Lesions
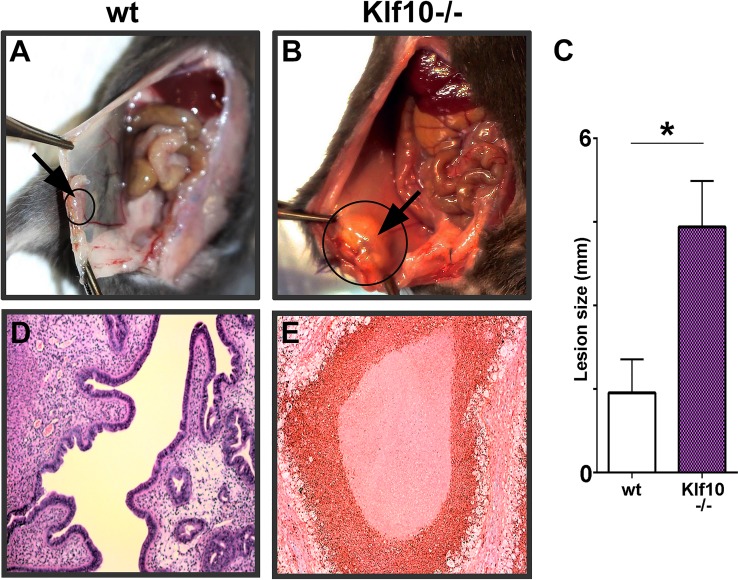
Although the presence of endometrial tissue in the peritoneal cavity elicits immune responses, the responses still remain to be coherently categorized [23, 33]. Failure of immune elimination likely results in miliary implantation and growth of ectopic endometrium that cause clinical endometriosis. To determine the role of KLF10 in the pathogenesis of endometriosis, we evaluated lesion progression in a Klf10−/− mouse disease model. Endometriosis was surgically induced in whole body Klf10−/− knockout mice. These Klf10−/− mice appeared phenotypically normal with no obvious reproductive tract defects although average litter size was significantly different in Klf10−/− animals compared to wt (5.1 ± 0.48 compared to 7.6 ± 0.4, respectively; P < 0.05, n = 30 litters/genotype). In contrast to wt animals, loss of Klf10 was associated with lesion enlargement with a >2-fold increase in size (P < 0.05; Fig. 2, A–C). Further, these lesions were associated with a massive, predominantly polymorphonuclear cell infiltrate unlike wt lesions in wt animals (Fig. 2, D and E).
FIG. 2.
Role of KLF10 on disease phenotype and lesion size. A, B, C) Endometriosis was surgically induced in 8-wk-old wt and Klf10−/− female mice (n = 7/group). At induction, 0.5 cm endometrial implants were sutured onto the parietal peritoneum of both groups. Lesion size, morphometry, and disease phenotype were evaluated 3 wk later at necropsy. Peritoneal lesions (circles and arrows) in Klf10−/− animals were larger, cystic, and associated with an inflammatory exudate (B). In contrast, wt mice showed lesion regression and minimal to no fibrosis or inflammatory exudate (A). Average lesion size of Klf10−/− animals was significantly greater (>2 fold) than wt (C). Error bars represent standard deviation (*P < 0.05). D, E) Histochemical analysis using hematoxylin and eosin revealed Klf10−/− lesions to be associated with a large neutrophilic infiltrate and complete obliteration of normal glands and stroma. In contrast, wt lesions had normal appearing glands and stroma. Magnification ×100 (A, B, D, E).
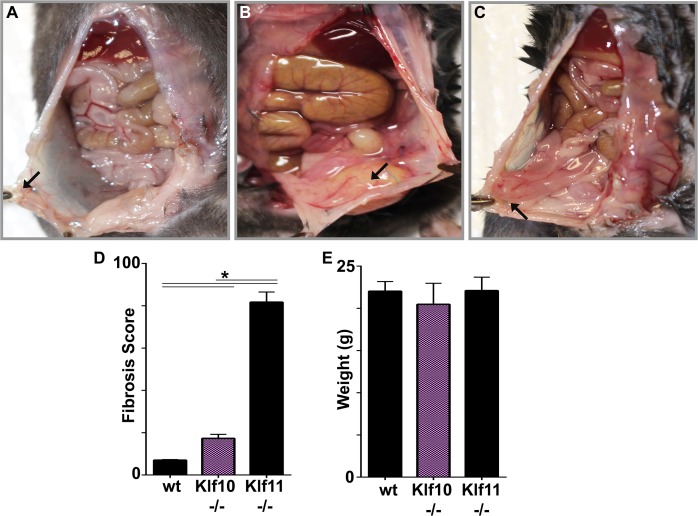
As previously shown, wt animals exhibited lesional regression and no fibrosis (Fig. 3, A and D). Despite the observed prolific inflammatory response, Klf10−/− animals unexpectedly did not display any significant fibrosis in contrast to that observed in Klf11−/− mice (Fig. 3, B–D) [3]. Lack of fibrosis in wt and Klf10−/− mice was evident by the presence of only discrete peritoneal lesions that were not adherent to any surrounding abdominal viscera (Fig. 3, A and B). In contrast, lesions in Klf11−/− animals were densely adherent to abdominal viscera with progression of scarring into connective tissues beyond the lesion (Fig. 3C) [3, 20]. Genotype-specific discrepancy in fibrotic extent was correspondingly reflected in their significantly divergent fibrosis scores (Fig. 3D). Specifically, Klf11−/− animals have a fibrosis score 2.3-fold greater than the Klf10−/− animals and 8-fold greater than wt controls (P < 0.05). Despite significant inflammation, in contrast with clinical findings in humans under comparable inflammatory conditions, Klf10−/− mice, like their wt and Klf11−/− counterparts, displayed continued well-being and no weight loss (Fig. 3E).
FIG. 3.
Loss of KLF10−/− demonstrates a predominately inflammatory response with minimal fibrosis. Endometriotic lesions in Klf10−/− mice were associated with prolific inflammation with associated infiltrate and minimal associated fibrosis. A) Wild-type animal demonstrating endometrial implant regression (arrow). Lesions were discrete and not physically adherent to surrounding peritoneum or abdominal viscera at time of necropsy. B) Klf10−/− animal demonstrating significant inflammatory infiltrate and minimal fibrosis (arrow: lesion). These lesions were associated with no or minimal easily ruptured adhesions to surrounding structures. There was also no associated mesenteric shortening. C) Klf11−/− animal demonstrating dense fibrosis of bowel to peritoneal lesion (arrow). In contrast to wt and Klf10−/− animals, lesions in Klf11−/− were nondiscrete and encased in adhesions to surrounding peritoneum and abdominal viscera. D) A murine fibrosis adhesion score was utilized to evaluate overall fibrosis in the endometriosis model. Klf11−/− animals demonstrate significantly higher fibrosis scores (>8 fold when compared to wt and >4 fold when compared to Klf10−/−). Error bars represent standard deviation (*P < 0.05). E) Induction of endometriosis, despite varying phenotypes, did not alter animal weights. Error bars represent standard deviation.
Lesion Rather Than Host Genotype Predominantly Drove Disease Progression in Klf10−/− Mice
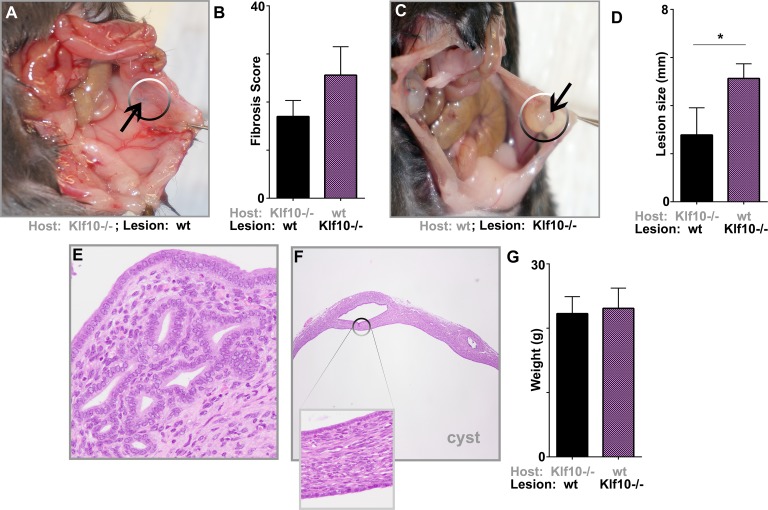
To distinguish lesion from host effects on the role of inflammation in disease pathogenesis, we transplanted Klf10−/− uterine lesions into wt control animals and vice versa. Lesions in Klf10−/− animals allografted with wt uterine implants regressed over the experimental duration (Fig. 4A). Moreover, these wt allografts in Klf10−/− mice displayed neither any significant inflammatory infiltrate nor increased fibrosis as reflected by their low fibrosis scores (Fig. 4, A, B, and E). In contrast, in wt animals allografted with Klf10−/− implants, the lesions were large, cystic, and phenotypically resembled cystic human endometriosis lesions. Klf10−/− implants in wt animals were 1.8-fold greater in size than wt lesions in Klf10−/− animals (P < 0.05; Fig. 4, C and D). These lesions remained cystic and large, demonstrating no regression over the study duration. Unlike Klf10−/− allografts in Klf10−/− animals, Klf10−/− allografts in wt animals were not heavily infiltrated with polymorphonuclear leukocytes (Fig. 4F). Despite the lack of a significant inflammatory infiltrate, Klf10−/− lesions retained their cystic phenotype in contrast to regression of wt lesions. Animals in both experimental groups displayed continued well-being with no significant weight loss (Fig. 4G).
FIG. 4.
Progression to endometriosis is lesion rather than host driven. Endometriosis was surgically induced in 8-wk-old wt and Klf10−/− mice (n = 5/group) and transplants were performed by suturing the Klf10−/−implants into wt control animals and vice versa. Lesion size, morphometry, and disease phenotype were evaluated 3 wk later. A) A Klf10−/− animal with wt implants demonstrating lesional regression. B) Klf10−/− animals with wt lesions had a trend toward lower fibrosis scores than wt animals with Klf10−/− lesions. Error bars represent standard deviation. C) Comparatively wt animals with Klf10−/− implants developed larger, cystic lesions. There was, however, no lesional inflammatory exudate as found in the Klf10−/− animals transplanted with Klf10−/−lesions. D) Graph depicts average lesion size for each group. Error bars represent standard deviation (*P < 0.05). E, F) Histochemical analysis of lesions using hematoxylin and eosin stain was performed on both lesion types. Klf10−/− animals with wt-transplanted lesion demonstrated either normal appearing glands and stroma or complete lesional regression (magnification ×200) (E). Wild-type animals with Klf10−/− implants demonstrated large cystic lesions (magnification ×200; inset ×400) (F). G) Animal weights did not differ across experimental groups. Average weight for each group is depicted with error bars representing standard deviation.
KLF10 Bound and Regulated CD154/CD40 in Endometrial Cells
The phenotype we obtained in the Klf10−/− animal endometriosis model suggested an underlying dysregulated immune response. Inflammation in endometriosis is determined by the qualitative interaction of displaced endometrial epithelial and stromal cells with host tissues. Recent evidence indicates that epithelial cells can impact immune function [34]. Because the lesion genotype was sufficient to drive disease progression, we evaluated the effect of diminished KLF10 expression on an array of immune-related genes in the model endometrial epithelial adenocarcinoma cell line (Ishikawa).
To investigate the role of diminished KLF10 expression (as in human endometriosis lesions and in the animal model), we transfected Ishikawa cells with KLF10 siRNA and evaluated the effect of diminished KLF10 expression on a profile of immune response genes (Fig. 5A and Table S1). Whereas diminished KLF10 activated genes that mediate diverse immune mechanisms, up-regulation of the ligand CD154 was most consistent and significant. Expression of CD154 was 5.3-fold greater in cells transfected with KLF10 siRNA compared to cells transfected with Scr control (P < 0.05) (Fig. 5, A and E, and Table S1). KLF10 siRNA also activated the CD154 receptor (CD40) significantly but to a lesser extent (1.36 fold, P < 0.05; Fig. 5A and Table S1).
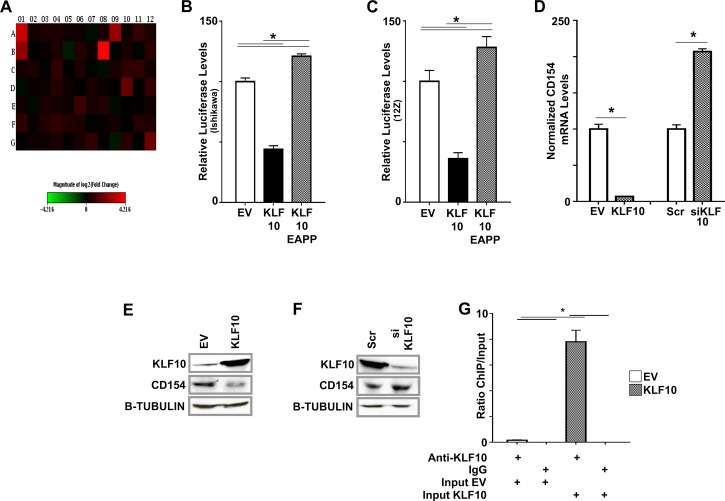
FIG. 5.
KLF10 binds to and regulates CD154. A) Heat map showing expression of 84 innate and acquired immune-related genes determined using PCR arrays in Ishikawa cells transfected with Scr and KLF10 siKLF10. The results were normalized to the expression of five housekeeping genes and represent the average fold change from three independent biological replicates. Decrease in KLF10 resulted in a statistically significant elevation in expression of CD40 (A12) and CD154 (B1) at 1.36 and 5.3 fold, respectively (P < 0.05 based on three biological replicates). Complete list of altered genes is provided in Supplemental Table S1. B, C) Ishikawa cells (B) and 12Z cells (C) were cotransfected with pcDNA3/HIS (EV), pcDNA3/HIS-KLF10, or pcDNA3/HIS-KLF10EAPP and a pGL4/CD154-promoter-reporter construct. KLF10 repressed CD154-promoter luciferase activity compared to EV (*P < 0.05 compared to EV). In contrast, KLF10EAPP derepressed and thus activated CD154 promoter luciferase expression (*P < 0.05, compared to KLF10 and EV as indicated). Luciferase levels were normalized to total lysate protein concentration. Assays were repeated in triplicate three times. D) Overexpression of KLF10 in Ishikawa cells decreased normalized CD154 mRNA expression 9 fold compared to EV. In contrast, CD154 mRNA expression levels were significantly increased in Ishikawa cells transfected with KLF10 siRNA compared to Scr control. CD154 mRNA expression was normalized to five housekeeping genes (mean expression levels ± SEM shown, *P < 0.05). E) KLF10 overexpression in Ishikawa cells transfected with pcDNA3/HIS-KLF10 cognately suppressed CD154 protein expression compared to corresponding EV. Beta-TUBULIN was used as a loading control. F) Conversely, suppressed transcription factor expression in cells transfected with KLF10siRNA was associated with increased CD154 protein expression compared to that in cells transfected with scrambled control (Scr). Beta-TUBULIN was used as a loading control. G) Chromatin immunoprecipitation (ChIP) assay was used to determine direct KLF10 binding to the region −600 to −401 of the CD154 promoter in Ishikawa cells. Promoter binding was detected by anti-KLF10 but not a control species- and isotype-specific IgG. Promoter-transcription factor binding was increased nearly 8 fold in cells transfected with pcDNA3/His KLF10 compared to EV. Levels were normalized to input (diluted 1:100). *P < 0.05 for comparisons of normalized binding levels in EV or KLF10-transfected cells analyzed by ChIP using anti-KLF10 or IgG as well as for comparison of normalized binding levels in EV and KLF10-transfected cells analyzed by ChIP using anti-KLF10 as indicated.
We focused on CD154 regulation and performed luciferase reporter assays using deletional constructs spanning the region −600 to +6. KLF10 repressed CD154 promoter/luciferase expression from the region −600 to −401 in the Ishikawa endometrial adenocarcinoma cell line and in 12Z primary endometriotic cells (Fig. 5, B and C). CD154 promoter activity was repressed >50% in both cell types transfected with KLF10 compared to controls transfected with the corresponding EV, (P < 0.05; Fig. 5, B and C). When the N-terminal SIN3A/HDAC-binding region of KLF10 was mutagenized in vitro to abrogate corepressor binding (KLF10EAPP), CD154 promoter activity was comparably derepressed in both cell types (P < 0.05; Fig. 5, B and C).
To determine the effect of promoter regulation on target gene expression, we measured CD154 mRNA and protein expression in Ishikawa cells under differential KLF10 expression levels. Overexpression of KLF10 cognately decreased CD154 expression at both RNA and protein levels corresponding to its effect on the promoter (P < 0.05; Fig. 5, D–F). In contrast, KLF10 siRNA transfection increased CD154 expression at both RNA and protein levels (P < 0.05; Fig. 5, D–F). KLF10 also specifically bound the region spanning −600 to −401 of the CD154 promoter (Fig. 5G) wherein it drove promoter activity, indicating direct transcriptional regulation (Fig. 5, B and C). KLF10 binding was detected using anti-KLF10 compared to species-specific IgG control. Transfection of cells with KLF10 further augmented transcription factor-promoter binding nearly 8 fold compared to that observed in EV-transfected controls (Fig. 5G). KLF10 was thus associated with repression of CD40/154 in Ishikawa cells although CD154 appeared to be more robustly regulated by KLF10 than its receptor CD40.
Endometrial Epithelial KLF10/CD154 Is a Novel Pathway Mediating Inflammatory Dysregulation and Disease Progression
To determine the disease relevance of KLF10-mediated CD154 repression in endometriosis, we evaluated CD154 expression in the murine endometrial implants as well as in human endometriotic lesions. Accordingly, CD154 mRNA expression in Klf10−/− murine implants was significantly elevated >4 fold compared to levels in wt implants (P < 0.05; Fig. 6A). Compared to wt, Klf10−/− lesions also demonstrated significantly greater CD154 protein expression (Fig. 6, B and C)
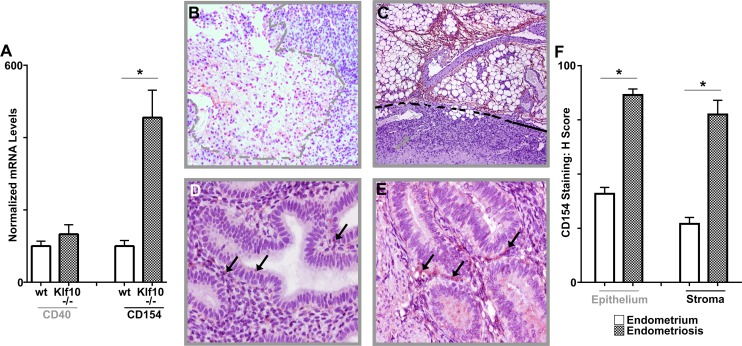
FIG. 6.
Role of CD154 in murine endometriotic implants and human endometriosis. A) CD154 mRNA expression levels were determined from one of two endometriotic implants in each animal (Klf10−/− and wt, n = 7). CD154 expression levels were increased >4 fold in implants from Klf10−/− animals compared to wt (*P < 0.05; error bars represent SEM). B, C) CD154 expression in wt (B) and Klf10−/− (C) implants demonstrate increased expression in Klf10−/− lesions particularly in vascular areas surrounding the neutrophilic infiltrate. Dashed lines indicate lesion extent and region of neutrophilic infiltrate. Magnification ×100. D, E) CD154 expression was also evaluated by immunohistochemistry in a tissue microarray (TMA) of 28 patients comparing eutopic (D) and ectopic endometria (E). CD154 expression was higher in ectopic compared to corresponding eutopic endometrium (arrows). Magnification ×400. F) Differences in CD154 expression were reflected in their H scores of 81 ± 6 and 163 ± 5.5 for epithelial CD154 expression in eutopic and ectopic endometrium, respectively, and 54 ± 6 and 155 ± 13 for stromal CD154 expression in eutopic and ectopic endometrium, respectively (*P < 0.05).
In human endometriosis lesions, diminished KLF10 was also associated with correspondingly increased CD154 protein expression. These differences were reflected in their corresponding H scores: 81 ± 6 and 163 ± 5.5 for epithelial CD154 expression in eutopic and ectopic endometrium, respectively (P < 0.05), and 54 ± 6 and 155 ± 13 for stromal CD154 expression in eutopic and ectopic endometrium, respectively (P < 0.05; Fig. 6, D–F).
DISCUSSION
Inflammation and fibrosis are critical pathogenic mechanisms in systemic- and organ-specific diseases. Clinically, both mechanisms are progressive and highly recalcitrant to conventional therapy; translational research that addresses inflammation and fibrosis is thus likely to have a significant positive therapeutic impact. We investigated the role of KLF10 in uterine endometrium and in endometriosis, a highly prevalent disease characterized by inflammation and fibrosis. KLF10 was expressed in endometrial epithelial and stromal cells with physiological variation throughout the menstrual cycle. As with its paralog KLF11, levels of KLF10 were selectively decreased in extra-uterine, peritoneal endometriosis disease lesions compared to corresponding levels in intrauterine endometrium. Endometriosis in humans was therefore associated with lesion-specific diminished KLF10 expression.
Selective and specific loss of Klf10−/− in our animal disease model was associated with disease progression characterized by large, cystic lesions and prominent inflammation compared to either Klf11−/− or wt animals. Unlike Klf11−/− mice, Klf10−/− animals demonstrated minimally increased fibrosis compared to wt. Because inflammation is believed to result in scarring, the evident phenotype was thus significantly discrepant from the expected. The relationship between inflammation and fibrosis is therefore likely to be nonlinear and/or nonimplicit, which explains disease heterogeneity as well as divergent responses to conventional therapy.
The endometriosis lesion consists of ectopically implanted uterine endometrial epithelial and stromal cells. Although it is likely that the host tissue mounts an inflammatory response to refluxed, spontaneously invading human, or surgically implanted murine endometrial fragments, the discrepant phenotypes evident in wt, Klf10−/−, and Klf11−/− animals does not support this paradigm. To further delineate discrepancies between host- and implant-driven tissue responses, we transplanted Klf10−/− uterine implants into wt hosts and vice versa. Cystic lesional development was only seen in wt animals with Klf10−/− endometrial implants. The genotype of the lesion therefore played a key role in determining disease progression.
Interaction with the host immune system is also critical in determining disease progression. This is evident from the divergence in tissue response ranging from highly inflammatory (Klf10−/− lesions in Klf10−/− hosts) or intermediate (Klf10−/− lesions in wt hosts) to none (wt lesions in Klf10−/− hosts). Of further interest is also the divergence in fibrotic extent amongst genotypically distinct animals. Accordingly, the closely related paralogs Klf10 and Klf11 display wide divergence in their respective pro-inflammatory and profibrotic disease phenotypes. Evidence from wound-healing mechanisms suggests that inflammation precedes fibrotic healing and that these are often proportionally related. When applied to chronic diseases such as endometriosis, postsurgical healing or chronic systemic diseases such as scleroderma or fibromyalgia, this translates into the use of anti-inflammatory agents and/or surgical lysis of scar tissue. Such conventional treatment options have an empiric rather than etiological basis and hence are commonly ineffective. In this study, in Klf10−/− animals with known immune dysfunction, lesion implantation resulted in massive inflammatory cell infiltration and necrosis with only modest fibrosis. In contrast, Klf11−/− animals had minimal lesional inflammatory infiltration although they developed prolific fibrosis that extended well beyond the peri-lesional region. Both the Klf10−/− and Klf11−/− phenotypes are commonly associated with chronic fibro-inflammatory systemic diseases such as endometriosis. Moreover, although the respective patterns of disease progression with loss of Klf10 or Klf11 are divergent, they still are phenotypically conformant with the heterogeneous presentations of human disease. Our findings suggest that divergence in activation of underlying signaling pathways induces differential disease phenotypes, progression patterns, and clinical presentation, which remains an ongoing area of investigation. Although phenotypically similar to infectious pathology, Klf10−/− animals did not display any clinical evidence associated with overwhelming sepsis. The immune response in endometriosis is therefore likely to be qualitatively different, resulting in chronic disease progression rather than in an acute inflammatory crisis.
KLF10 is a regulator of genes mediating diverse immune pathways [16, 21]. KLF10 was diminished in human endometriosis lesions, and lesional loss of Klf10 drove disease progression in mice. We therefore determined the effect of diminished KLF10 levels on an array of immune pathway-related genes in the Ishikawa endometrial epithelial adenocarcinoma cell line. Diminished KLF10 levels significantly activated the expression of CD40 and more significantly its ligand CD154. KLF10 also directly bound the CD154 promoter and repressed promoter activity and gene expression in these cells. In human endometriosis lesions, diminished KLF10 levels were also associated with increased CD154 expression, suggestive of a role for the KLF10/CD154 pathway in human endometriosis.
The CD40/CD154 co-stimulatory system belongs to the TNFα receptor family and was discovered as a critical link between T and B cell-mediated immune responses [35, 36]. CD40 is expressed on a variety of epithelial cells in the lung, thymus, and kidney [37–40]. Recently, increased CD40 and soluble CD154 have been associated with chronic inflammatory diseases such as asthma, chronic renal insufficiency, cardiovascular risk, and inflammatory bowel disease [34, 40–42]. Here, we show for the first time that KLF10 regulates CD154 expression in vitro in the Ishikawa uterine epithelial adenocarcinoma cell line, in 12Z primary endometriotic epithelial cells, in vivo in human endometriosis lesions, as well as in the mouse model in ectopic endometrial lesions.
KLF10 recruits nuclear cofactors to epigenetically regulate target genes [21]. Depending on the cofactor complex recruited, Klf10 functions either as a transcriptional activator or repressor of cognate target. KLF10-recruited cofactor complexes enzymatically induce specific posttranslational histone modifications on its target gene promoters [21, 43, 44]. Such histone modifications result in alterations of local chromatin configuration that critically determines gene activation or repression. KLF10 has been previously shown to interact with the JARID1B/KDM5B histone demethylase corepressor [45]. KLF10 shares an N-terminal repressor domain with KLF11 that binds to SIN3A/HDAC corepressor to affect target gene regulation [31, 46–48]. We show here for the first time that as with KLF11, this region in KLF10 is critical for epigenetic repression of CD154. The clinical significance of recruitment of epigenetic-signaling pathways by KLF transcription factors is evidenced by human diseases such as neonatal diabetes in individuals with SIN3-binding domain mutations [2]. We have shown that epigenetic dysregulation as a consequence of diminished KLF11 expression can be pharmacologically overcome with targeted use of epigenetic inhibitors and reverse disease progression in the endometriosis disease model; we are investigating pharmacological modulation of inflammation therein [48]. Uterine gene therapy is technically, but not medically or ethically, feasible [49–52]. Epigenetic inhibitors bypass this limitation to offer novel, individualized therapeutic options that effectively target underlying molecular dysregulation while obviating the need for gene therapy.
Our study is the first to show that KLF10, like its paralog KLF11, has a role in human endometriosis, a highly prevalent heterogeneous disease. Most significantly, despite high homology and concordant expression, KLF10 and KLF11 regulate distinct gene repertoires. Both are associated with diverse human diseases; their dysregulation therefore affects fundamental mechanisms of disease pathogenesis such as inflammation and fibrosis. The endometriosis model is significant given the clinical relevance of the disease and its association with other chronic systemic diseases [53, 54]. In addition, it offers a cogent scientific tool for mechanistic research on the biological relationship of inflammation and fibrosis with translational capability.
Footnotes
Funded by the Center of Biomedical Discovery Award (G.S.D.), Mayo Clinic Institutional Funds and Fraternal Order of Eagles Pilot Award (G.S.D.), and NIHR01 DE14036 (J.R.H., M.S.).
REFERENCES
- Giudice LC, Kao LC. Endometriosis. Lancet. 2004;364:1789–1799. doi: 10.1016/S0140-6736(04)17403-5. [DOI] [PubMed] [Google Scholar]
- Bonnefond A, Lomberk G, Buttar N, Busiah K, Vaillant E, Lobbens S, Yengo L, Dechaume A, Mignot B, Simon A, Scharfmann R, Neve B, et al. Disruption of a novel Kruppel-like transcription factor p300-regulated pathway for insulin biosynthesis revealed by studies of the c.-331 INS mutation found in neonatal diabetes mellitus. J Biol Chem. 2011;286:28414–28424. doi: 10.1074/jbc.M110.215822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daftary GS, Zheng Y, Tabbaa ZM, Schoolmeester JK, Gada RP, Grzenda AL, Mathison AJ, Keeney GL, Lomberk GA, Urrutia R. A novel role of the Sp/KLF transcription factor KLF11 in arresting progression of endometriosis. Plos One. 2013;8:e60165. doi: 10.1371/journal.pone.0060165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathison A, Grzenda A, Lomberk G, Velez G, Buttar N, Tietz P, Hendrickson H, Liebl A, Xiong YY, Gores G, Fernandez-Zapico M, Larusso NF, et al. Role for Krüppel-like transcription factor 11 in mesenchymal cell function and fibrosis. PLoS One. 2013;8:e75311. doi: 10.1371/journal.pone.0075311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Lin Z, Reierstad S, Wu J, Ishikawa H, Marsh EE, Innes J, Cheng Y, Pearson K, Coon JS V, Kim JJ, Chakravarti D, Bulun SE. Transcription factor KLF11 integrates progesterone receptor signaling and proliferation in uterine leiomyoma cells. Cancer Res. 2010;70:1722–1730. doi: 10.1158/0008-5472.CAN-09-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song KD, Kim DJ, Lee JE, Yun CH, Lee WK. KLF10, transforming growth factor-β-inducible early gene 1, acts as a tumor suppressor. Biochem Biophys Res Commun. 2012;419:388–394. doi: 10.1016/j.bbrc.2012.02.032. [DOI] [PubMed] [Google Scholar]
- Subramaniam M, Hawse JR, Rajamannan NM, Ingle JN, Spelsberg TC. Functional role of KLF10 in multiple disease processes. Biofactors. 2010;36:8–18. doi: 10.1002/biof.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinholz MM, An MW, Johnsen SA, Subramaniam M, Suman VJ, Ingle JN, Roche PC, Spelsberg TC. Differential gene expression of TGF beta inducible early gene (TIEG), Smad7, Smad2 and Bard1 in normal and malignant breast tissue. Breast Cancer Res Treat. 2004;86:75–88. doi: 10.1023/B:BREA.0000032926.74216.7d. [DOI] [PubMed] [Google Scholar]
- Subramaniam M, Hefferan TE, Tau K, Peus D, Pittelkow M, Jalal S, Riggs BL, Roche P, Spelsberg TC. Tissue, cell type, and breast cancer stage-specific expression of a TGF-beta inducible early transcription factor gene. J Cell Biochem. 1998;68:226–236. [PubMed] [Google Scholar]
- Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- Klevit RE. Recognition of DNA by Cys2,His2 zinc fingers. Science. 1991;253:1367. doi: 10.1126/science.1896847. [DOI] [PubMed] [Google Scholar]
- Cook T, Gebelein B, Urrutia R. Sp1 and its likes: biochemical and functional predictions for a growing family of zinc finger transcription factors. Ann N Y Acad Sci. 1999;880:94–102. doi: 10.1111/j.1749-6632.1999.tb09513.x. [DOI] [PubMed] [Google Scholar]
- Cook T, Gebelein B, Belal M, Mesa K, Urrutia R. Three conserved transcriptional repressor domains are a defining feature of the TIEG subfamily of Sp1-like zinc finger proteins. J Biol Chem. 1999;274:29500–29504. doi: 10.1074/jbc.274.41.29500. [DOI] [PubMed] [Google Scholar]
- Cook T, Gebelein B, Mesa K, Mladek A, Urrutia R. Molecular cloning and characterization of TIEG2 reveals a new subfamily of transforming growth factor-beta-inducible Sp1-like zinc finger-encoding genes involved in the regulation of cell growth. J Biol Chem. 1998;273:25929–25936. doi: 10.1074/jbc.273.40.25929. [DOI] [PubMed] [Google Scholar]
- Cao Z, Wara AK, Icli B, Sun X, Packard RR, Esen F, Stapleton CJ, Subramaniam M, Kretschmer K, Apostolou I, von Boehmer H, Hansson GK, et al. Kruppel-like factor KLF10 targets transforming growth factor-beta1 to regulate CD4(+)CD25(-) T cells and T regulatory cells. J Biol Chem. 2009;284:24914–24924. doi: 10.1074/jbc.M109.000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spittau B, Krieglstein K. Klf10 and Klf11 as mediators of TGF-beta superfamily signaling. Cell Tissue Res. 2012;347:65–72. doi: 10.1007/s00441-011-1186-6. [DOI] [PubMed] [Google Scholar]
- Subramaniam M, Gorny G, Johnsen SA, Monroe DG, Evans GL, Fraser DG, Rickard DJ, Rasmussen K, van Deursen JM, Turner RT, Oursler MJ, Spelsberg TC. TIEG1 null mouse-derived osteoblasts are defective in mineralization and in support of osteoclast differentiation in vitro. Mol Cell Biol. 2005;25:1191–1199. doi: 10.1128/MCB.25.3.1191-1199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook T, Urrutia R. TIEG proteins join the Smads as TGF-beta-regulated transcription factors that control pancreatic cell growth. Am J Physiol Gastrointest Liver Physiol. 2000;278:G513–G521. doi: 10.1152/ajpgi.2000.278.4.G513. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Khan Z, Zanfagnin V, Correa LF, Delaney AA, Daftary GS. Epigenetic modulation of collagen 1A1: therapeutic implications in fibrosis and endometriosis. Biol Reprod. 2016;94:87. doi: 10.1095/biolreprod.115.138115. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Khanna S, Grzenda AL, Sarmento OF, Svingen PA, Lomberk GA, Urrutia RA, Faubion WA. Polycomb antagonizes p300/CREB-binding protein-associated factor to silence FOXP3 in a Kruppel-like factor-dependent manner. J Biol Chem. 2012;287:34372–34385. doi: 10.1074/jbc.M111.325332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadakis KA, Krempski J, Svingen P, Xiong Y, Sarmento OF, Lomberk GA, Urrutia RA, Faubion WA. Krüppel-like factor KLF10 deficiency predisposes to colitis through colonic macrophage dysregulation. Am J Physiol Gastrointest Liver Physiol. 2015;309:G900–G909. doi: 10.1152/ajpgi.00309.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers PA, D'Hooghe TM, Fazleabas A, Giudice LC, Montgomery GW, Petraglia F, Taylor RN. Defining future directions for endometriosis research: workshop report from the 2011 World Congress of Endometriosis in Montpellier, France. Reprod Sci. 2013;20:483–499. doi: 10.1177/1933719113477495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang PC, Wu MH, Shoji Y, Tsai SJ. Downregulation of CD36 results in reduced phagocytic ability of peritoneal macrophages of women with endometriosis. J Pathol. 2009;219:232–241. doi: 10.1002/path.2588. [DOI] [PubMed] [Google Scholar]
- Schulke L, Berbic M, Manconi F, Tokushige N, Markham R, Fraser IS. Dendritic cell populations in the eutopic and ectopic endometrium of women with endometriosis. Hum Reprod. 2009;24:1695–1703. doi: 10.1093/humrep/dep071. [DOI] [PubMed] [Google Scholar]
- Rochman Y, Leonard WJ. The role of thymic stromal lymphopoietin in CD8+ T cell homeostasis. J Immunol. 2008;181:7699–7705. doi: 10.4049/jimmunol.181.11.7699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Myers AC, Chen L, Pardoll DM, Truong-Tran QA, Lane AP, McDyer JF, Fortuno L, Schleimer RP. Constitutive and inducible expression of b7 family of ligands by human airway epithelial cells. Am J Respir Cell Mol Biol. 2005;33:280–289. doi: 10.1165/rcmb.2004-0129OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salik E, Tyorkin M, Mohan S, George I, Becker K, Oei E, Kalb T, Sperber K. Antigen trafficking and accessory cell function in respiratory epithelial cells. Am J Respir Cell Mol Biol. 1999;21:365–379. doi: 10.1165/ajrcmb.21.3.3529. [DOI] [PubMed] [Google Scholar]
- Krikun G, Mor G, Alvero A, Guller S, Schatz F, Sapi E, Rahman M, Caze R, Qumsiyeh M, Lockwood CJ. A novel immortalized human endometrial stromal cell line with normal progestational response. Endocrinology. 2004;145:2291–2296. doi: 10.1210/en.2003-1606. [DOI] [PubMed] [Google Scholar]
- Zeitvogel A, Baumann R, Starzinski-Powitz A. Identification of an invasive, N-cadherin-expressing epithelial cell type in endometriosis using a new cell culture model. Am J Pathol. 2001;159:1839–1852. doi: 10.1016/S0002-9440(10)63030-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daftary GS, Lomberk GA, Buttar NS, Allen TW, Grzenda A, Zhang J, Zheng Y, Mathison AJ, Gada RP, Calvo E, Iovanna JL, Billadeau DD, et al. Detailed structural-functional analysis of the Kruppel-like factor 16 (KLF16) transcription factor reveals novel mechanisms for silencing Sp/KLF sites involved in metabolism and endocrinology. J Biol Chem. 2012;287:7010–7025. doi: 10.1074/jbc.M111.266007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JS, Moncrieffe MC, Kaczynski J, Ellenrieder V, Prendergast FG, Urrutia R. A conserved alpha-helical motif mediates the interaction of Sp1-like transcriptional repressors with the corepressor mSin3A. Mol Cell Biol. 2001;21:5041–5049. doi: 10.1128/MCB.21.15.5041-5049.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjabi S, Zenewicz LA, Kamanaka M, Flavell RA. Anti-inflammatory and pro-inflammatory roles of TGF-beta, IL-10, and IL-22 in immunity and autoimmunity. Curr Opin Pharmacol. 2009;9:447–453. doi: 10.1016/j.coph.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugger K, Lowder TW, Tucker TA, Schwiebert LM. Epithelial cells as immune effector cells: the role of CD40. Semin Immunol. 2009;21:289–292. doi: 10.1016/j.smim.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noelle RJ, Roy M, Shepherd DM, Stamenkovic I, Ledbetter JA, Aruffo AA. 39-kDa protein on activated helper T cells binds CD40 and transduces the signal for cognate activation of B cells. Proc Natl Acad Sci U S A. 1992;89:6550–6554. doi: 10.1073/pnas.89.14.6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönbeck U, Libby P. The CD40/CD154 receptor/ligand dyad. Cell Mol Life Sci. 2001;58:4–43. doi: 10.1007/PL00000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kooten C, Banchereau J. CD40-CD40 ligand. J Leukoc Biol. 2000;67:2–17. doi: 10.1002/jlb.67.1.2. [DOI] [PubMed] [Google Scholar]
- van Kooten C. Immune regulation by CD40-CD40-l interactions - 2; Y2K update. Front Biosci. 2000;5:D880–693. doi: 10.2741/kooten. [DOI] [PubMed] [Google Scholar]
- Grewal IS, Flavell RA. CD40 and CD154 in cell-mediated immunity. Annu Rev Immunol. 1998;16:111–135. doi: 10.1146/annurev.immunol.16.1.111. [DOI] [PubMed] [Google Scholar]
- Schönbeck U, Varo N, Libby P, Buring J, Ridker PM. Soluble CD40L and cardiovascular risk in women. Circulation. 2001;104:2266–2268. doi: 10.1161/hc4401.099447. [DOI] [PubMed] [Google Scholar]
- Danese S, Scaldaferri F, Papa A, Pola R, Gasbarrini A, Sgambato A, Cittadini A. CD40L-positive platelets induce CD40L expression de novo in endothelial cells: adding a loop to microvascular inflammation. Arterioscler Thromb Vasc Biol. 2004;24:e162. doi: 10.1161/01.ATV.0000138073.91195.70. [DOI] [PubMed] [Google Scholar]
- Danese S, Sans M, Fiocchi C. The CD40/CD40L costimulatory pathway in inflammatory bowel disease. Gut. 2004;53:1035–1043. doi: 10.1136/gut.2003.026278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaumond F, Gréchez-Cassiau A, Subramaniam M, Brangolo S, Peteri-Brünback B, Staels B, Fiévet C, Spelsberg TC, Delaunay F, Teboul M. Kruppel-like factor KLF10 is a link between the circadian clock and metabolism in liver. Mol Cell Biol. 2010;30:3059–3070. doi: 10.1128/MCB.01141-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Svingen PA, Sarmento OO, Smyrk TC, Dave M, Khanna S, Lomberk GA, Urrutia RA, Faubion WA. Differential coupling of KLF10 to Sin3-HDAC and PCAF regulates the inducibility of the FOXP3 gene. Am J Physiol Regul Integr Comp Physiol. 2014;307:R608–R620. doi: 10.1152/ajpregu.00085.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Shin S, Subramaniam M, Bruinsma E, Kim TD, Hawse JR, Spelsberg TC, Janknecht R. Histone demethylase JARID1B/KDM5B is a corepressor of TIEG1/KLF10. Biochem Biophys Res Commun. 2010;401:412–416. doi: 10.1016/j.bbrc.2010.09.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttar NS, DeMars CJ, Lomberk G, Rizvi S, Bonilla-Velez J, Achra S, Rashtak S, Wang KK, Fernandez-Zapico ME, Urrutia R. Distinct role of Kruppel-like factor 11 in the regulation of prostaglandin E2 biosynthesis. J Biol Chem. 2010;285:11433–11444. doi: 10.1074/jbc.M109.077065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabbaa ZM, Zheng Y, Daftary GS. KLF11 epigenetically regulates glycodelin-A, a marker of endometrial biology via histone-modifying chromatin mechanisms. Reprod Sci. 2014;21:319–328. doi: 10.1177/1933719113503407. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Tabbaa ZM, Khan Z, Schoolmeester JK, El-Nashar S, Famuyide A, Keeney GL, Daftary GS. Epigenetic regulation of uterine biology by transcription factor KLF11 via posttranslational histone deacetylation of cytochrome p450 metabolic enzymes. Endocrinology. 2014;155:4507–4520. doi: 10.1210/en.2014-1139. [DOI] [PubMed] [Google Scholar]
- Daftary GS, Taylor HS. Efficient liposome-mediated gene transfection and expression in the intact human uterus. Human Gene Therapy. 2001;12:2121–2127. doi: 10.1089/10430340152677458. [DOI] [PubMed] [Google Scholar]
- Daftary GS, Taylor HS. Reproductive tract gene transfer. Fertil Steril. 2003;80:475–484. doi: 10.1016/s0015-0282(03)00970-1. [DOI] [PubMed] [Google Scholar]
- Lehrman S. Virus treatment questioned after gene therapy death. Nature. 1999;401:517–518. doi: 10.1038/43977. [DOI] [PubMed] [Google Scholar]
- Marshall E. Gene therapy death prompts review of adenovirus vector. Science. 1999;286:2244–2245. doi: 10.1126/science.286.5448.2244. [DOI] [PubMed] [Google Scholar]
- Nunes FR, Ferreira JM, Bahamondes L. Prevalence of fibromyalgia and quality of life in women with and without endometriosis. Gynecol Endocrinol. 2014;30:307–310. doi: 10.3109/09513590.2013.876401. [DOI] [PubMed] [Google Scholar]
- Pasoto SG, Abrao MS, Viana VS, Bueno C, Leon EP, Bonfa E. Endometriosis and systemic lupus erythematosus: a comparative evaluation of clinical manifestations and serological autoimmune phenomena. Am J Reprod Immunol. 2005;53:85–93. doi: 10.1111/j.1600-0897.2005.00252.x. [DOI] [PubMed] [Google Scholar]