Abstract
Immaturity of the immune system of human fetuses and neonates is often invoked to explain their increased susceptibility to infection; however, the development of the fetal innate immune system in early life remains incompletely explored. We now show that the most mature NK cells found in adult (or postnatal) human circulation (CD94–CD16+) are absent during ontogeny. Human fetal NK cells were found to express the 2 signature T-box transcription factors essential for the development of all murine NK and NK-like cells, eomesodermin (Eomes) and T-bet. The single-cell pattern of Eomes and T-bet expression during ontogeny, however, revealed a stereotyped pattern of reciprocal dominance, with immature NK cells expressing higher amounts of Eomes and more mature NK cells marked by greater abundance of T-bet. We also observed a stereotyped pattern of tissue-specific NK cell maturation during human ontogeny, with fetal liver being more restrictive to NK cell maturity than fetal bone barrow, spleen, or lung. These results support the hypothesis that maturation of human NK cells has a discrete restriction until postnatal life, and provide a framework to better understand the increased susceptibility of fetuses and newborns to infection.
Human fetal natural killer cells are defined by reciprocal expression of T-box transcription factors with distinct developmental and tissue-specific distribution.
Introduction
Congenital and neonatal infections cause substantial morbidity and mortality worldwide. Why human fetal immune system development and function enhances susceptibility to infectious diseases is incompletely understood (1). Our understanding of the development of the human immune system in the fetus and newborn is limited by availability of specimens, and thus relies heavily on insights from appropriate animal models and more readily accessible peripheral blood samples. However, mouse models of immune development are limited by species-specific differences in the timing of key developmental events, such as the coincidence of the transition of hepatic to medullary hematopoiesis with parturition. Furthermore, recent studies (2, 3) have highlighted the diversity of the immune system profile in tissues versus the bloodstream, arguing that studying human immune development using only peripheral blood is necessarily incomplete.
NK cells, members of the innate lymphoid cell (ILC) family, play important roles in cytotoxicity and cytokine production (4, 5). Human fetal (6–8) and umbilical cord blood (UCB) (9–14) NK cells have been suggested to be phenotypically and functionally immature compared with adult NK cells. In the mouse, 2 major subsets of NK cells (or ILC1s) have been defined based on their requirement for transcription factors of the T-box family; helper (hILC1s) require T-bet, whereas cytotoxic (cILC1s) require Eomes (15, 16). Murine T-bet–dependent hILC1s are enriched in liver and other mucosal organs and are the predominant ILC1 population found in the neonate, whereas postnatal expression of Eomes directs differentiation of the cILC1 subset, which increases in frequency as neonatal pups reach weaning and young adulthood (15, 17). In contrast, recent studies have demonstrated that tissue-resident NK cells in the human are Eomes+, while the majority of circulating NK cells are T-bet+ (18, 19). Whether there is also differential distribution of Eomes+ and T-bet+ NK cells according to age in the human remains to be established.
In both mice and humans, the presence of NK cell precursors in multiple tissues suggests that NK cell development can occur both in the bone marrow and in extramedullary tissues (14, 20). Human NK cell development has been characterized according to surface expression of CD34, CD117, CD94, and CD16 on cells lacking T cell, B cell, DC, and myeloid markers (21): stage 1 are CD34+CD117–CD94–CD16–; stage 2 are CD34+CD117+CD94–CD16–; stage 3 are CD34–CD117+CD94–CD16–; stage 4 are CD34–CD117+/–CD94+CD16–; and stage 5 are CD34–CD117+/–CD94+CD16+. A proposed stage 6 refers to the most terminally mature NK cells that express CD57 and possess memory-like features (20). Based on ex vivo profiling and in vitro culture assays, it has been suggested that these NK cell populations represent sequential stages of NK cell development, with the exception of stage 6 memory NK cells, which may arise directly from either stage 4 or stage 5 cells. Loss of DC and T cell lineage potential occurs at stage 3 and restriction to the NK cell lineage was thought to occur at that point; however, it is now appreciated that stage 3 cells thus defined are a heterogeneous population that contains precursors capable of differentiating into other ILC lineages. Similarly, stage 4 cells are not NK lineage restricted, as recent evidence suggests that they can be further dissected based on expression of NKp80, with the NKp80– stage 4a cells exhibiting features consistent with ILC3 cells and the NKp80+ 4b cells appearing more consistent with classical NK cells based on expression of T-bet, Eomes, and IFN-γ (22). Continued differentiation after acquisition of CD16 (stage 5) is suggested by several lines of evidence demonstrating that NK cell functional maturation is characterized by downregulation of CD94/NKG2A and acquisition of killer immunoglobulin receptors (KIRs) (14, 23, 24). This basic framework of human NK cell development is largely accepted, but how anatomical location and appearance during ontogenesis shape the NK cell repertoire remains underexplored.
Results
To investigate human NK cell ontogeny, we established a collection of de-identified discarded fetal tissue from elective terminations of pregnancy at 14 to 22 weeks. We adopted the conventional scheme of adult human NK cell development (20) with the refinement to stage 5 suggested by Eissens et al. (14), and defined stages 3 to 5b as lineage-negative (Linneg: depleted of T cells, B cells, DCs, monocytes, granulocytes, erythroid cells, and CD34+ precursor cells) CD161-expressing cells that were additionally CD94–CD16– (stage 3); CD94+CD16– (stage 4); CD94+CD16+ (stage 5a); and CD94–CD16+ (stage 5b) (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.90063DS1). CD117 staining was inconsistent on our specimens, possibly as a result of the enzymatic digestion necessary to process whole organs, so we used CD161 expression to approximate the stages of NK cell development, as all CD117+ NK cells are also CD161+, and CD161 is expressed on all NK cell stages from stage 3 on (21). As with the CD34–CD117+CD94–CD16– population, defining stage 3 cells as LinnegCD161+CD94–CD16– includes other ILC precursors and is thus a heterogeneous population (see below and Supplemental Figure 2). CD56 expression is conventionally used to define human NK cells; we found that unlike adult NK cells that express differential levels of CD56 (i.e., bright vs. dim), fetal NK cells were uniformly CD56bright (Supplemental Figure 1C), and thus we did not routinely include it in the staining panel. We did not include quantification of memory-like stage 6 cells in our analyses because although the adult peripheral blood mononuclear cells (PBMCs) and spleen specimens were variably CD57+, all fetal specimens were consistently CD57 negative (data not shown).
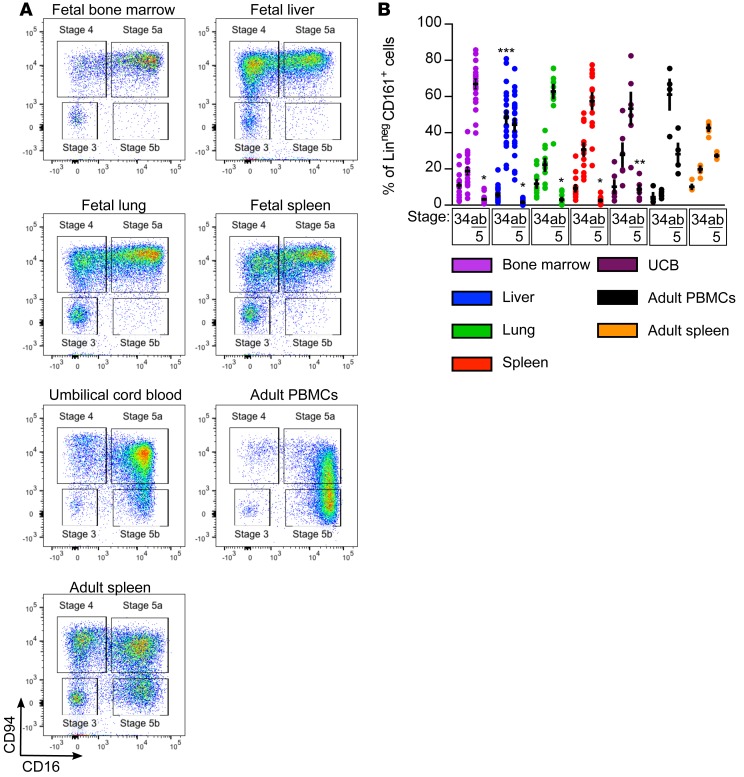
We first defined the range of NK cells present in fetal tissues (liver, bone marrow [BM], spleen, and lung), UCB, and adult samples. In adult spleen and blood, NK cells of all 4 developmental stages are evident, with the majority being the most mature stages 5a and 5b. In all fetal tissues examined, by contrast, stage 5b cells were generally absent (Figure 1, A and B). UCB from full-term pregnancies, however, appeared to contain a small but detectable subpopulation of stage 5b cells, suggesting that CD94–CD16+ NK cells arise either during the last trimester of gestation or at the time of parturition. The low frequency of stage 5b cells at the 2 time points for which a direct comparison can be made (UCB compared with adult blood and fetal spleen compared with adult spleen) indicates that a substantial portion of human NK cell maturation occurs postnatally. In fetal BM, lung, and spleen, stage 5a cells are the predominant subset; however, in fetal liver there is an enrichment of the more immature stage 4 cells. These inter-organ differences in the fetus reveal dynamic tissue-specific regulation of NK cell development and suggest a role for the microenvironment in shaping the local NK cell repertoire.
Figure 1. Fetal tissues contain fewer mature NK cells than umbilical cord blood and adult samples.
(A) Representative flow cytometry plots of CD94 and CD16 expression on lineage-negative (Linneg: depleted of T cells, B cells, DCs, monocytes, granulocytes, erythroid cells, and CD34+ precursor cells) CD161+ lymphocytes from indicated tissues. Fetal samples are from an 18-week gestational age specimen. (B) Mean frequencies ± SEM of LinnegCD161+ cells of indicated stages from indicated tissues. Fetal bone marrow, n = 20; fetal liver, n = 23; fetal lung, n = 11; fetal spleen, n = 15; umbilical cord blood (UCB), n = 6; adult peripheral blood mononuclear cells (PBMCs), n = 4; adult spleen, 6 independent measurements made on n = 2 specimens. Significance determined using 2-way ANOVA with Bonferroni post-hoc tests. *P < 0.001, **P < 0.05 compared with adult PBMCs; ***P < 0.001 compared with all other tissues.
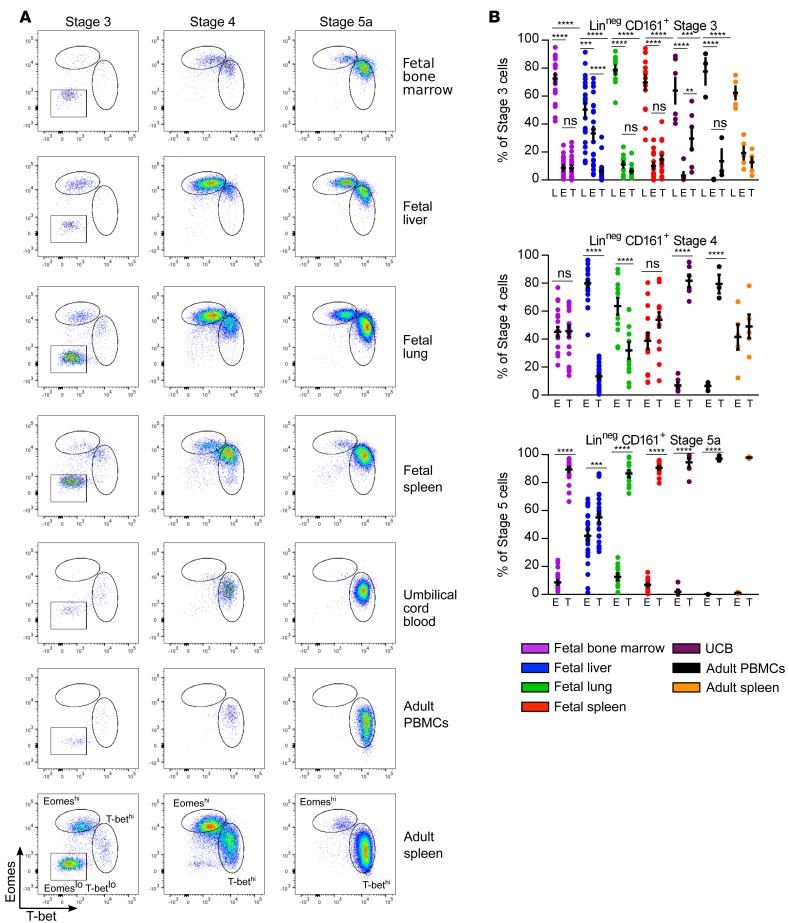
To examine the transcriptional regulation of human NK cell development, we analyzed T-bet and Eomes expression in all developmental stages. In contrast to the mouse, both T-bet and Eomes expression is evident in human fetal NK cells (Figure 2). Stage 3 cells in all tissues are mainly EomesloT-betlo, consistent with stage 3 representing a heterogeneous population of both NK and other ILC precursors (20), and confirmed by NKp80 and CD127 staining (Supplemental Figure 2A). Stage 4 contains 2 populations with reciprocal levels of Eomes and T-bet expression. All fetal tissue and adult spleen stage 4 cells exhibit either an EomeshiT-betint (referred to as Eomeshi) or an EomesintT-bethi (referred to as T-bethi) profile. Fetal liver stage 4 cells are almost exclusively Eomeshi, fetal lung stage 4 contain more Eomeshi than T-bethi cells, and fetal BM, fetal spleen, and adult spleen stage 4 have equal proportions of Eomeshi and T-bethi cells. In the periphery (UCB and adult peripheral blood), stage 4 cells are almost exclusively T-bethi. NKp80 staining, which has been shown to dissect stage 4 NK cells, was similar in Eomeshi and Tbethi populations, with both containing NKp80– and NKp80+ cells (Supplemental Figure 2B). As cells progress to stage 5a, in all tissues except fetal liver, T-bethi cells are the predominant population; in the fetal liver there are equal proportions of Eomeshi and T-bethi cells. Likewise, stage 5b cells in fetal liver are equally divided between Eomeshi and T-bethi populations (data not shown), whereas other fetal, UCB, and adult stage 5b cells are T-bethi. Prenatal immaturity of human NK cell development, therefore, does not appear to be restricted at the level of Eomes expression, which might have been predicted from the absence of Eomes-expressing NK cells in the murine fetus.
Figure 2. Both Eomes- and T-bet–expressing NK cells are found in fetal tissues and Eomeshi cells are enriched in stage 4 and fetal liver.
(A) Representative flow cytometry plots of intracellular Eomes and T-bet expression on lineage-negative (Linneg: depleted of T cells, B cells, DCs, monocytes, granulocytes, erythroid cells, and CD34+ precursor cells) CD161+ cells gated on indicated stages based on CD94 and CD16 expression from indicated tissues. (B) Mean frequencies ± SEM of stage 3 (top), stage 4 (middle), and stage 5a (bottom) EomesloT-betlo (L), Eomeshi (E) and T-bethi (T) NK cells in indicated samples. Fetal bone marrow, n = 20; fetal liver, n = 23; fetal lung, n = 11; fetal spleen, n = 15; umbilical cord blood (UCB), n = 6; adult peripheral blood mononuclear cells (PBMCs), n = 3; adult spleen, 5 independent measurements made on n = 2 specimens. Significance determined using 2-way ANOVA with Bonferroni post-hoc tests. **P < 0.01, ***P < 0.001, ****P < 0.0001. ns, not significant).
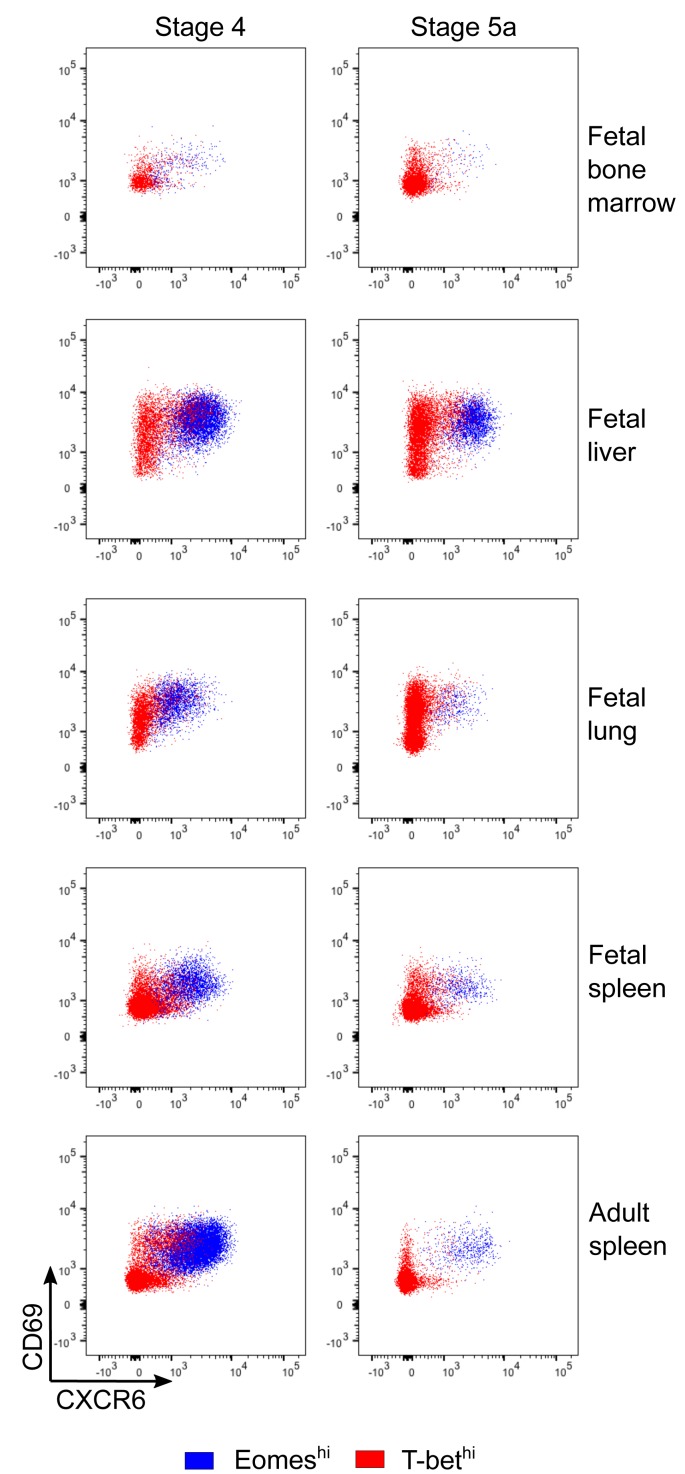
The pattern of Eomes and T-bet staining recapitulates what has been shown in adult human liver samples (18, 19), where a distinct population of EomeshiT-betlo cells has been identified. This Eomeshi population is described as coexpressing CXCR6 and CD69 as markers of tissue residency, with reports differing on whether the Eomeshi cells are more or less cytotoxic than peripheral blood T-bethi NK cells (18, 19, 25). A similar CXCR6+CD69+ population is also described in human lymphoid tissue (26), although the relative expression of Eomes and T-bet was not examined. We therefore examined CXCR6 and CD69 expression in our samples, and found that all Eomeshi NK cells, regardless of tissue or stage, coexpress CXCR6 and CD69 (Figure 3). T-bethi NK cells are uniformly CXCR6 negative, but have variable CD69 expression, with T-bethi cells in the fetal liver and fetal lung being evenly divided between CD69– and CD69+. The majority of T-bethi cells in fetal bone marrow, fetal spleen, and adult spleen are CD69–. These data demonstrate that this distinct subset of EomeshiCXCR6+ NK cells is established prenatally. As we were able to analyze organs in the fetus (bone marrow, lung, and spleen) that were not included in the adult analyses, our results further suggest that the EomeshiCXCR6+ NK cell population is not specific to any particular organ but rather represents tissue-resident NK cells as opposed to circulating NK cells.
Figure 3. Eomeshi cells are CXCR6+CD69+.
Representative flow cytometry plots of Eomeshi and T-bethi NK cells showing CXCR6 and CD69 staining. Experiment was performed 3 times; n = 4 for fetal specimens and n = 2 for adult spleen.
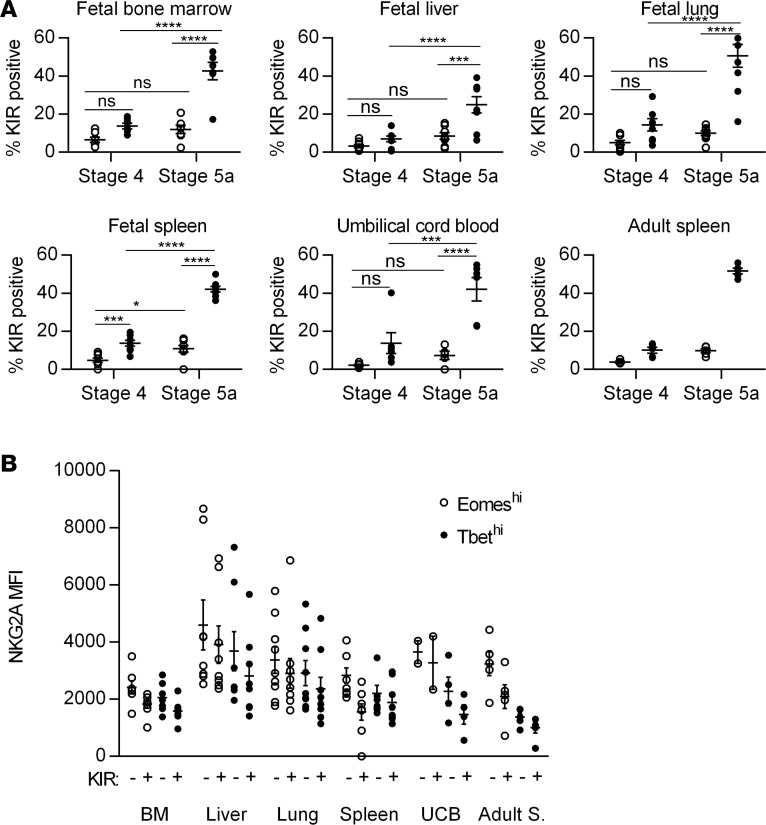
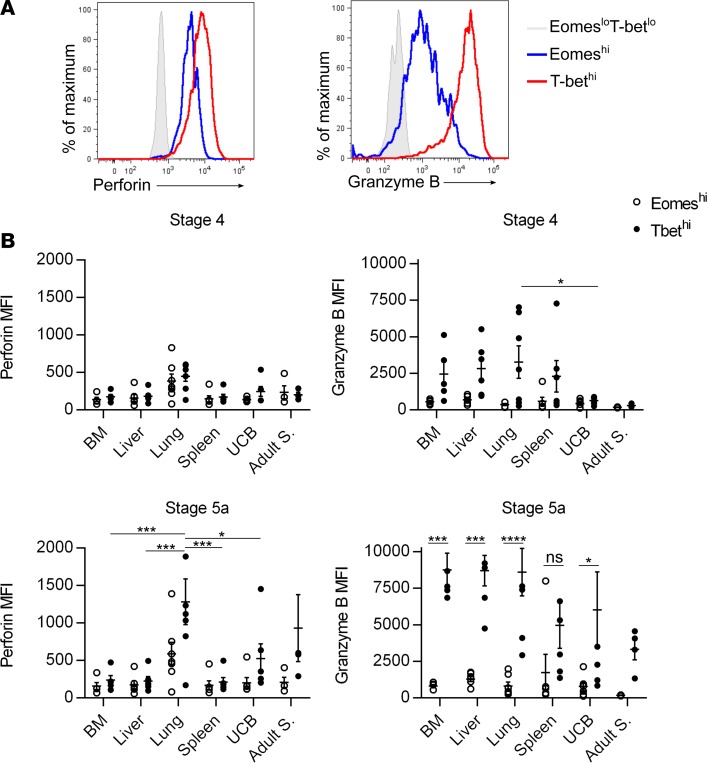
The enrichment of Eomeshi cells in fetal liver and stage 4 suggests that these cells represent a more immature stage than T-bethi cells. We therefore examined developmentally regulated surface and intracellular proteins amongst Eomeshi and T-bethi cells. KIRs are the major activating/inhibitory receptors expressed by NK cells and their expression is developmentally acquired (24). There are higher percentages of KIR-expressing stage 5a cells than stage 4 cells in all tissues (Figure 4A). Within each stage, more T-bethi cells express KIRs than do Eomeshi cells. The proportion of T-bethi cells that express KIRs is similar in all tissues, pre- and postnatal, with the exception of fetal liver, which contains significantly fewer T-bethi cells that express KIRs (Figure 4A). NKG2A is a major inhibitory receptor on NK cells and its downregulation has been shown to correlate with terminal NK cell maturation (24). NKG2A median fluorescence intensity (MFI) decreases with acquisition of KIRs, and Eomeshi cells have higher NKG2A MFI than T-bethi cells (Figure 4B). Granzyme B and perforin are intracellular proteins required for NK cell cytotoxicity (27). Granzyme B and perforin expression is generally limited to stage 5 cells (Figure 5). Granzyme B is significantly higher in T-bethi cells vs. Eomeshi cells in most tissues. The only fetal tissue with perforin expression comparable to adult NK cell levels is fetal lung, and herein T-bethi cells express greater levels of perforin than Eomeshi. These data suggest that T-bethi cells may be more mature than Eomeshi cells, but that there is dynamic tissue-specific regulation within fetal life.
Figure 4. Eomeshi cells are less mature than T-bethi cells based on killer immunoglobulin receptor (KIR) and NKG2A expression.
(A) Mean frequencies ± SEM of KIR expression (based on pan-KIR staining for KIR2DL1, KIR2DS1, KIR2DL2, KIR2DS2, KIR2DL3, KIR2DS3, KIR2DS4, and KIR2DS5) on lineage-negative (Linneg: depleted of T cells, B cells, DCs, monocytes, granulocytes, erythroid cells, and CD34+ precursor cells) CD161+ stage 4 and 5 Eomeshi (open circles) and T-bethi (closed circles) cells from indicated tissues. Fetal bone marrow (BM), n = 7; fetal liver, n = 8; fetal lung, n = 9; fetal spleen, n = 8; umbilical cord blood (UCB), n = 6; adult spleen, 5 independent measurements made on n = 2 specimens. (B) Median fluorescence intensity (MFI) ± SEM of NKG2A on KIR-negative and KIR-positive Eomeshi (open circles) and T-bethi (closed circles) stage 5a (CD94+CD16+) cells from indicated tissues. Fetal BM, n = 7; fetal liver, n = 8; fetal lung, n = 9; fetal spleen, n = 8; UCB, n = 2; adult spleen (Adult S.), 5 independent measurements made on n = 2 specimens. Significance determined using 2-way ANOVA with Bonferroni post-hoc tests. *P < 0.05, ***P < 0.001, ****P < 0.0001. ns, not significant.
Figure 5. Eomeshi cells are less mature than T-bethi cells based on granzyme B and perforin expression.
(A) Representative flow cytometry histogram showing median fluorescence intensity (MFI) of granzyme B and perforin expression on 15-week gestational age lung specimen stage 5a cells gated on Eomeshi (blue line) and T-bethi (red line) cells compared to Eomeslo T-betlo (L) stage 3 cells (shaded gray histogram). (B) Mean frequencies ± SEM of granzyme B or perforin MFI on Eomeshi (open circles) and T-bethi (closed circles) cells from indicated tissues and stages. Fetal bone marrow (BM), n = 5; fetal liver, n = 6; fetal lung, n = 7; fetal spleen, n = 6; umbilical cord blood (UCB), n = 6; adult spleen (Adult S.), 4 independent measurements made on n = 2 specimens. Significance determined using 2-way ANOVA with Bonferroni post-hoc tests. *P < 0.05, ***P < 0.001, ****P < 0.0001. ns, not significant.
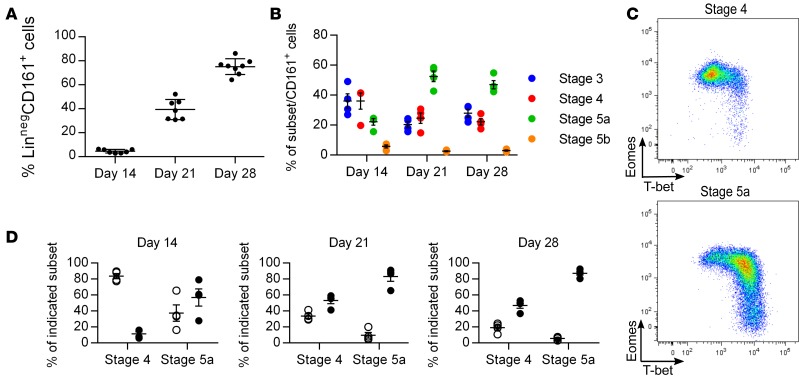
To further test the hypothesis that Eomeshi cells are less mature than T-bethi cells, we studied their developmental kinetics using an established in vitro culture system (28, 29). Starting from hematopoietic stem cells, LinnegCD161+ cells arise at approximately 14 days of culture and continue to increase in frequency over time (Figure 6A). The more immature stages 3 and 4 are evident at early time points, while the more mature stage 5a cells arise later (Figure 6B). There is no significant appearance of the most mature stage 5b cells in this culture system. At the earliest time points when LinnegCD161+ cells are evident (day 14), stage 4 cells are almost exclusively Eomeshi, whereas the few stage 5a cells present contain equal percentages of Eomeshi and T-bethi cells (Figure 6, C and D). By day 21, stage 4 cells contain an equal proportion of Eomeshi and T-bethi cells, while stage 5a cells are almost exclusively T-bethi. By day 28, both stages contain significantly more T-bethi cells than Eomeshi cells. At all time points examined, stage 3 cells are almost exclusively EomesloT-betlo (data not shown). These data are consistent with a model in which Eomeshi cells are more immature than T-bethi cells, although direct precursor-product assessment could not be addressed. It is possible that the depletion of Eomeshi NK cells at the later time points in this culture system reflects decreased survival, increased apoptosis, or the absence of specific microenvironmental signals rather than continued differentiation to the T-bethi stage.
Figure 6. Eomeshi cells appear before T-bethi cells in an in vitro NK cell development culture system.
(A) Mean frequencies ± SEM of lineage-negative (Linneg: depleted of T cells, B cells, DCs, monocytes, granulocytes, erythroid cells, and CD34+ precursor cells) CD161+ cells on days 14, 21, and 28 of culture on EL08.ID2 cells. Each data point represents cells derived from a unique hematopoietic stem cell (HSC) source performed in 2 independent experiments. (B) Mean frequencies ± SEM of indicated stages of NK cell development on days 14, 21, and 28 of culture on EL08.ID2 cells; n = 4, representative of 2 independent experiments each performed with unique HSC sources. (C) Representative flow cytometry plot of intracellular Eomes and T-bet staining on indicated stages from day 14 of culture on EL08.ID2 cells. (D) Mean frequencies ± SEM of Eomeshi (open circles) and T-bethi (closed circles) cells from indicated stages on days 14, 21, and 28 of culture on EL08.ID2 cells; n = 4, representative of 2 independent experiments each performed with unique HSC sources.
Discussion
We demonstrate that a major difference between early-life and adult NK cells lies in the distribution of stages 3–5b, with significant restriction of the development of stage 5b cells until after birth. One could have hypothesized that early-life NK cells contribute to the relative immune deficiency of fetuses and neonates by being less functional. Our data instead support a model where, on a per-population basis, the NK cells present contain the same functional machinery as their adult counterparts, but the full complement of all NK cell subsets has yet to develop. Further studies aimed at understanding how the microenvironment of the fetus/neonate restricts the final maturation steps of NK-poiesis will be crucial to helping us understand the limitations of fetal/neonatal immune responses.
Our finding that human NK cells appear to utilize T-box family transcription factors in a reciprocal manner to what has been described in the mouse underscores the importance of performing studies in human samples. We confirm the presence of an EomeshiCXCR6+ NK cell population in the human liver, extend the description of this population to other organs not examined in adult specimens, and establish the development of this population prenatally. We show that the Eomeshi NK cells express fewer KIRs, perforin, and granzyme B, and more of the inhibitory receptor NKG2A than the T-bethi NK cells. Using an in vitro culture assay, we further show that the Eomeshi NK cells arise earlier than the T-bethi NK cells. Together, these data support the notion that the Eomeshi NK cells represent a more immature NK cell population than the T-bethi NK cells. This is consistent with the finding that graded levels of T-bet and Eomes correlate with advancing stages of human NK cell maturation, with Eomes expression decreasing and T-bet expression increasing as NK cells acquire functional maturity (30). However, these data do not exclude an alternative model in which Eomeshi NK cells represent an alternative lineage, whose development may occur preferentially within tissues, and whose function may be uniquely suited to the microenvironment of those tissues. Further experiments will be necessary to distinguish between these models.
Our data also demonstrate significant tissue-specific variation within the fetal NK cell compartment, with regard to distribution of NK cell stages, distribution of Eomeshi and T-bethi NK cells, and expression of proteins involved in NK cell cytotoxicity. The fetal liver is enriched in immature NK cells compared with other fetal organs; similarly the adult liver contains fewer CD16+ NK cells and the CD16+ NK cells present have reduced cytotoxicity compared with their peripheral blood counterparts (31). In the adult liver, that immune responses are more likely to result in tolerance is hypothesized to be a result of the immunosuppressive environment that results from constant low-level LPS exposure and food antigens from the gut (32). Whether the mechanisms that lead to an immunosuppressive environment and those that appear to restrict development are similar is an unanswered question, but our data suggest that this unique hepatic microenvironment may be established in utero, prior to any significant exposure to food antigens from the gut and presumably to intestinal bacterial colonization. In contrast, fetal lung NK cells express higher levels of perforin than in any other fetal tissues, at a level comparable to adult NK cells. It is possible that this tissue-specific regulation reflects exposure of fetal lung to amniotic fluid, but further work remains to be done. Continued investigation into the mechanisms that regulate human NK cell development in situ will be critical to understanding and ultimately treating the increased susceptibility to infection that fetuses and newborns exhibit.
Methods
Acquisition of tissue.
Fetal tissues (liver, femurs, spleen, lung) were obtained from de-identified discarded elective terminations of pregnancy up to 24 weeks of gestational age. Products of conception with known karyotype or structural abnormalities, known maternal or fetal infection, or the result of miscarriage were not included in the study. Umbilical cord and adult peripheral blood were obtained from the New York Blood Bank. Adult spleen was obtained as discarded surgical samples.
Lymphocyte isolation from tissues.
Tissue samples were maintained in cold sterile PBS and brought to the laboratory within 4 hours of procurement. Samples were processed as described (2). Briefly, fetal liver and lung were first minced and incubated at 37°C in enzymatic digestion media: complete RPMI 1640 (Thermo Fisher) containing 10% FBS (Serum Source International), penicillin/streptomycin (Thermo Fisher), L-glutamine (Thermo Fisher), nonessential amino acids (Thermo Fisher), and HEPES (Thermo Fisher) with collagenase D (1 mg/ml, Roche), DNase I (0.1 mg/ml, Roche), and trypsin inhibitor (1 mg/ml, Roche). Digested tissue was further mechanically dissociated by pushing through a 70-μm filter and pelleted by centrifugation. Fetal spleen was mechanically dissociated using the back of a syringe and pushing through a 70-μm filter. Femurs were washed extensively and flushed with complete RPMI as above. We did not detect CD9 staining in any fetal tissues (data not shown), confirming that our specimens were not contaminated with maternal uterine NK cells. Adult spleen was minced and incubated at 37°C in digestion media (RPMI with 2% FCS, collagenase D (0.1 mg/ml, Roche), and DNase I (0.1 mg/ml, Roche); digested tissue was pushed through a 70-μm strainer and washed with complete RPMI without enzymes. The cell pellets were washed 3 times with complete RPMI (without enzymes) and red blood cells were lysed with ACK lysis buffer (Lonza). Lymphocytes were isolated via centrifugation using lymphocyte separation media (LymphoPrep, Stemcell Technologies). Lymphocytes were isolated from umbilical cord and adult peripheral blood using ACK lysis buffer and centrifugation over LymphoPrep. Cells were counted and cryopreserved at 10 × 106/ml in 10% DMSO (Sigma-Aldrich)/90% FCS.
NK cell isolation and flow cytometric analysis.
Cryopreserved cells were thawed in warmed complete RPMI media and immediately washed. They were then incubated for 10 minutes in complete RPMI with DNase I (0.1 mg/ml, Roche). Cells were pelleted by centrifugation and counted with trypan blue exclusion dye (Sigma-Aldrich) to determine viability. Cell recovery ranged from 50% to 100% of initial number and viability was greater than 90%. Pilot experiments were performed on fresh unfrozen cells in comparison to thawed cryopreserved cells with no differences noted in cell surface markers examined, so cryopreserved cells were used in all experiments. As our interest was focused on the later stages of development when T-bet and Eomes are expressed, and to facilitate identification of rare NK cell populations from fetal tissues, we enriched for NK cells by negative selection using the Human NK Cell Isolation Kit (Miltenyi Biotec) before staining for flow cytometric analysis, thus depleting our samples of CD34+ precursors as well as T cells, B cells, DCs, monocytes, granulocytes, and erythroid cells (data not shown). The following fluorochrome-conjugated anti-human antibodies were used for surface staining: CD3 (eF450, clone UCHT1; PerCPeF710, clone OKT3), CD9 (PE, clone eBioSN4), CD14 (eF450 or PerCP-Cy5.5, clone 61D3), CD19 (eF450 or PerCP-Cy5.5, clone HIB19), CD38 (PE, clone HB7), CD127 (FITC, clone eBioRDR5), CD158a/h/g (FITC, clone HP-MA4), CD161 (eF450 or PerCPeF710, clone HP-3G10) (all eBioscience); CD16 (APC-Cy7, clone 3G8), CD34 (APC, clone 581), CD94 (PerCP-Cy5.5, clone HP-3D9) (all 3 from BD Biosciences); CD69 (FITC, clone FN50), CXCR6 (PE, clone K041E5) (both BioLegend); CD158 (FITC, clone 180704), NK2GA (PE, clone 131411) (both R&D Systems). Dead cells were excluded using Live/Dead (AmCyan, Molecular Probes). For intracellular/intranuclear staining, after surface staining cells were fixed and permeabilized according to the manufacturer’s instructions using an Intracellular Fixation and Permeabilization Kit (eBiosciences) and the following fluorochrome-conjugated anti-human intracellular antibodies were used: Eomes (PEeF610, clone WD1928), perforin (FITC, clone dG9), T-bet (eF660, clone 4B10) (all eBioscience); and granzyme B (PE, clone GB11, BD Biosciences). Stained cells were acquired on an LSRII or LSR Fortessa (BD Biosciences) and analyzed using FlowJo software (Treestar). For sorting, stained cells were acquired and sorted on an LSR Aria (BD Biosciences); a small sample of post-sort cells was always retained and acquired on an LSRII or LSR Fortessa to evaluate for post-sort purity.
HSC isolation and EL08.ID2 in vitro culture system.
Hematopoietic stem cells (HSCs) were isolated from cryopreserved fetal liver or bone marrow, and first purified via a single round of positive selection using a human CD34 Microbead Kit (Miltenyi Biotec) followed by surface staining and sorting on an LSR Aria (BD Biosciences). A small sample of cells were always retained and acquired on LSRII or Fortessa to evaluate for post-sort purity, which was routinely greater than 97%. HSCs were defined as Linneg (CD3–CD14–CD19–CD161–), CD34+, and CD38–. The murine embryonic liver cell line EL08.1D2 (a gift from Jeffrey Miller (University of Minnesota, Minneapolis, Minnesota, USA) and Elaine Dzierzak (University of Edinburgh, Edinburgh, United Kingdom) was cultured on gelatinized plates at 32°C in 41.5% α–minimum essential medium (α-MEM; Thermo Fisher), 50% Myelocult (M5300; Stemcell Technologies), 7.5% FBS, with 50 μM β-mercaptoethanol (Sigma-Aldrich), penicillin/streptomycin (Thermo Fisher), and hydrocortisone (1 μM, Sigma-Aldrich). Before coculture with progenitor cells, EL08.1D2 cells were irradiated (3,000 rads). Sorted HSCs (50 per well) were plated in a 96-well plate on an irradiated confluent monolayer of EL08.1D2 cells in Ham’s F12 (Thermo Fisher) plus DMEM (1:2 ratio, Thermo Fisher) with 10% human male AB serum (Sigma-Aldrich), 25 μM β-mercaptoethanol, ethanolamine (50 μM, Sigma-Aldrich), sodium selenite (50 ng/ml, Sigma-Aldrich), ascorbic acid (2 mg/ml, Sigma-Aldrich), and penicillin/streptomycin. At the start of cultures, IL-3 (5 ng/ml), IL-7 (10 ng/ml), IL-15 (10 ng/ml), stem cell factor (10 ng/ml), and Flt3-Ligand (10 ng/ml) were added (all human recombinant cytokines from Peprotech). Cultures were refed weekly by 50% volume change and supplemented with the previously mentioned cytokines except IL-3. Cells were harvested at indicated time points for analysis by flow cytometry.
Statistics.
Statistical significance was calculated using 2-way ANOVA and adjusted for multiple comparisons by Bonferroni post-hoc tests using Prism (GraphPad Software). A P value less than 0.05 was considered significant.
Study approval.
For fetal tissue acquisition, members of the Division of Family Planning at Columbia University Medical Center identified products of conception from pregnancies that were eligible for inclusion but had no role in study design or execution. The study does not qualify as human subjects research, as confirmed by the Columbia University IRB who reviewed and assigned an approval number to the protocol (AAAK3206), because tissue samples were obtained from nonviable, deceased fetuses and the study authors had no identifying information about the products of conception except gestational age. Adult spleen was obtained under IRB protocol AAAN7655.
Author contributions
AC codesigned the study, collected and processed fetal specimens, performed all experiments, analyzed the data, prepared the figures, and cowrote the manuscript. NR assisted with fetal specimen processing. KL contributed adult spleen tissue. SLR codesigned the study, analyzed the data, and cowrote the manuscript.
Supplementary Material
Acknowledgments
The authors would like to thank the Family Planning Division at Columbia University Medical Center for assistance with fetal specimen collection, and Jeffrey Miller (University of Minnesota) and Elaine Dzierzak (University of Edinburgh) for the gift of EL08.ID2 stromal cells. This work was supported by the US NIH grant R01 AI061699 to S.L.R.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:JCI Insight. 2017;2(5):e90063. https://doi.org/10.1172/jci.insight.90063.
Contributor Information
Amélie Collins, Email: ac3310@cumc.columbia.edu.
Nyanza Rothman, Email: nr2433@cumc.columbia.edu.
Kang Liu, Email: kl2529@columbia.edu.
References
- 1.PrabhuDas M, et al. Challenges in infant immunity: implications for responses to infection and vaccines. Nat Immunol. 2011;12(3):189–194. doi: 10.1038/ni0311-189. [DOI] [PubMed] [Google Scholar]
- 2.Thome JJ, et al. Early-life compartmentalization of human T cell differentiation and regulatory function in mucosal and lymphoid tissues. Nat Med. 2016;22(1):72–77. doi: 10.1038/nm.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thome JJ, Farber DL. Emerging concepts in tissue-resident T cells: lessons from humans. Trends Immunol. 2015;36(7):428–435. doi: 10.1016/j.it.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vivier E, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331(6013):44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9(5):503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 6.Uksila J, Lassila O, Hirvonen T, Toivanen P. Development of natural killer cell function in the human fetus. J Immunol. 1983;130(1):153–156. [PubMed] [Google Scholar]
- 7.Phillips JH, Hori T, Nagler A, Bhat N, Spits H, Lanier LL. Ontogeny of human natural killer (NK) cells: fetal NK cells mediate cytolytic function and express cytoplasmic CD3 epsilon,delta proteins. J Exp Med. 1992;175(4):1055–1066. doi: 10.1084/jem.175.4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivarsson MA, et al. Differentiation and functional regulation of human fetal NK cells. J Clin Invest. 2013;123(9):3889–3901. doi: 10.1172/JCI68989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luevano M, et al. The unique profile of cord blood natural killer cells balances incomplete maturation and effective killing function upon activation. Hum Immunol. 2012;73(3):248–257. doi: 10.1016/j.humimm.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 10.Dalle JH, et al. Characterization of cord blood natural killer cells: implications for transplantation and neonatal infections. Pediatr Res. 2005;57(5 Pt 1):649–655. doi: 10.1203/01.PDR.0000156501.55431.20. [DOI] [PubMed] [Google Scholar]
- 11.Sundström Y, Nilsson C, Lilja G, Kärre K, Troye-Blomberg M, Berg L. The expression of human natural killer cell receptors in early life. Scand J Immunol. 2007;66(2–3):335–344. doi: 10.1111/j.1365-3083.2007.01980.x. [DOI] [PubMed] [Google Scholar]
- 12.Le Garff-Tavernier M, et al. Human NK cells display major phenotypic and functional changes over the life span. Aging Cell. 2010;9(4):527–535. doi: 10.1111/j.1474-9726.2010.00584.x. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Xu H, Zheng X, Wei H, Sun R, Tian Z. High expression of NKG2A/CD94 and low expression of granzyme B are associated with reduced cord blood NK cell activity. Cell Mol Immunol. 2007;4(5):377–382. [PubMed] [Google Scholar]
- 14.Eissens DN, et al. Defining early human NK cell developmental stages in primary and secondary lymphoid tissues. PLoS ONE. 2012;7(2):e30930. doi: 10.1371/journal.pone.0030930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon SM, et al. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36(1):55–67. doi: 10.1016/j.immuni.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daussy C, et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J Exp Med. 2014;211(3):563–577. doi: 10.1084/jem.20131560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeda K, et al. TRAIL identifies immature natural killer cells in newborn mice and adult mouse liver. Blood. 2005;105(5):2082–2089. doi: 10.1182/blood-2004-08-3262. [DOI] [PubMed] [Google Scholar]
- 18.Stegmann KA, et al. CXCR6 marks a novel subset of T-bet(lo)Eomes(hi) natural killer cells residing in human liver. Sci Rep. 2016;6:26157. doi: 10.1038/srep26157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harmon C, et al. Tissue-resident Eomes(hi) T-bet(lo) CD56(bright) NK cells with reduced proinflammatory potential are enriched in the adult human liver. Eur J Immunol. 2016;46(9):2111–2120. doi: 10.1002/eji.201646559. [DOI] [PubMed] [Google Scholar]
- 20.Yu J, Freud AG, Caligiuri MA. Location and cellular stages of natural killer cell development. Trends Immunol. 2013;34(12):573–582. doi: 10.1016/j.it.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freud AG, et al. Evidence for discrete stages of human natural killer cell differentiation in vivo. JEM. 2006;203(4):1033–433. doi: 10.1084/jem.20052507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freud AG, et al. NKp80 defines a critical step during human natural killer cell development. Cell Rep. 2016;16(2):379–391. doi: 10.1016/j.celrep.2016.05.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu J, et al. CD94 surface density identifies a functional intermediary between the CD56bright and CD56dim human NK-cell subsets. Blood. 2010;115(2):274–281. doi: 10.1182/blood-2009-04-215491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Béziat V, Descours B, Parizot C, Debré P, Vieillard V. NK cell terminal differentiation: correlated stepwise decrease of NKG2A and acquisition of KIRs. PLoS One. 2010;5(8):e11966. doi: 10.1371/journal.pone.0011966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hudspeth K, et al. Human liver-resident CD56(bright)/CD16(neg) NK cells are retained within hepatic sinusoids via the engagement of CCR5 and CXCR6 pathways. J Autoimmun. 2016;66:40–50. doi: 10.1016/j.jaut.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lugthart G, et al. Human lymphoid tissues harbor a distinct CD69+CXCR6+ NK cell population. J Immunol. 2016;197(1):78–84. doi: 10.4049/jimmunol.1502603. [DOI] [PubMed] [Google Scholar]
- 27.Lieberman J. The ABCs of granule-mediated cytotoxicity: new weapons in the arsenal. Nat Rev Immunol. 2003;3(5):361–370. doi: 10.1038/nri1083. [DOI] [PubMed] [Google Scholar]
- 28.Grzywacz B, et al. Coordinated acquisition of inhibitory and activating receptors and functional properties by developing human natural killer cells. Blood. 2006;108(12):3824–3833. doi: 10.1182/blood-2006-04-020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cichocki F, Miller JS. In vitro development of human killer-immunoglobulin receptor-positive NK cells. Methods Mol Biol. 2010;612:15–26. doi: 10.1007/978-1-60761-362-6_2. [DOI] [PubMed] [Google Scholar]
- 30.Luetke-Eversloh M, et al. NK cells gain higher IFN-γ competence during terminal differentiation. Eur J Immunol. 2014;44(7):2074–2084. doi: 10.1002/eji.201344072. [DOI] [PubMed] [Google Scholar]
- 31.Burt BM, et al. The lytic potential of human liver NK cells is restricted by their limited expression of inhibitory killer Ig-like receptors. J Immunol. 2009;183(3):1789–1796. doi: 10.4049/jimmunol.0900541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crispe IN. The liver as a lymphoid organ. Annu Rev Immunol. 2009;27:147–163. doi: 10.1146/annurev.immunol.021908.132629. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.