Abstract
Cytochrome P450 enzyme (CYP450s) assays are critical enzymes in early-stage lead discovery and optimization in drug development. Currently available fluorescence-based reaction assays provide a rapid and reliable method for monitoring CYP450 enzyme activity but are confined to medium-throughput well-plate systems. The authors present a high-throughput, integrated screening platform for CYP450 assays combining enzyme encapsulation techniques, microarraying methods, and wide-field imaging. Alginate-containing microarrays consisting of up to 1134 CYP450 reaction elements were fabricated on functionalized glass slides (reaction volumes 20 to 80 nL, total enzyme content in pg) and imaged to yield endpoint activity, stability, and kinetic data. A charge-coupled device imager acquired quantitative, high-resolution images of a 20 × 20 mm area/snapshot using custom-built wide-field optics with telecentric lenses and easily interchangeable filter sets. The imaging system offered a broad dynamic intensity range (linear over 3 orders of magnitude) and sensitivity down to fluorochrome quantities of <5 fmols, with read accuracy similar to a laser scanner or a fluorescence plate reader but with higher throughput. Rapid image acquisition enabled analysis of CYP450 kinetics. Fluorogenic assays with CYP3A4, CYP2C9, and CYP2D6 on the alginate microarrays exhibited Z′ factors ranging from 0.75 to 0.85, sensitive detection of inhibitory compounds, and reactivity comparable to that in solution, thereby demonstrating the reliability and accuracy of the microarray platform. This system enables for the first time a significant miniaturization of CYP enzyme assays with significant conservation of assay reagents, greatly increased throughput, and no apparent loss of enzyme activity or assay sensitivity.
Keywords: cytochromes P450, microarrays, high-throughput screening, wide-field imaging
INTRODUCTION
Recent Advances in Genomics, proteomics, combinatorial chemistry, and structural biology have sparked a tremendous increase in the number of screenable therapeutic targets as well as novel compounds that can be tested against these targets.1,2 This situation has necessitated the development of high-throughput, in vitro screening systems that can be used to probe the function of clinically relevant enzymes rapidly and at minimal cost. One particularly important superfamily of enzymes is the cytochromes P450 (CYP450), which are found throughout the body but exist most prevalently in the liver. CYP450s serve as the most important class of drug-metabolizing enzymes, catalyzing first-pass metabolism of xenobiotics.3,4 Nearly 60 distinct human CYP450 genes have been identified, and 3 major CYP isoforms—CYP3A4, CYP2C9, and CYP2D6—are responsible for the metabolism and clearance of nearly 75% of all drugs.5,6 In addition to metabolic clearance, CYP450s are also required for the metabolism-aided bioactivation of many prodrugs to therapeutically active products.7,8 Inhibition of CYP450s blocks the metabolism of several drugs, which may result in adverse drug reactions (ADRs) and systemic toxicity. ADRs are particularly problematic now that multidrug therapeutic regimens have become more common, with unsafe drug-drug interactions estimated to be the fourth to sixth leading cause of death in hospitalized patients in the United States.9 Furthermore, the pharmaceutical industry incurs costs totaling billions of dollars in the form of drugs withdrawn from market after late detection of CYP450 inhibition.10 It is therefore imperative that CYP450-catalyzed metabolism and inhibition be studied early and as efficiently as possible in the drug development process.
Several in vitro screening techniques have been developed to assess the potential of new chemical entities to inhibit CYP450s, including radiometric, fluorogenic, and rapid liquid chromatography/mass spectrometry (LC/MS)–based assays.11,12 Despite recent advances in LC/MS-based inhibition assays, such as the use of probe substrate cocktails for increased throughput,12–14 and advances in automation and miniaturization,14 these methods still remain largely medium throughput and require postreaction separation steps prior to analysis. Fluorescence-based assays, on the other hand, are the most amenable to high-throughput screening and miniaturization because of their inherent ease of operation and lack of multiple separation and processing steps.11,12,15 The development of highly sensitive and specific fluorogenic CYP450 substrates has facilitated CYP450 inhibition assays in 96-, 384-, and 1536-well microplates.15,16 However, well plates require 5 to 250 µL of reagents per well and are medium throughput in nature. Microarrays, on the other hand, offer the ability to perform CYP450 assays in nanoliter volumes on array platforms consisting of thousands of reaction spots, thereby greatly increasing throughput and reducing reagent costs. Development of miniaturized arrays employing CYP450s remains difficult, however, because of the inherent instability of the multienzyme human CYP450 reaction system. To overcome this instability, several immobilization techniques have been used to stabilize CYP450s, including covalent bonding onto various supports17 and entrapment in polyacrylamide gels18 and sol-gels.19 However, these approaches have been largely limited to low- or medium-throughput, well-plate– based platforms. Recently, we described a method for encapsulating CYP450s in nanoliter-scale alginate microarrays for the high-throughput, cell-based screening of drug compounds and their CYP450-generated metabolites.20 We now expand the application of alginate as an immobilization matrix to CYP450 activity assays.
Although advances in microarray technology (e.g., increased printing speeds) enable fabrication of complex, high-throughput enzyme-based microarrays, the lack of simple imaging technologies impedes analysis of these arrays, particularly for realtime analysis. Several laser-based scanner and charge-coupled device (CCD) imaging systems have been developed for DNA microarray analysis.21,22 Indeed, enzymatic microarrays have been imaged and analyzed using commercially available chip scanners.23,24 Scanners interrogate samples point by point, thereby making the time taken to image a slide (even at a single wavelength) too long to capture the data required for real-time enzyme activity assays. CCD cameras, however, have been used with some degree of success in enzyme microarray imaging. Gosalia and others25 complexed a 12-bit CCD camera with a fluorescence microscope for the high-throughput screening of proteases in fluid phase nanoliter-scale arrays. Rupcich and others26 obtained images of enzymatic reactions in sol-gel microarrays using a bright-field microscope equipped with a CCD camera. Similarly, Lu and Yeung27 used a plano objective lens attached to a CCD camera for 10× magnification of horseradish peroxidase arrays in high-throughput kinetic studies. Although these systems allow for rapid imaging of enzyme arrays, the optical resolutions are much higher than required, and the small field of view severely limits the number of spots in the array that can be imaged simultaneously. As a result, an X-Y moving stage with multiple stage movements and numerous camera exposures must be used to cover the entire slide, and the resultant images must be extensively assembled to yield a composite image.
In the present study, we use a novel wide-field fluorescence-based detection platform to extend the applicability of alginate microarrays to real-time CYP450 enzyme assays, resulting in dramatic scale down and reagent conservation when compared with conventional well-plate–based assays. In a broader context, this platform can be further extended to other applications such as enzyme inhibition assays and assays for assessing the metabolic stability of drug candidates.
MATERIALS AND METHODS
Reagents
Vivid® CYP450 enzyme screening kits were purchased from Invitrogen Inc. (Carslbad, CA). Kits for CYP3A4, CYP2C9, and CYP2D6, each containing Vivid® substrates, their corresponding fluorescent standards (fluorescein for CYP3A4 and CYP2C9, and coumarin for CYP2D6), CYP450 baculosomes (microsomes from baculovirus-infected cells coexpressing human CYP450s and NADPH-cytochrome P450 reductase), NADP+, and a regeneration system containing 333 mM glucose-6-phosphate and 30 U/ml glucose-6-phosphate dehydrogenase in 100 mM potassium phosphate buffer (pH 8.0) were used. Methyltrimethoxysilane (MTMOS), alginate, poly-L-lysine (PLL; 0.01%, w/v), barium chloride, ketoconazole, sulfaphenazole, and quinidine were purchased from Sigma-Aldrich (St. Louis, MO). All other reagents used were analytical grade.
Slide surface treatment
Borosilicate 25 × 75 mm glass slides from Fisher (Pittsburgh, PA) were cleaned with detergent and water, placed in a removable glass rack, and immersed in a tank containing concentrated H2SO4 for 2 days. The slides were then rinsed 3 times in deionized, distilled water; rinsed once in acetone; and then dried under nitrogen. The acid-cleaned slides were then spin coated with a solution of 15% (v/v) MTMOS in 0.5 mM HCl and 100 mM potassium phosphate buffer (pH 6.0, 1.5 mL) for 30 s at 3000 rpm. Treatment with MTMOS rendered the slide surface highly hydrophobic, facilitating the easy attachment of individual microarray spots and preventing local spreading and merging of printed spots. The MTMOS-coated slides were stored in the dark at 20 °C prior to printing.
Arraying and immobilization of CYP450 enzymes
Enzyme microarrays were fabricated on the MTMOS-coated glass slides using a MicroSys 5100-4SQ microarray spotter (Genomics Solutions, Ann Arbor, MI) equipped with a robotic pin spotter (100 µm orifice, 50 µL uptake). CYP450-alginate solutions (spotting volume 20 nL) were prepared by mixing 144 µL potassium phosphate buffer solution (pH 7.5) containing CYP450 baculosomes at 4-fold the final assay concentration, and 66 µL alginate (3% [w/v] in filtered distilled water) and were dispensed on top of 20 nL PLL-BaCl2 spots. The PLL-BaCl2 solution was prepared by mixing equal volumes of 0.01% (w/v) PLL and 0.1 M BaCl2, followed by sonication for 10 min. Final concentrations of the CYP450 enzymes were 10 nM for CYP3A4, 25 nM for CYP2C9, and 25 nM for CYP2D6. Enzyme printing was performed at room temperature under 90% relative humidity. Following printing, the slides were stored at −80 °C. For enzyme assays, the slides were thawed and placed in the microarray spotter, where the spots were overlaid with solution containing the fluorogenic substrate, NADP+, and the regeneration system.
CYP450 reaction assays on microarrays
Reaction kinetics were determined for the CYP450s by spotting 40 nL fluorogenic substrate solutions on top of the alginate-encapsulated enzymes spots. Vivid® green substrates, di(benzyloxymethoxy)fluorescein (DBOMF), and benzyloxymethoxyfluorescein (BOMF; ex: 485 nm; em: 530 nm) were used for CYP3A4 and CYP2C9 assays, respectively, and the Vivid® blue substrate 7-ethylmethoxy-3-cyanocoumarin (EOMCC) (ex: 405 nm; em: 460 nm) was used for CYP2D6 assays. The substrate solutions were prepared by mixing Vivid® substrate, 10 µL NADP+, and 10 µL regeneration system in 100 µL of potassium phosphate buffer, pH 8.0 (100 mM for CYP3A4 and CYP2D6 assays, and 50 mM for CYP2D6 assays). Final substrate concentrations were 10 µM for DBOMF and BOMF and 20 µM for EOMCC. Immediately following printing under 100% relative humidity, each slide was covered with a CoverWell perfusion chamber gasket (Invitrogen) to retard evaporation and prevent spot drying, and imaged continuously with the wide-field imaging system. Enzyme kinetics were evaluated with the Michaelis-Menten nonlinear regression model using GraphPad Prism 5.0 software (GraphPad, La Jolla, CA), and the kinetic constants were determined through Eadie-Hofstee plots.
Well-plate methods
All well-plate experiments were performed in 384-well plates (Fisher). The total reagent volume in each well was 28 µL, with 14 µL of reaction solution containing fluorogenic CYP substrate, NADP+, and regeneration system added to 14 µL of enzyme solution containing the CYP450 enzyme at twice the final assay concentration in potassium phosphate buffer (pH 8.0). For assays with inhibitory compounds, the enzyme solution also contained the inhibitory test compound prepared at twice the final assay concentration from a 100-mM stock solution. Final concentrations of enzyme, test compound, substrate, and all other assay components were identical to those used in the alginate microarrays. Increased fluorescence due to substrate oxidation was measured at 1-min intervals in the well plates using a PMT plate reader (PerkinElmer, Waltham, MA). Background fluorescence for each assay condition was measured in wells and microarrays containing all the assay components except the enzyme and subtracted prior to analysis.
Instrumentation: wide-field imaging system
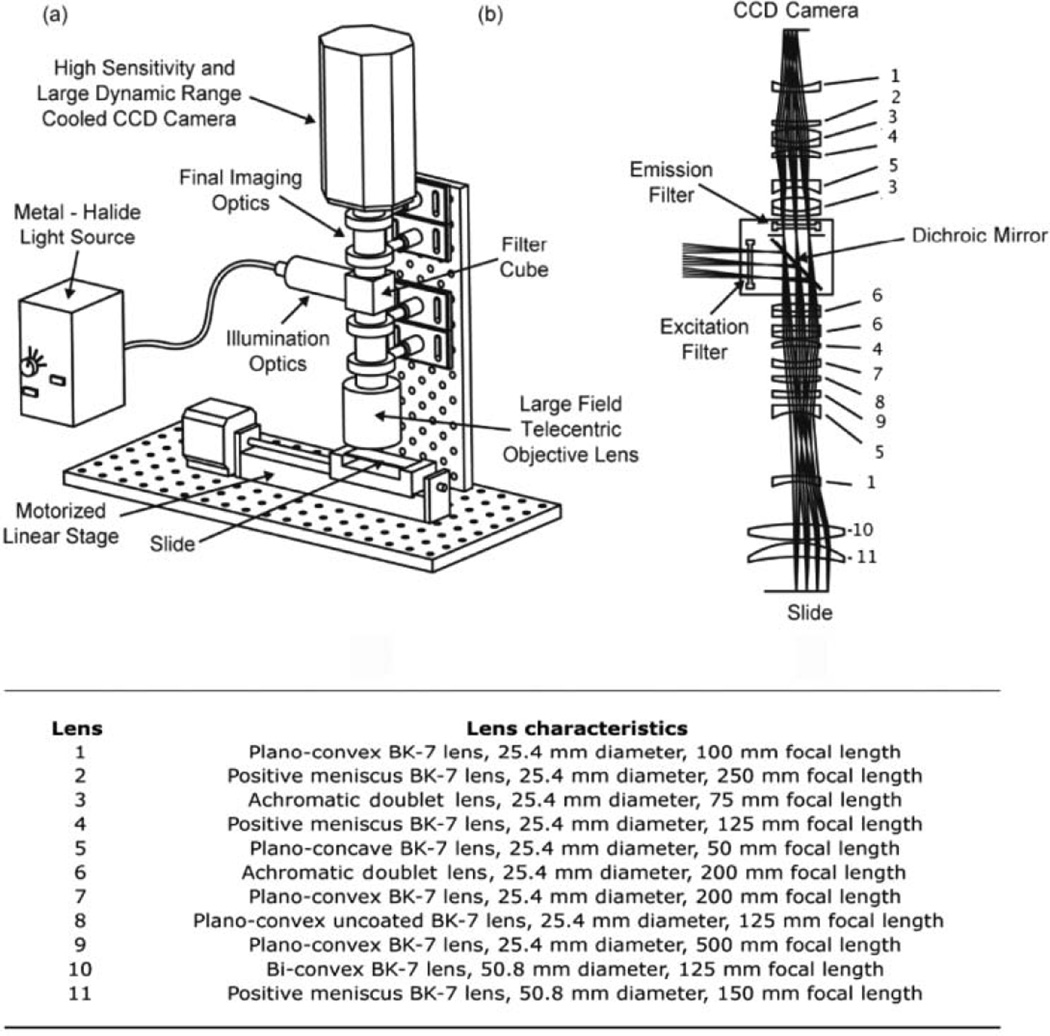
The wide-field imaging and detection system and its major components are shown schematically in Figure 1a. The optical design was simulated and optimized before construction using the ZEMAX optical design software (ZEMAX Development Corporation, Bellevue, WA). The composite lens system, shown in Figure 1b, was custom designed to enable the imaging of ~100-µm spots with high resolution while retaining a field of view large enough (~25 mm) to cover a large region of the microarray per image. All lenses used in the imager were purchased off the shelf from Thorlabs, Inc. (Newton, NJ). Light for excitation was supplied by an EL6000 metal halide light source (Leica Microsystems, Wetzlar, Germany) connected to the illumination optics with a liquid light guide. A filter cube consisting of an excitation filter, dichroic mirror, and emission filter (all from Chroma Technologies, Brattleboro, VT), was assembled and placed in the light path. The filter cube assembly was manually swapped for imaging probe assays requiring different filter characteristics. A custom infinity corrected objective lens system (0.02 numerical aperture) was used to perform telecentric illumination of the sample. Following excitation, light emitted from the sample was collected by the objective lens, collimated, and projected toward the dichroic mirror, which mainly transmitted only the light associated with the emission wavelength. An additional emission filter placed after the dichroic mirror was used to further filter out residual excitation light, allowing only the Stoke’s shifted emitted light to proceed to the detection system. Finally, light exiting the emission filter was passed through the final imaging optics and projected onto a CoolSnapK4 large format, 4-megapixel thermoelectrically cooled CCD camera (Photometrics, Pleasanton, CA). The slide to be imaged was placed on a motorized linear stage (Velmex, Inc., Broomfield, NY), which was programmed to take 6 separate images of the slide. The 6 images were combined to form 1 composite image, and the center 20 × 20 mm portion of each individual image was used for analysis. The fluorescence in each spot on the array was analyzed and quantified using the GenePixPro 6.0 software (Axon Instruments, Union City, CA).
FIG. 1.
(a) Overview of the wide-field optical imaging system with all major components labeled. (b) Lens assembly in the imaging system, showing the overall optical layout and detailing the lens design for the telecentric objective lens and the final imaging optics.
RESULTS
Performance of the imaging system
We initially investigated the imaging performance of the wide-field optical system. To minimize imaging artifacts, a plain MTMOS-coated slide was imaged prior to microarray imaging under assay conditions and used to subtract CCD fixed pattern noise and background fluorescence. The CCD detector acquired images of ~22-mm square fields on a chip with 2048 × 2048 pixels in a 12-bit gray-scale format. The imaging system provided a magnification of 18 µm per pixel with 2 pixels across the resolution, corresponding to a resolving power of 36 µm for the system. Six snapshots were taken and stitched together to yield a composite image of the microarray with a readout time of 3 s per image. The performance characteristics of the wide-field imaging optical detection system are summarized in Table 1. In all further experiments, a spot-by-spot protocol was used to correct for the ~45% drop off in the illumination intensity toward the edges of the image. The correction parameters were determined by scanning 5 slides (1134 spots per slide), with each spot on a slide containing 40 nL of the fluorescent standard fluorescein at concentrations ranging from 50 to 1000 nM.
Table 1.
Performance Figures for the Wide-Field Imaging System with CYP450 Alginate Arrays
|
Performance of the Optical Imaging System | |
|---|---|
| Analytical Figure of Merit | Value |
| Exposure time | 1200 ± 300 ms |
| Read timea | 18 s |
| Detection limit | 4 femtomoles fluorescein |
| S/Nmaxb | 600 ± 52 |
| S/Nassayc | 370.33 ± 15.99 |
| Average array-to-array reproducibilityd | 92% |
| Maximum signal drop off at edgese | 44% |
| Absolute spot-to-spot signal intensity variation following correction |
5.54% |
All experiments were performed with 3 separate batches of slides and 200 array elements per reaction condition.
Imaging a 25 × 75 mm slide in 6 snapshots (slide handling time excluded).
Maximum signal-to-noise ratio for 60 µM di(benzyloxymethoxy)fluorescein (DBOMF) substrate incubated with CYP3A4 for 45 min.
Signal-to-noise ratio obtained under assay conditions with 10 µM DBOMF incubated with CYP3A4 for 10 min.
Based on fluorescent readouts from 10 separate slides arrayed with different batches of fluorescein (concentrations of 100–1000 nM) over 1 month.
Based on fluorescent signal measurements from a 20 × 20 array printed with 10 µM DBOMF on a 25 × 25 region of a glass slide and incubated with CYP3A4 for 10 min.
Analysis of CYP450 microarrays
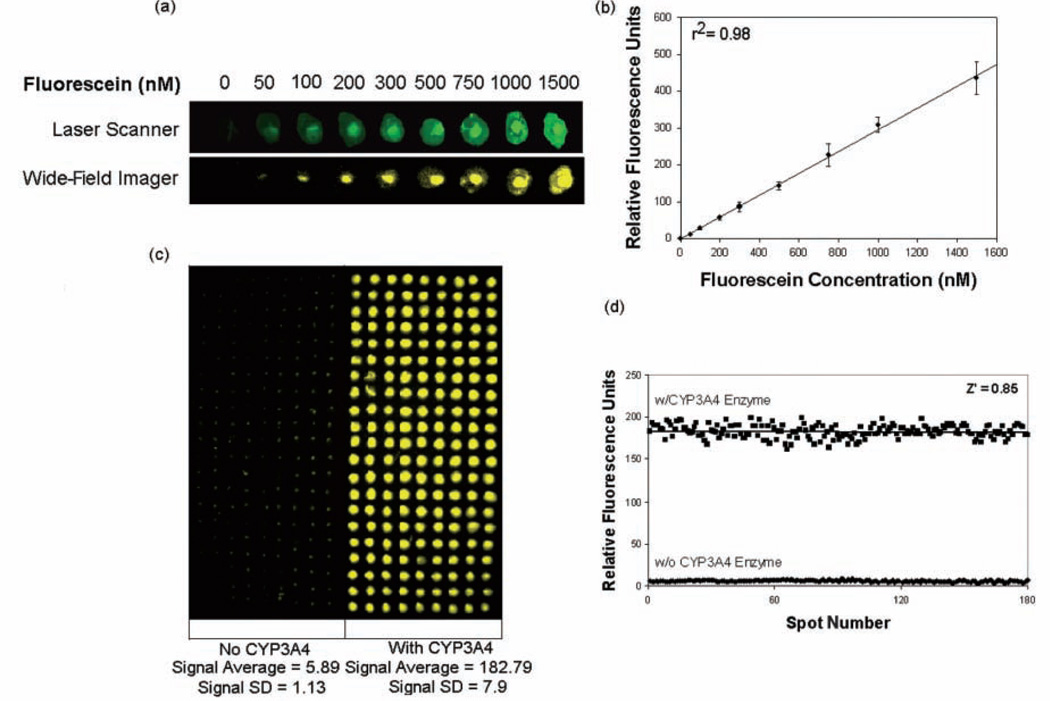
To determine the detection sensitivity of CYP450 microarrays with the imaging system, 40 nL of the CYP450 assay standard fluorescein (ex: 485 nm, em: 530 nm) was spotted from 1% (w/v) alginate solutions in concentrations ranging from 0 to 1200 nM on top of 20-nL spots containing PLL-BaCl2. Fluorescent data extracted from imaging this slide with the automated imaging system was plotted as a concentration-dependent calibration graph and compared to results obtained with the GenePix 4000B microarray scanner (Axon Instruments). Figure 2a compares the images of a 1 × 9 array obtained with both the scanner and the wide-field imaging system. The latter yielded a linear standard curve with a detection limit of 50 nM fluorescein, corresponding to 4 fmols of the compound in an 80-nL spot and a linear dynamic range of 50 to 1200 nm (r2 = 0.98), in excellent agreement with the results obtained with the laser scanner (Figure 2b). Experiments with another CYP450 assay standard, coumarin (ex: 405 nm, em: 460 nm), yielded similar results with a detection limit of 50 nM and a linear dynamic range of 50 to 1500 nm (r2 = 0.97, data not shown).
FIG. 2.
(a) Calibration assay with fluorescein. Representative 1×9 images of an array of 9 concentrations of the green fluorescent standard (ex: 485 nm, em: 535 nm; total spot volume 80 nL), as imaged with the wide-field imaging system and a laser scanner. (b) Calibration curve for fluorescein. Error bars on the graph represent standard deviations obtained from simultaneous measurements from 120 independent reaction spots per assay concentration (total spot content/microarray = 1080). (c) Wide-field image for 21×9 CYP3A4 reaction arrays with and without the enzyme present. (d) Z′ factor analysis for CYP3A4 alginate microarrays; 100 reaction elements (with enzyme) and 100 control elements (no enzyme) were analyzed simultaneously to yield a Z′ factor of 0.85 with a signal-to-noise ratio of ~31.
To evaluate the reliability of the CYP450 microarray platform, Z′ factors and signal-to-background ratios were determined following the method of Zhang and others.28 The Z′ factor is a statistical analysis parameter that reflects the performance of a screening platform. Assay platforms with Z′ values between 0.5 and 1.0 are considered to be robust and reliable. Alginate microarrays with and without a CYP450 enzyme were prepared and allowed to react with substrate solutions for CYP3A4, CYP2C9, and CYP2D6. The fluorescent output from these arrays was captured by the wide-field imaging system and was used to determine the Z′ factor. High signal-to-background ratios of 28 to 38 were obtained for all 3 CYP450 isoforms on the microarrays. Upon analysis of the microarray images, signal data with very little standard deviation were obtained. As an example, the CYP3A4 microarray is shown in Figure 2c, and Figure 2d shows the data obtained from the assay. Z′ values of 0.85, 0.75, and 0.79 were obtained from 21 × 9 arrays with and without enzyme for CYP3A4, CYP2D6, and CYP2C9, respectively. These results indicate that the alginate microarrays are excellent platforms for CYP450 assays and are capable of providing statistically relevant information in high throughput.
Stability and reactivity of CYP450 enzymes in alginate
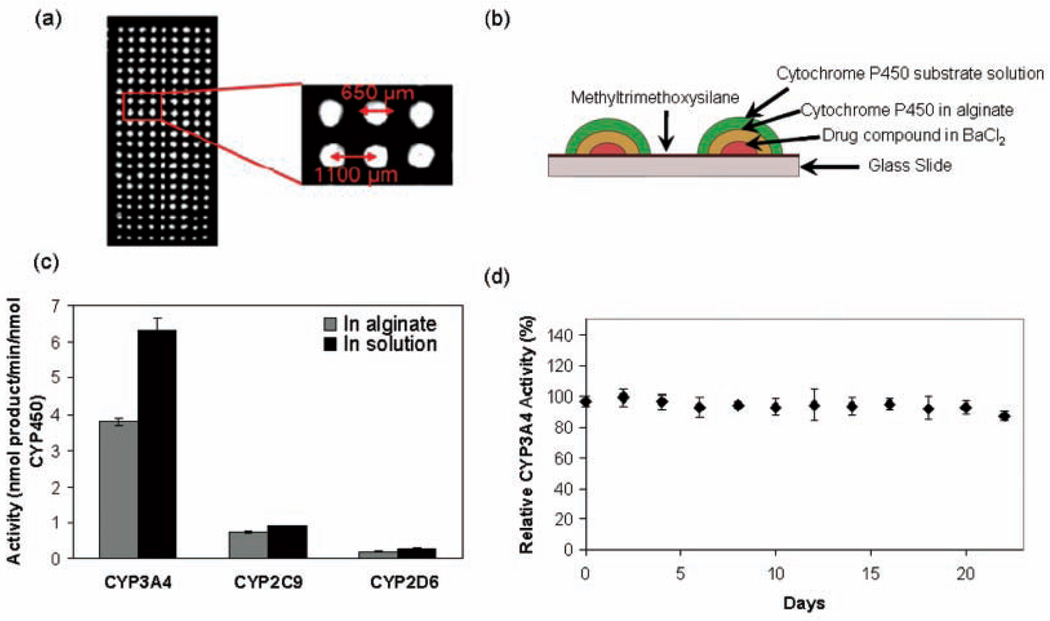
We have focused on the development of alginate-entrapped CYP450 microarrays for the high-throughput analysis of CYP450 function. Alginate serves as an ideal matrix because of its inertness and biocompatibility and is also known for its stabilizing effect on enzymes.29,30 In addition, alginate solutions are easily printable and do not gel until contacted with a divalent cation, with Ca2+ being used most extensively. Because phosphate buffer, which would rapidly sequester Ca2+, was used in our reactions, we chose Ba2+ as the divalent cation, as described in our previous studies.20 Each hemispherical spot on the resultant CYP450 alginate microarray had a volume of 80 nL with a diameter of 0.65 mm, and spot-to-spot contact was not observed (Fig. 3a). Each spot, therefore, served as a separate and independent enzyme reactor consisting of overlaid layers of the test compound, CYP450, and fluorogenic substrate, as shown in Figure 3b, with up to 1134 spots per microarray slide. CYP450 enzymes in 1% alginate retained 60% or more of their native solution activity (Fig. 3c), indicating that the immobilization process does not considerably alter enzyme function. Presumably, the high water content in alginate allows the enzyme to retain its conformation and activity within the matrix. The storage stability of CYP3A4 in alginate microarrays (spot volumes of 40 nL and 30 pg enzyme per spot) was also investigated. All 3 CYP450 isoforms exhibited excellent storage stability in alginate, with CYP3A4 retaining ~90% activity after storage for 3 weeks at −80 °C (Fig. 3d).
FIG. 3.
(a) Image of a 21 × 9 portion of a 1134-spot microarray (spot volume 80 nL) taken with the charge-coupled device system. (b) Construction of the CYP450 assay system on glass slide microarrays. (c) Reaction rates for CYP3A4, CYP2D6, and CYP2C9 in 1% alginate microarrays and in solution in 384-well plates. Error bars represent standard deviations from simultaneous measurements of 100 independent reaction elements (alginate microarrays) or 20 wells (384-well plate). (d) Storage stability of CYP3A4 alginate microarrays (100-spot arrays and a spot volume of 40 nL) as assayed with di(benzyloxymethoxy)fluorescein and expressed as percentage of enzyme activity observed with no storage. Error bars represent standard deviations.
Evaluation of enzyme kinetics on the microarray
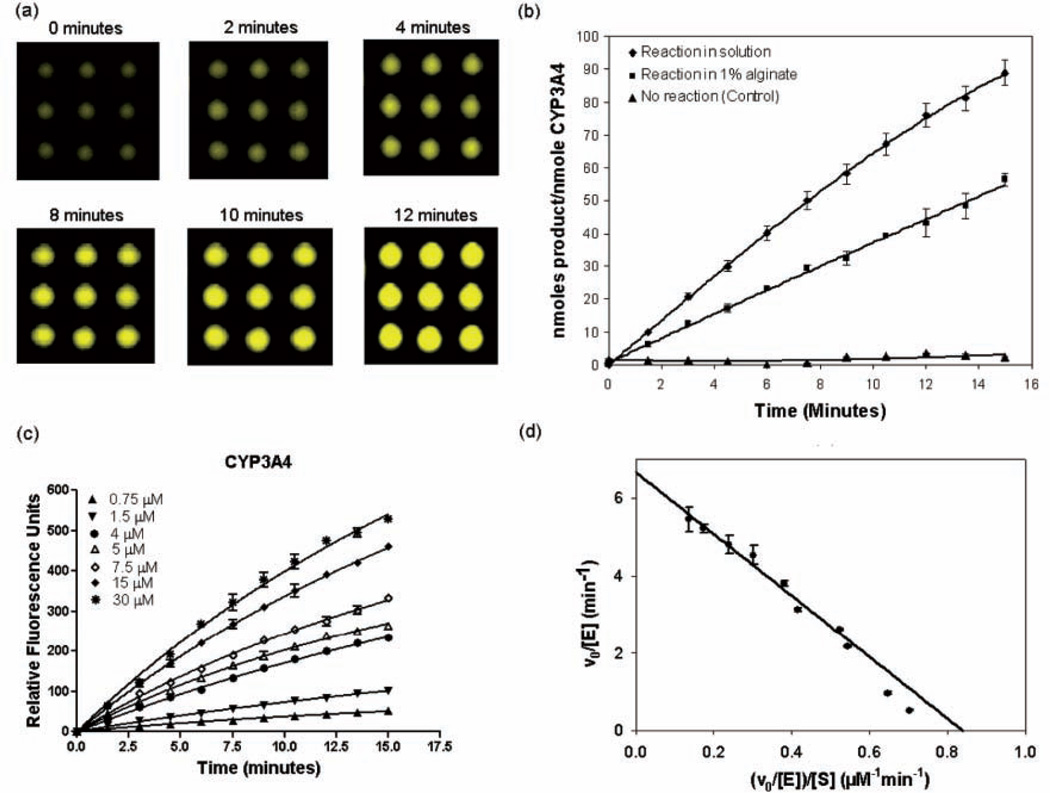
CYP3A4, CYP2C9, and CYP2D6 enzyme kinetics were investigated in alginate microarrays using the wide-field imaging system and compared with kinetic data obtained in solution in 384-well plates under identical enzyme and substrate concentrations. Representative images of a 3 × 3 CYP3A4 assay microarray (Fig. 4a) taken with the wide-field detection system showed a time-dependent increase in observed fluorescence with a linear reaction time of ~14 min, similar to the ~10-min linear reaction time obtained in solution (Fig. 4b). CYP450 kinetics were then evaluated at substrate concentrations ranging from 0 to 40 µM, and Michaelis-Menten behavior was observed (Fig. 4c, d) with linear Eadie-Hofstee plots. All 3 CYP450 isoforms on the microarray displayed values of kcat/Km within 2-fold of that obtained in solution (Table 2), thereby demonstrating that the alginate matrix and the dramatic scale down (375-fold v. that of the 384-well plate) did not adversely affect CYP450 activity. The Km values for CYP enzymes encapsulated in alginate on the microarray were nearly identical (within experimental error) to those obtained in free solution in the well plate, suggesting that the alginate matrix did not influence substrate partitioning or substrate diffusion. This is consistent with the very high water content of the alginate and its low polymer volume fraction.
FIG. 4.
Reaction kinetics for CYP3A4 (10 nM) with dibenzoxymethylfluorescein (DBOMF) substrate on 1134-spot alginate microarrays (spot volume 80 nL, spot diameter 280 µm). (a) Images of a representative 3 × 3 section of the alginate array (DBOMF concentration 10 µM) taken with the wide-field imaging system. (b) Comparison of the progress of DBOMF substrate oxidation (concentration 10 µM) as a function of time in solution in 384-well plates (experiments conducted in triplicate) and on chip (100 independent spots arrayed and analyzed) in 1% alginate. Error bars represent standard deviations. (c) Initial rates obtained with different DBOMF concentrations (0.75-30 µM) for CYP3A4-catalyzed oxidation on the alginate chip. Error bars represent standard deviations from a 21 × 9 element array for each substrate concentration. (d) Eadie-Hofstee plot showing the fit to Michaelis-Menten kinetics for CYP3A4 encapsulated in 1% alginate with DBOMF substrate.
Table 2.
Kinetic Constants for CYP3A4, CYP2C9, and CYP2D6 Assays
| CYP Isoform | Reaction Condition | Enzyme Concentration | Km (µM) | kcat (min−1) | kcat/Km (M−1 min−1) |
|---|---|---|---|---|---|
| CYP3A4 | Solution | 10 nM | 8.4 ± 1.7 | 10 ± 0.7 | (11.9 ± 1.3) × 105 |
| Alginate | 10 nM | 7.9 ± 1.4 | 6.7 ± 0.19 | (8.3 ± 0.8) × 105 | |
| CYP2C9 | Solution | 25 nM | 2.7 ± 0.4 | 1.1 ± 0.1 | (3.9 ± 0.45) × 105 |
| Alginate | 25 nM | 2.6 ± 0.4 | 0.86 ± 0.04 | (3.3 ± 0.8) × 105 | |
| CYP2D6 | Solution | 25 nM | 16 ± 3.4 | 0.62 ± 0.04 | (0.39 ± 1.3) × 105 |
| Alginate | 25 nM | 19 ± 4.2 | 0.40 ± 0.02 | (0.21 ± 0.3) × 105 |
Initial rates were obtained in triplicate with different substrate concentrations (0 to 40 µM) on alginate microarrays (alginate content 1%, 100 spots assayed per substrate concentration, spot volume 80 nL) and in solution in 384-well plates.
CYP450 inhibition on the microarray platform
The microarray platform coupled with the wide-field imaging system was ideally suited to evaluate CYP450 inhibition. We assayed 3 potent, enzyme-selective inhibitory compounds (sulfaphenazole, a CYP2C9 inhibitor; ketoconazole, a CYP3A4 inhibitor; and quinidine, a CYP2D6 inhibitor) against CYP3A4, CYP2C9, and CYP2D6 on the 1134-spot microarray. Stock solutions of 100-mM test compounds were prepared in dimethylsulfoxide, further diluted in 1:1 PLL-BaCl2, and printed in 20-nL spots on an MTMOS-coated slide. The final assay concentrations of 10 nM for quinidine, 100 nM for ketoconazole, and 250 nM for sulfaphenazole were chosen to approximate the published IC50 values against CYP2D6, CY3A4, and CYP2C9, respectively.12,15,16 The final DMSO concentration was 0.5% (v/v). The bottom spots were overlaid with enzyme-alginate solution, which immediately gelled on the slide, preincubated for 20 min with the inhibitor compounds, and then overprinted with the substrate solution. Enzyme and substrate assay components were similar to those used in the aforementioned kinetic assays. The reactions was allowed to proceed for 15 min (to remain in the linear reaction rate regime) and terminated by dispensing 10% (v/v) aqueous acetonitrile to each spot. The slide was then dried under nitrogen and imaged with the wide-field imaging system.
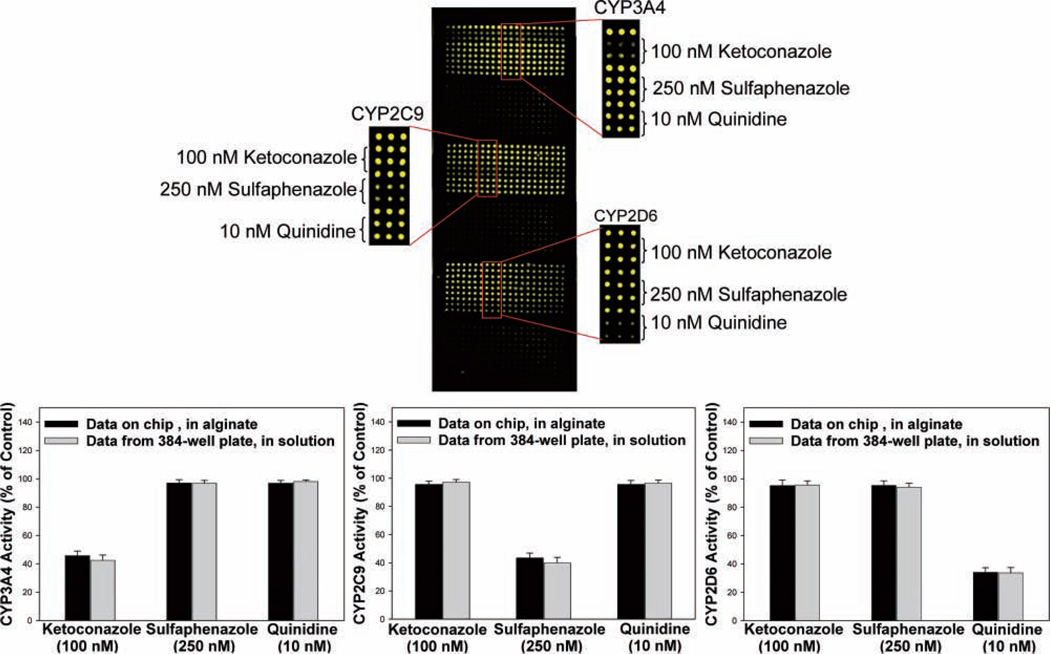
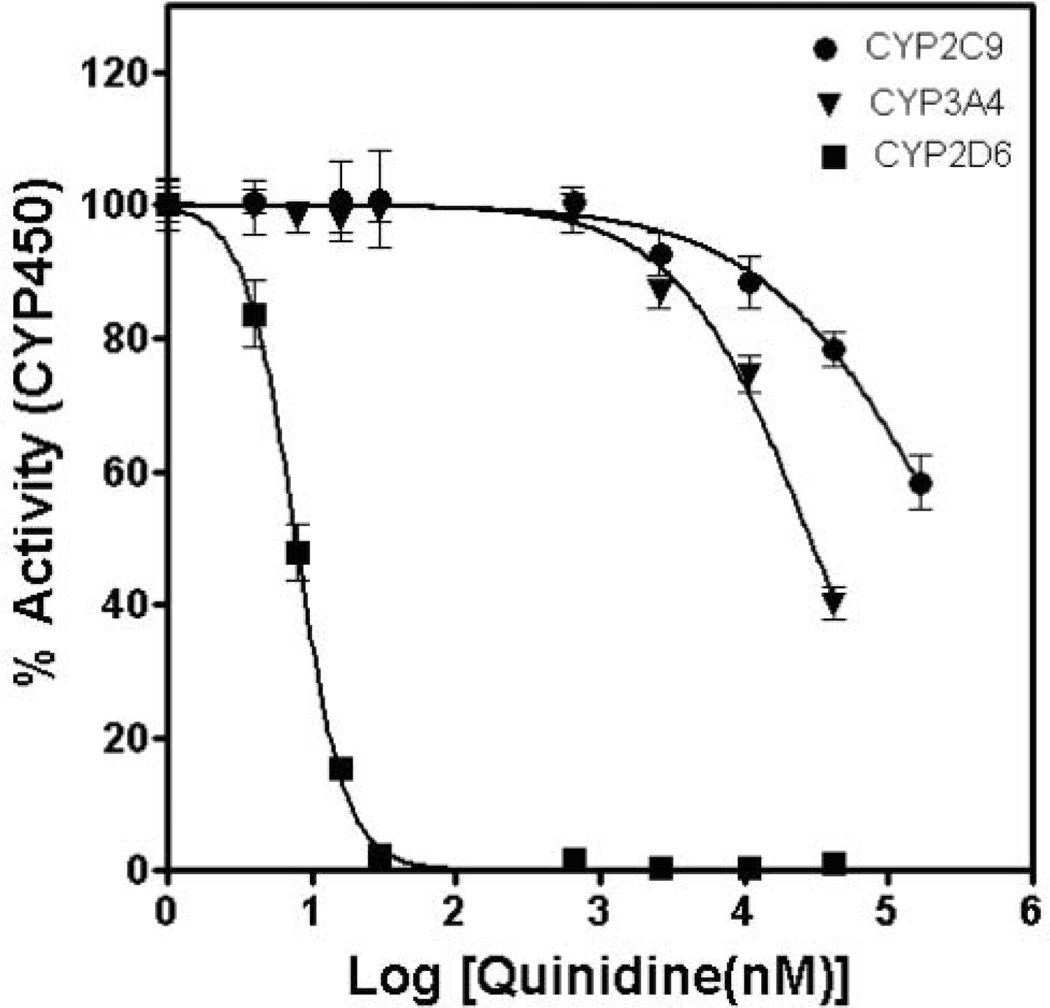
Figure 5a shows the resultant microarray imaged for the 3 CYP450s. For each isoform, control spots containing no inhibitor exhibited bright fluorescence produced by the CYP450-catalyzed oxidation of the substrate. In contrast, in the presence of an inhibitor, a decrease in fluorescence is observed, indicating successful detection of enzyme inhibition. Ketoconazole potently and selectively inhibited CYP3A4 at a concentration of 100 nM, reducing substrate oxidation to less than 40%, but it did not appreciably inhibit CYP2C9 or CYP2D6 (Fig. 5b). Similarly, with 250 nM of the CYP2C9 inhibitor sulfaphenazole, CYP2C9 activity was reduced to nearly 50%, whereas CYP3A4 and CYP2D6 activities remained largely unaffected. Quinindine (10 nM) reduced CYP2D6 activity to 40% of that without the inhibitor, with virtually no inhibition of CYP3A4 or CYP2C9 activity observed. In addition, the relative fluorescence values obtained in the presence of test compounds on microarrays (expressed as a percentage of fluorescence observed in control spots) were in excellent agreement with those obtained in 384-well plates in solution with the plate reader (Fig. 5b). We further proceeded to obtain a complete inhibitory profile with dose-response curves for 1 of the test compounds, quinidine, against CYP3A4, CYP2C9, and CYP2D6 on the alginate microarray platform (Fig. 6). The IC50 value obtained for quinidine against CYP2D6 was 8 nM, whereas little inhibitory activity was observed with CYP3A4 (IC50 ~30 µM) and CYP2C9 (IC50 ~300 µM), in excellent agreement with literature values.12,15,16 These results demonstrate the potential of the alginate microarrays to detect the loss of CYP450 activity in the presence of inhibitors.
FIG. 5.
CYP450 inhibition on the microarray platform. (a) CYP3A4, CYP2C9, and CYP2D6 were immobilized on 3 different regions of the slide. For each of the 3 enzymes, a 21 × 9 array consisting of both reaction control rows (no inhibitor) and rows containing the inhibitory compounds (ketoconazole, sulfaphenazole, and quinidine) was printed. Interspersing the reaction blocks, three 21 × 9 arrays containing only buffer were printed to account for background fluorescence. Six snapshots with appropriate filters were taken of the slide, false colored in green, and reconstituted to give a single image. (b) Activity obtained for CYP3A4, CYP2C9, and CYP2D6 in the presence of inhibitors on the alginate microarray and in solution in 384-well plates, expressed as percentage of enzyme activity observed in the absence of any inhibitor. Standard deviations in fluorescent intensity across 21 independent reaction spots (alginate arrays) or triplicate wells (in solution) are represented as error bars.
FIG. 6.
IC50 curves generated from CYP3A4, CYP2C9, and CYP2D6 inhibition assays with quinidine. Nine concentrations of quinidine (0–166 µM) were assayed against each isoform (21 reaction elements per concentration, spot volume 80 nL) on an alginate microarray. Error bars represent standard deviations. Background signals from negative controls (no reaction) were subtracted, and the data were normalized to the maximum fluorescent intensity obtained in the absence of quinidine.
DISCUSSION
Enzyme-containing microarrays represent an emerging assay platform for drug discovery. The ability to carry out multiple reactions in manifold, miniaturized replicates on a single glass slide with limited manual labor and significant savings in reagents makes microarrays ideal for high-throughput, early-stage testing of compounds in the pharmaceutical industry. However, microarrays have shown little success in completely replacing the well-plate platform in enzyme kinetics and enzyme inhibition assays. The limited success to date can be ascribed to 2 significant hurdles: the need for a suitable immobilization method for delicate enzymes (e.g., CYP450s) and the lack of an optical system suited to endpoint and real-time array-based imaging and analysis. Stabilizing enzyme matrices, such as alginate, can be used to address the first hurdle and provide ease of use due to the solution properties of alginate. With respect to the second hurdle, the limited range of wavelengths and the long scanning times typically associated with laser scanners make them unsuitable for most enzyme assays. In addition, no wide-field imaging system has been previously used to image enzyme microarrays with minimal camera exposures, sufficient resolution, and short exposure times.
Despite the significant miniaturization (nearly 2000-fold from a 96-well plate) and enzyme immobilization associated with the CYP450 microarrays, important assay parameters such as the signal-to-background ratio, minimum detectable signal, and the linear dynamic range of fluorescence detection remained statistically excellent and similar to data obtained in solution. Assays with the 3 most important drug-metabolizing enzymes (CYP3A4, CYP2C9, and CYP2D6) yielded Z′ values higher than those reported in solution with 1536-well plates for CYP3A4 and CYP2D6 and similar to the Z′ value reported with 1536-well plate data for CYP2C9.15 Km and kcat values obtained with alginate microarrays showed slight deviation from data obtained in solution with 384-well plates, with the most marked differences between microarray and well-plate data observed for CYP2D6. However, the maximum variation was less than 2-fold, and the reaction kinetics on the chip were sufficient to accurately detect isoform-specific inhibition of enzyme activity with nanomolar concentrations of potent CYP inhibitors.
One potential concern with fluorogenic assays for CYP450 inhibition is the often poor correlation between data obtained with fluorogenic probes and those obtained with actual drugs in LC/MS-based assays.11,12 However, substrate-dependent variation of inhibition data is observed, even among different drugs for CYP450 isoforms, in particular for CYP3A4.11 Indeed, fluorogenic assays still represent robust, reliable preliminary assays for detecting the inhibitory potential of test compounds in drug discovery.11,12,15 Moreover, the ability to perform kinetic experiments with the microarray platform makes it possible to investigate the mechanism of enzyme inhibition, as well as to determine the Ki value for an inhibitor using fluorogenic substrates in high throughput. This would enable an efficient determination of intrinsic and comprehensive CYP450 inhibitory profiles for lead compounds.
In conclusion, with a nearly 2000-fold scale down from 96-well plates and a 100-fold scale down from 1536-well plates, the CYP450 alginate-based microarray together with the wide-field imaging CCD system represents an ultra high-throughput screening tool for the rapid, accurate, and sensitive assessment of CYP450 activity and inhibition. This approach may provide a route to the further miniaturization of conventional well-plate–based assays that can be used in concert with other ultra high-throughput assays in early-stage drug discovery. The applicability of this platform can logically be extended to the simultaneous determination of complete inhibition profiles for multiple test compounds against multiple CYP450 isoforms. In addition, this approach is expected to be applicable to any enzyme assay as long as suitable fluorescent substrates are available.
Acknowledgments
The authors thank Glenn Saunders and John Wen for helpful discussions and engineering assistance in constructing the imaging system. This work was supported by the National Institutes of Health (ES012619) and the New York State Office of Science and Technology (NYSTAR).
REFERENCES
- 1.Dobson CM. Chemical space and biology. Nature. 2004;432:824–828. doi: 10.1038/nature03192. [DOI] [PubMed] [Google Scholar]
- 2.Geysen HM, Schoenen F, Wagner D, Wagner R. Combinatorial compound libraries for drug discovery: an ongoing challenge. Nat Rev Drug Discov. 2003;2:222–230. doi: 10.1038/nrd1035. [DOI] [PubMed] [Google Scholar]
- 3.Guengerich FP. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol. 2001;14:611–650. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- 4.Wrighton SA, Stevens JC. The human hepatic cytochromes-P450 involved in drug metabolism. Crit Rev Toxicol. 1992;22:1–21. doi: 10.3109/10408449209145319. [DOI] [PubMed] [Google Scholar]
- 5.Wienkers LC, Heath TG. Predicting in vivo drug interactions from in vitro drug discovery data. Nat Rev Drug Discov. 2005;4:825–833. doi: 10.1038/nrd1851. [DOI] [PubMed] [Google Scholar]
- 6.Ekins S, Stresser DM, Williams JA. In vitro and pharmacophore insights into CYP3A enzymes. Trends Pharmacol Sci. 2003;24:161–166. doi: 10.1016/s0165-6147(03)00049-x. [DOI] [PubMed] [Google Scholar]
- 7.Roy P, Waxman DJ. Activation of oxazaphosphorines by cytochrome P450: application to gene-directed enzyme prodrug therapy for cancer. Toxicol In Vitro. 2006;20:176–186. doi: 10.1016/j.tiv.2005.06.046. [DOI] [PubMed] [Google Scholar]
- 8.Aghi M, Hochberg F, Breakefield XO. Prodrug activation enzymes in cancer gene therapy. J Gene Med. 2000;2:148–164. doi: 10.1002/(SICI)1521-2254(200005/06)2:3<148::AID-JGM105>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 9.Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients—a meta-analysis of prospective studies. JAMA. 1998;279:1200–1205. doi: 10.1001/jama.279.15.1200. [DOI] [PubMed] [Google Scholar]
- 10.Bjornsson TD, Callaghan JT, Einolf HJ, Fischer V, Gan L, Grimm S, et al. The conduct of in vitro and in vivo drug-drug interaction studies: a Pharmaceutical Research and Manufacturers of America (PhRMA) perspective. Drug Metab Dispos. 2003;31:815–832. doi: 10.1124/dmd.31.7.815. [DOI] [PubMed] [Google Scholar]
- 11.Cohen LH, Remley MJ, Raunig D, Vaz ADN. In vitro drug interactions of cytochrome P450: an evaluation of fluorogenic to conventional substrates. Drug Metab Dispos. 2003;31:1005–1015. doi: 10.1124/dmd.31.8.1005. [DOI] [PubMed] [Google Scholar]
- 12.Di L, Kerns EH, Li SQ, Carter GT. Comparison of cytochrome P450 inhibition assays for drug discovery using human liver microsomes with LC-MS, rhCYP450 isozymes with fluorescence, and double cocktail with LC-MS. Int J Pharm. 2007;335:1–11. doi: 10.1016/j.ijpharm.2006.10.039. [DOI] [PubMed] [Google Scholar]
- 13.Dierks EA, Stams KR, Lim HK, Cornelius G, Zhang HL, Ball SE. A method for the simultaneous evaluation of the activities of seven major human drug-metabolizing cytochrome P450s using an in vitro cocktail of probe substrates and fast gradient liquid chromatography tandem mass spectrometry. Drug Metab Dispos. 2001;29:23–29. [PubMed] [Google Scholar]
- 14.Youdim KA, Lyons R, Payne L, Jones BC, Saunders K. An automated, high-throughput, 384 well cytochrome P450 cocktail IC50 assay using a rapid resolution LC-MS/MS end-point. J Pharm Biomed Anal. 2008;48:92–99. doi: 10.1016/j.jpba.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 15.Trubetskoy OV, Gibson JR, Marks BD. Highly miniaturized formats for in vitro drug metabolism assays using Vivid® fluorescent substrates and recombinant human cytochrome P450 enzymes. J Biomol Screen. 2005;10:56–66. doi: 10.1177/1087057104269731. [DOI] [PubMed] [Google Scholar]
- 16.Crespi CL, Miller VP, Penman BW. Microtiter plate assays for inhibition of human, drug-metabolizing cytochromes P450. Anal Biochem. 1997;248:188–190. doi: 10.1006/abio.1997.2145. [DOI] [PubMed] [Google Scholar]
- 17.Lamb SB, Lamb DC, Kelly SL, Stuckey DC. Cytochrome P450 immobilisation as a route to bioremediation/biocatalysis. FEBS Lett. 1998;431:343–346. doi: 10.1016/s0014-5793(98)00771-6. [DOI] [PubMed] [Google Scholar]
- 18.Yawetz A, Perry AS, Freeman A, Katchalski-Katzir E. Monooxygenase activity of rat liver microsomes immobilized by entrapment in a cross-linked prepolymerized polyacrylamide hydrazide. Biochim Biophys Acta. 1984;798:204–209. doi: 10.1016/0304-4165(84)90305-2. [DOI] [PubMed] [Google Scholar]
- 19.Sakai-Kato K, Kato M, Homma H, Toyo’oka T, Utsunomiya-Tate N. Creation of a P450 array toward high-throughput analysis. Anal Chem. 2005;77:7080–7083. doi: 10.1021/ac050714y. [DOI] [PubMed] [Google Scholar]
- 20.Lee MY, Kumar RA, Sukumaran SM, Hogg MG, Clark DS, Dordick JS. Three-dimensional cellular microarray for high-throughput toxicology assays. Proc Natl Acad Sci U S A. 2008;105:59–63. doi: 10.1073/pnas.0708756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Che DP, Bao YJ, Muller UR. Novel surface and multicolor charge coupled device-based fluorescent imaging system for DNA microarrays. J Biomed Optic. 2001;6:450–456. doi: 10.1117/1.1412437. [DOI] [PubMed] [Google Scholar]
- 22.Hamilton G, Brown N, Oseroff V, Huey B, Segraves R, Sudar D, et al. A large field CCD system for quantitative imaging of microarrays. Nucleic Acids Res. 2006;34:e58. doi: 10.1093/nar/gkl160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horiuchi KY, Wang Y, Diamond SL, Ma HC. Microarrays for the functional analysis of the chemical-kinase interactome. J Biomol Screen. 2006;11:48–56. doi: 10.1177/1087057105282097. [DOI] [PubMed] [Google Scholar]
- 24.Funeriu DP, Eppinger J, Denizot L, Miyake M, Miyake J. Enzyme family-specific and activity-based screening of chemical libraries using enzyme microarrays. Nat Biotechnol. 2005;23:622–627. doi: 10.1038/nbt1090. [DOI] [PubMed] [Google Scholar]
- 25.Gosalia DN, Diamond SL. Printing chemical libraries on microarrays for fluid phase nanoliter reactions. Proc Natl Acad Sci U S A. 2003;100:8721–8726. doi: 10.1073/pnas.1530261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rupcich N, Goldstein A, Brennan JD. Optimization of sol-gel formulations and surface treatments for the development of pin-printed protein microarrays. Chem Mater. 2003;15:1803–1811. [Google Scholar]
- 27.Lu GX, Yeung ES. High-throughput enzyme kinetics using microarrays. Isr J Chem. 2007;47:141–147. [Google Scholar]
- 28.Zhang JH, Chung TDY, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 29.Chung TW, Yang J, Akaike T, Cho KY, Nah JW, Kim SI, et al. Preparation of alginate/galactosylated chitosan scaffold for hepatocyte attachment. Biomaterials. 2002;23:2827–2834. doi: 10.1016/s0142-9612(01)00399-4. [DOI] [PubMed] [Google Scholar]
- 30.Smidsrod O, Skjakbraek G. Alginate as immobilization matrix for cells. Trends Biotechnol. 1990;8:71–78. doi: 10.1016/0167-7799(90)90139-o. [DOI] [PubMed] [Google Scholar]