Abstract
An estimated 40,000 deaths will be attributed to breast cancer in 2016, underscoring the need for improved therapies. Evading cell death is a major hallmark of cancer, driving tumor progression and therapeutic resistance. To evade apoptosis, cancers use anti-apoptotic Bcl-2 proteins to bind to and neutralize apoptotic activators, such as Bim. Investigation of anti-apoptotic Bcl-2 family members in clinical breast cancer datasets, revealed greater expression and more frequent gene amplification of MCL1 as compared to BCL2 or BCL2L1 (Bcl-xL) across three major molecular breast cancer subtypes, Luminal (A and B), HER2-enriched, and Basal-like. While Mcl-1 protein expression was elevated in estrogen receptor α (ERα)-positive and ERα-negative tumors as compared to normal breast, Mcl-1 staining was higher in ERα+ tumors. Targeted Mcl-1 blockade using RNAi increased caspase-mediated cell death in ERα+ breast cancer cells, resulting in sustained growth inhibition. In contrast, combined blockade of Bcl-2 and Bcl-xL only transiently induced apoptosis, as cells rapidly acclimated through Mcl-1 upregulation and enhanced Mcl-1 activity, as measured in situ using Mcl-1/Bim proximity ligation assays. Importantly, MCL1 gene expression levels correlated inversely with sensitivity to pharmacological Bcl-2/Bcl-xL inhibition in luminal breast cancer cells, whereas no relationship was seen between gene expression of BCL2 or BCL2L1 and sensitivity to Bcl-2/Bcl-xL inhibition. These results demonstrate that breast cancers rapidly deploy Mcl-1 to promote cell survival, particularly when challenged with blockade of other Bcl-2 family members, warranting the continued development of Mcl-1 selective inhibitors for targeted tumor cell killing.
Keywords: Mcl-1, ABT-263 resistance, Bcl-2 family proteins, luminal breast cancers
Introduction
The intrinsic apoptotic pathway is tightly regulated by Bcl-2 family members to support developmental processes and proper physiological function (1). Apoptosis dysregulation often produces pathological consequences, including cancer formation, progression and therapeutic resistance (2). At the center of the intrinsic apoptotic pathway are the ‘effectors’ Bax and Bak, which oligomerize at the outer mitochondrial membrane (OMM) by binding to ‘activators’ (Bim, Bid, and Puma) (3). Bak/Bax oligomerization promotes pore formation in the OMM, resulting in mitochondrial depolarization, disruption of oxidative phosphorylation, mitochondrial cytochrome-c release into the cytoplasm, and apoptosome activation, thus initiating caspase-dependent apoptosis (4-6). Anti-apoptotic Bcl-2 family proteins (Bcl-2, Bcl-A1, Bcl-xL, Bcl-w, and Mcl-1) restrain the intrinsic apoptotic pathway by binding and sequestering effectors (7) and/or activators (8), thus favoring cell survival. Pro-apoptotic ‘sensitizers’ (Bad, Hrk, and Noxa) bind and saturate anti-apoptotic Bcl-2 proteins, thus favoring apoptosis (9-11). A delicate balance in the relative ratio of anti-apoptotic Bcl-2 proteins to apoptotic activators or sensitizers is necessary for cell survival regulation. Cancers can exploit this pathway to evade apoptosis, often through increased levels of anti-apoptotic Bcl-2 factors (12-14).
Nearly 250,000 new breast cancer cases will be diagnosed in the U.S. in 2016 (15). Despite advances in detection and treatment of breast cancers, it is estimated that up to 40,000 patients will die from breast cancer in the U.S. each year, often due to recurrent metastatic disease, highlighting the need for improved treatments that promote tumor cell killing. Several studies suggest that anti-apoptotic Bcl-2 family proteins may be particularly attractive therapeutic targets in breast cancers. For example, Bcl-2 expression observed in up to 70% of ERα+ breast cancers (16). While Bcl-xL remains less studied in primary breast tumors, Bcl-xL expression was increased in ductal carcinoma in situ (DCIS) compared to normal breast in some breast cancer subtypes (17). Interestingly, many breast cancers increase levels of Bcl-2 family proteins following treatment with tamoxifen (18), fulvestrant (19), and neoadjuvant chemotherapy (NAC) (20). Bcl-2 and Bcl-xL levels reportedly predict poor response to taxanes (21), Adriamycin (22) and the HER2-monocolonal antibody Herceptin (23). Additionally, Bcl-2 family proteins are often upregulated in endocrine-resistant cancers (24, 25). These findings support an intense interest in research strategies to block activity of Bcl-2 family proteins as a means to enhance tumor cell killing.
Pharmacological inhibition of Bcl-2 family proteins has been achieved using compounds that bind to the BH3-domain binding pocket of Bcl-2 family members. These BH3 ‘mimetics’ block the interaction of Bcl-2 family proteins with BH3-domain containing pro-apoptotic factors (i.e. Bcl-2 activators or sensitizers), including Bim (26). ABT-199/vinitoclax, which specifically blocks Bcl-2 from interacting with BH3-domain containing pro-apoptotic factors, is currently approved for clinical use in chronic lymphocytic leukemia (CLL) (27), and is in clinical trial in several additional cancers, including breast cancers (28). ABT-263/novitoclax binds to and blocks activity of Bcl-2 and Bcl-xL and is showing efficacy in early-phase clinical trials in hematological malignancies (29-31). Pre-clinical studies in luminal breast cancers show that ABT-263 (targeting Bcl-2 and Bcl-xL) or ABT-199 (targeting Bcl-2) may effectively increase tumor killing when cells were first ‘primed’ with tamoxifen (32-34).
These advances in Bcl-2 family targeting warrant a greater understanding of the molecular characteristics of breast cancers that might benefit from this treatment strategy. Examination of large clinical datasets identified expression and gene amplification of BCL2 and BCL2L1 (Bcl-xL). However, we found that MCL1 gene expression occurred more frequently in breast cancers that other Bcl-2 family members. Disruption of Mcl-1 activity increased caspase activated apoptosis and impaired cell growth to a greater extent than combined disruption of Bcl-2 and Bcl-xL. Importantly, expression levels of MCL1 predicted sensitivity to ABT-263 in a panel of breast cancer cell lines, which may inform results in ongoing clinical trials, or guide patient selection for future trials.
Materials and Methods
Expression analysis of publically available cancer cell line and breast cancer datasets
mRNA expression of MCL1, BCL2, and BCL2L1 (Bcl-xL) were curate using cBio Portal (www.cbio.org) for cancer cell lines (CCLE) and breast tumor specimens (TCGA). Breast cancer specimens were stratified based on PAM50 molecular markers (TCGA), and CGH analysis was used to observed alterations at the genetic level (amplifications). mRNA expression of MCL1, BCL2, and BCL2L1 in breast cancer cell liens (CCLE) were correlated to the IC50 of ABT-263 as determined by the Sanger Institute (http://www.cancerrxgene.org/), data was fit to a linear regression.
Western Blotting
Cells and tumor tissue were homogenized in ice-cold lysis buffer [50mM Tris pH 7.4, 100mM NaF, 120mM NaCl, 0.5% NP-40, 100 μM Na3VO4, 1× protease inhibitor cocktail (Roche), 0.5μM proteasome inhibitor (Santa Cruz Technologies)]. Proteins were resolved on 4-12% SDS-PAGE gels and transferred to nitrocellulose membranes, which were blocked in 3% gelatin in TBS-T [Tris-buffered saline, 0.1% Tween-20], incubated in primary antibody [Mcl-1 S19, Bim, Bcl-2, Bcl-xL (Santa Cruz 1:500); β-Actin, E-Cadherin (Cell Signaling, 1:10,000)], secondary antibody [Rabbit, Goat, Mouse (Santa Cruz, 1:5,000-10,000)], and developed with ECL substrate (Thermo Scientific).
Proximity Ligation Assay
Cells cultured in 96-well plates were fixed with methanol, stained with the Duolink (Sigma) PLA protocol according to manufacturer's directions using Mcl-1 (Santa Cruz, 1:25) and Bim (Santa Cruz, 1:25) antibodies, counterstained with Hoescht and scanned by ImageXpress Micro XL Automated Microscope. PLA fluorescent puncta and Hoescht-stained cells were enumerated using ImageJ software.
Caspase Activity Assay
5,000 cells/well or 10,000 cells/well were seeded in 96-well plates in Growth Media and were treated with ABT-263 or DMSO for 4-48 hours. Caspase-Glo 3/7 Assay (Promega) was used according to manufacturer's directions. Luminescence was measured on a Glomax Mutli+ Detection System (Promega) luminometer and was standardized to protein values.
Cell Culture
Cell lines were purchased directly from American Tissue Type Collection (Homo sapiens ATCC CRL 2327; HTB-22; HTB-133; CRL-1500), and cultured in Growth Media (DMEM, 10% fetal bovine serum, 1× antibiotics/anti-mycotics). Cells were transduced with lentiviral particles expressing three distinct shControl or shMCL1 sequences (Santa Cruz Biotechnologies) and kept under constant Puromycin selection (1μg/mL, Life Technologies). For cell growth analyses, 2,500 cells/well [growth 3D Matrigel (BD Bioscience)] or 5,000 cells/well [growth monolayer] were seeded in a 96-well or 12-well plate, respectively. Media, antibiotic and/or drug were changed every 3 days. For analysis, 3D colonies were imaged after 14d (Motic AE3, ProRes CapturePro v2.8.0) and enumerated using ImageJ software. Colonies in monolayer were stained with 0.01% w/v crystal violet (Sigma Life Sciences) and measured using ImageJ. Trypan blue-excluding cells were counted after seeding 50,000 cells/well in 12-well plates and treating with drug for 48h.
Statistical Analysis
Statistical significance (P<0.05) was determined using Student's unpaired 2-tailed T-Test or ANOVA with Bonferroni post hoc tests followed by Student's unpaired 2-tailed T-test using Graphpad Prism5 software.
Results
Mcl-1 is highly expressed in breast cancers
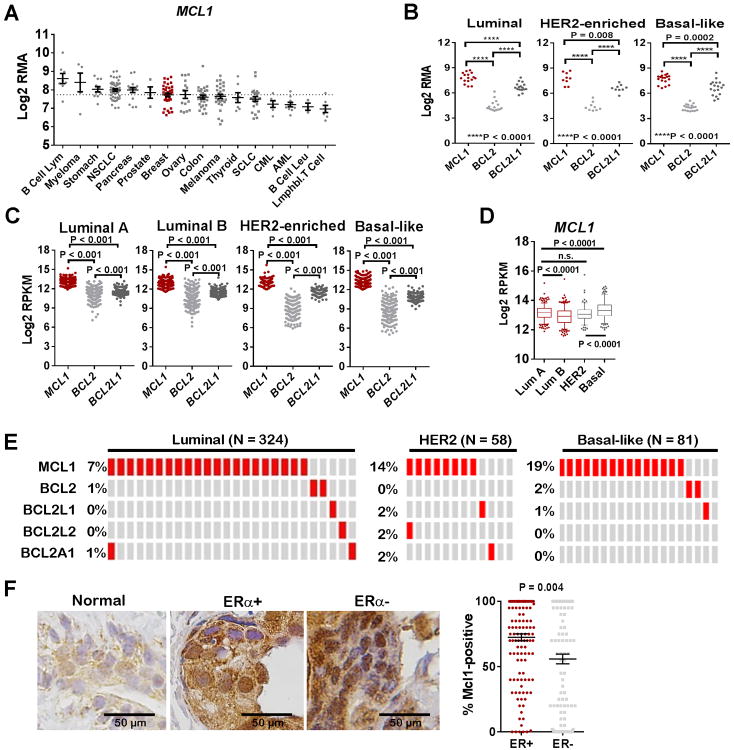
Anti-apoptotic Bcl-2 family member transcripts were assessed in Cancer Cell Line Encyclopedia (CCLE) tumor cell line expression datasets (35). BCL2 transcripts were high in tumors of hematological origin, but were relatively low in epithelial tumor cells, including breast, while BCL2L1 (Bcl-xL) transcripts were higher in tumors of epithelial origin (Supplemental Figure S1). MCL1 levels were relatively high across several cancers of epithelial (lung, breast, ovary, pancreas, prostate, and stomach) and hematological (B-cell lymphomas, myelomas) origin, and in melanomas (Figure 1A). Focusing specifically on breast cancer, we assessed MCL1, BCL2, and BCL2L1 in cell lines stratified by PAM50 molecular subtypes (36-38). MCL1 was detected at higher levels than BCL2 and BCL2L1 across all breast cancer molecular subtypes (Figure 1B). This observation was confirmed by western analysis in a smaller panel of breast cancer lines, showing abundant Mcl-1 expression across most lines, and variable levels of Bcl-2 and Bcl-xL (Supplemental Figure S2).
Figure 1. Mcl-1 expression is highest in ER+ breast cancers.
A. CCLE-curated cancer cells were grouped according to tumor type and assessed for mRNA expression of MCL1 using cBio Portal (www.cbio.org).
B. CCLE-curated breast cancer cells were grouped according to PAM50 molecular subtype and assessed for mRNA expression of MCL1, BCL2, and BCL2L1 using cBio Portal (www.cbio.org).
C-D. Clinical breast tumor specimens curated by TCGA and subjected to RNA-Seq analysis were grouped according to PAM50 molecular subtype, then assessed for absolute read for transcripts encoding MCL1, BCL2 and BCL2L1 using cBio Portal (www.cbio.org). Unpaired 2-tailed Student's T-test.
E. Clinical breast tumor specimens curated by TCGA and subjected to CGH analysis were grouped according to PAM50 molecular subtype, then assessed for amplifications of genes encoding anti-apoptotic Bcl-2 family members.
F. Breast tumor tissue microarray (N = 266) was stained for Mcl-1 and scored for percent of tumor cells that are Mcl1-positive by a breast pathologist. Samples were stratified by ER status, ER+ N =132/ER– N =134. The average % Mcl1-positive cells (±S.E.) is shown as the midline Individual samples are shown as datapoints. Student's unpaired 2-tailed T-test.
Next, we queried RNA-Seq data from The Cancer Genome Atlas (TCGA)-curated clinical breast cancer datasets for total MCL1, BCL2, and BCL2L1 transcript counts, finding more MCL1 in Luminal A (N = 333), Luminal B (N = 325), HER2-enriched (N = 150) and Basal-like (N = 211) samples than BCL2 and BCL2L1 (Figure 1C). Basal-like tumors harbored highest MCL1 transcripts, followed by Luminal A, HER2-enriched, and finally Luminal B (Figure 1D). Comparative genomic hybridization (CGH) analysis of TCGA Luminal (A and B) breast cancers (N = 324) demonstrated that MCL1 gene amplifications were found in 21/324 tumor samples (7%), which was more frequent than amplifications in BCL2 (2/324) and BCL2L1 (1/324) (Figure 1E). Gene amplifications in MCL1 also occurred in HER2-enriched and Basal-like breast cancer specimens at rates higher than what was seen for other Bcl-2 family-encoding genes. Immunohistochemical analysis of clinical breast tumor specimens (N = 266) showed little Mcl-1 staining in normal breast epithelium, but substantial Mcl-1 upregulation in breast tumor specimens, with the highest expression seen in ERα+ tumors (Figure 1F). These results suggest a role for Mcl-1 in breast cancer biology, motivating our continued investigation of Mcl-1 in ERα+ breast cancers.
Mcl-1 inhibition decreases luminal breast cancer tumor cell growth
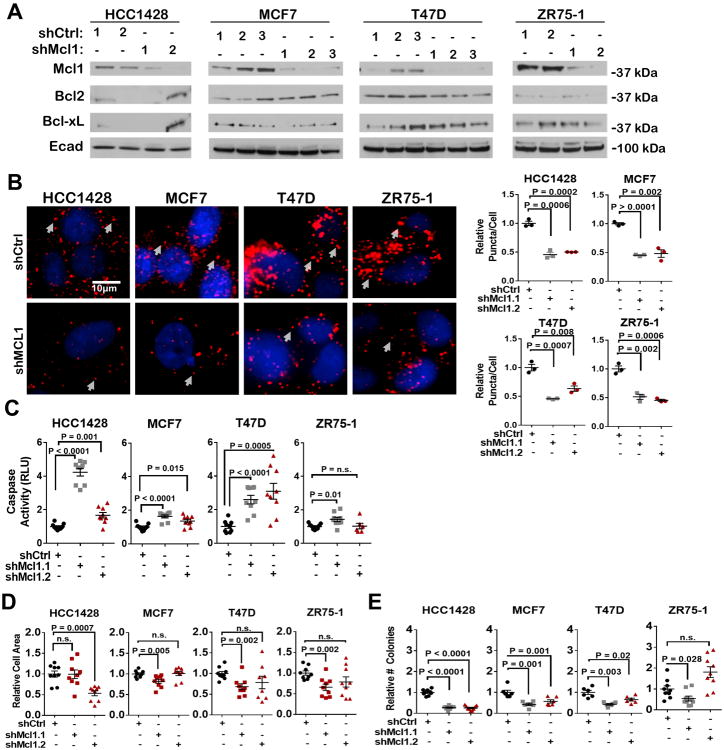
We used MCL1 shRNA sequences (shMCL1) (2-3 sequences per cell line) in the luminal breast cancer cell lines HCC1428, MCF7, T47D and ZR75-1 to knock down Mcl-1 expression. Western analysis of polyclonal puromycin-selected cells demonstrated decreased Mcl-1 protein expression in each cell line, with no change in Bcl-2 and Bcl-xL expression (Figure 2A). Proximity ligation assay [PLA,(39)] showed a decreased association between Mcl-1 and Bim in cells expressing shMCL1 as compared to shControl non-targeting sequences (Figure 2B). Mcl-1 knockdown increased caspase-3/7 activity in three of four cells lines (Figure 2C), and decreased cell growth in three of four lines grown in monolayer (Figure 2D) or in 3D Matrigel (Figure 2E). These results confirm that specific Mcl-1 inhibition increases cell death in some, but not all ERα+ breast cancer cells.
Figure 2. Decreased activity and expression of Mcl-1 induces tumor cell killing.
A-E. Polyclonal pools of cells stably expressing shCtrl (shControl) or shMcl1 sequences were assessed.
A. Western analysis of whole cell lysates.
B. Cells were assessed by PLA to detect Mcl-1/Bim interactions. Right: Representative images: red = Mcl-1/Bim proximity, blue = Hoescht. Left: Average cytosolic puncta/cell (±S.E.) was quantitated for 3 cells/sample, N = 3 (assessed in triplicate), Student's unpaired 2-tailed T-test.
C. Cells were assessed for caspase activity by Caspase-3/7 Glo assay. Average RLU (±S.E.) is shown, N=6-9, Student's un paired 2-tailed T-test.
D. Cells grown for 7d in monolayer. Average cell area (±S.E.) was determined using crystal violet, setting the number of shCtrl colonies equal to 1 as a common reference of comparison for each cell line (N = 6-9 per group). Student's unpaired 2-tailed T-test.
E. Cells embedded in Matrigel were cultured for 14d. Number of colonies/well was measured, setting the number of shCtrl colonies equal to 1 as a common reference of comparison for each cell line (N = 6-9 per group). Student's unpaired 2-tailed T-test.
ERα+ breast cancer cell lines have limited sensitivity to ABT-263
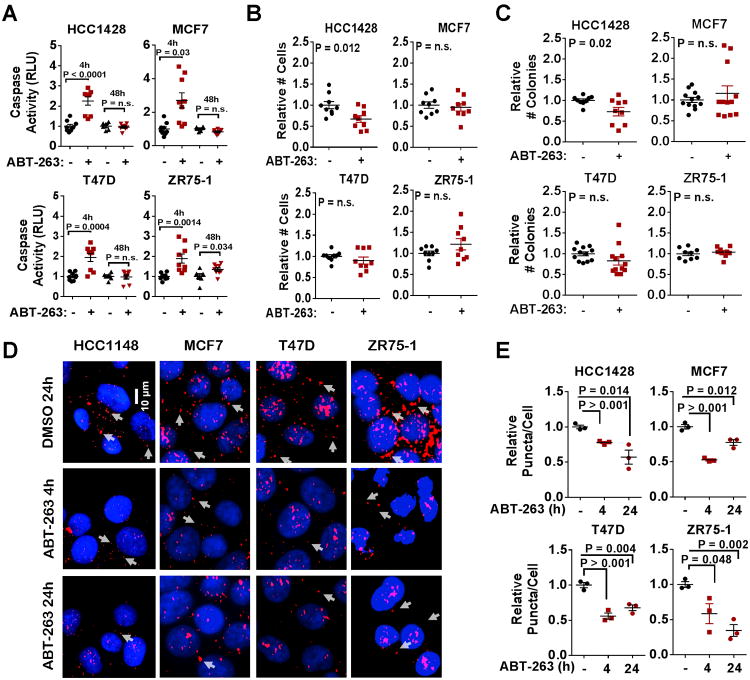
To determine how blockade of other Bcl-2 family members impacts the growth and survival of ERα+ breast cancer cells, we treated HCC1428, MCF7, T47D, and ZR75-1 with ABT-263, a compound that inhibits three Bcl-2 family members, Bcl-2 and Bcl-xL. Although each cell line showed increased caspase-3/7 activity at 4 hours exposure to ABT-263 (1 μM), caspase activation was not sustained through 48 hours in three of the four cell lines (Figure 3A), suggesting a rapid loss of sensitivity to ABT-263. Consistent with these findings, ABT-263 did not affect the total number of MCF7, T47D, or ZR75-1 cells grown in monolayer for 48 hours (Figure 3B), or in three-dimensional (3D) Matrigel for 14 days (Figure 3C and Supplemental Figure S3). PLA (39) confirmed that ABT-263 disrupted interaction of its target protein Bcl-2 with Bim (Figure 3D-3E), confirming on-target activity of ABT-263 at 4 and 24 hours treatment, despite the lack of caspase activation at distal time points in three of the four cells tested.
Figure 3. Luminal breast cancer cell lines have limited sensitivity to ABT-263.
A. Caspase activity was measured in cells treated for 4h or 48h with 1.0μM ABT-263 using a luminescent Cleaved Caspase-3/7-Glo assay. Average (±S.E.) relative luminescence is shown, Student's unpaired 2-tailed T-test, N=3.
B. Cells were treated 48h with 1.0μM ABT-263, and live cells (trypan blue-negative) were counted. Values shown are average cell number (±S.E.), Student's unpaired 2-tailed T-test, N=3.
C. Cells were Matrigel embedded and cultured 14d. Average number of colonies/well (±S.E.) is shown. T-test, N=3.
D-E. PLA of cells treated 4h or 24h ±1.0μM ABT-263. Representative images are shown. red = Bcl-2/Bim proximity, blue = Hoechst (D). Values shown are average cytosolic puncta/nuclei (±S.E.), N=3 (E).
ABT-263 induces Mcl-1 expression and activity
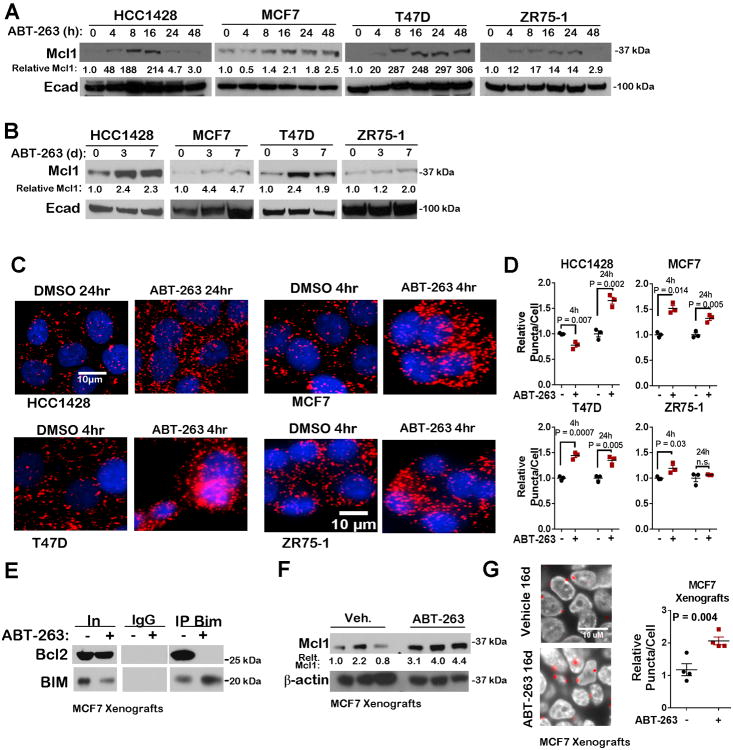
Cells were treated with a time course of ABT-263 to determine whether rapid upregulation of Bcl-2 family proteins could desensitize cells to ABT-263. Although Bcl-xL expression remained relatively unchanged after 24 hours treatment with ABT-263 (Supplemental Figure S4A), Bcl-2 expression was elevated at this time point. Despite an increase in Bcl-2 levels, Bcl-2/Bim interactions remain inhibited at 24 hours treatment (Figure 3D-E), suggesting that elevated Bcl-2 expression does not enhance the ability for Bcl-2 to sequester Bim in the presence of ABT-263. Alternatively, Mcl-1 expression was promptly increased within 4-8 hours treatment with ABT-263 (Figure 4A). This rapid upregulation was also sustained, as Mcl-1 levels were increased at 7 days treatment with ABT-263 in three of four cell lines (Figure 4A-4B). Increased Mcl-1/Bim interactions were observed in each cell line upon treatment with ABT-263 (Figure 4C-4D). While Mcl-1/Bim interaction increased in three of four cell lines at 4 hours treatment, these interactions decreased or returned to baseline at 24h in MCF7 and ZR75-1 cells, possibly due to decreased Bim expression at this time point (c S4B). Additionally, the delayed upregulation of Mcl-1/Bim interactions in HCC1428 cells (24h) relative to the other three cell lines (4h) may explain the relatively increased sensitivity of HCC1428 cells to ABT-263 (as shown in Figure 3), suggesting that complex interactions between Bcl-2 family proteins may determine sensitivity to ABT-263.
Figure 4. Increased Mcl-1 expression and activity in luminal breast cancer cells treated with ABT-263.
A-B. Western analysis of whole cell lysates after treatment with 1.0μM ABT-263 or DMSO for 0-48h (A) or 0, 3, or 7d (B).
C-D. PLA of cells treated 4 or 24h ±1.0μM ABT-263. Representative images are shown. red = Bim/Mcl-1 proximity, blue = Hoechst (C). Average cytosolic puncta/cell (±S.E.) was quantitated for 3 cells/sample, N = 3 (assessed in triplicate), Student's unpaired 2-tailed T-test (D).
- E. Immunoprecipitation (IP) of Bim or IgG control was conducted on whole tumor lysates.
- F. Whole tumor lysates were assessed by western analysis.
- G. PLA of MCF7 xenografts treated daily with ABT-263 for 16d. Average puncta/cell was quantitated for three fields/tumor, N = 4.
We assessed MCF7 xenografts treated in vivo with ABT-263 (20 mg/kg) for 16 days. Co-precipitation of Bim with Bcl-2 was seen in control-treated tumors, but was absent in MCF7 xenografts treated with ABT-263 (Figure 4E), confirming on-target activity of ABT-263 within tumors. Increased Mcl-1 was seen in ABT-263-treated MCF7 whole tumor lysates (Figure 4F), and PLA measured increased Mcl-1/Bim interactions in tumors treated with ABT-263 as compared to vehicle-treated tumors (Figure 4G). Although decreased recovery of signal was achieved in FFPE as compared to what was seen in methanol fixed MCF7 cell cultures (comparing Figure 4C to Figure 4G), these results are consistent with the idea that tumors in vivo upregulate Mcl-1 activity in response to blockade of other Bcl-2 family members similar to what was seen in cell culture.
MCL1 expression correlates inversely with sensitivity to Bcl-2/Bcl-xL inhibition
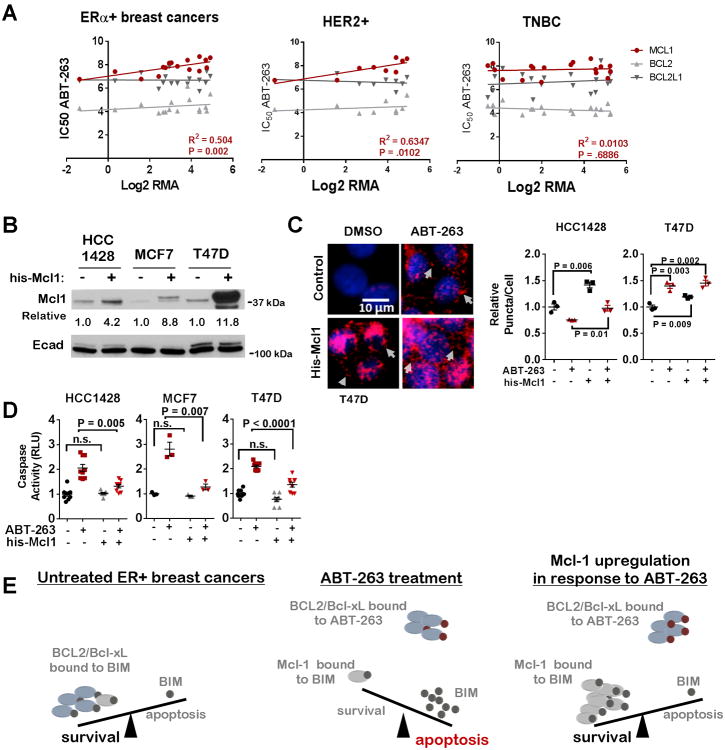
Bcl-2 and/or Bcl-xL inhibition is currently under clinical investigation for treatment of breast cancers. Given the high Mcl-1 expression levels in breast cancers, and the capacity for rapid Mcl-1 upregulation following Bcl-2/Bcl-xL using ABT-263, we assessed the relationship between MCL1 and sensitivity to ABT-263 across a panel of CCLE-curated ERα+ (N = 16), HER2-amplified (N = 9) and triple negative (N = 18) breast cancer cell lines [using datasets published in (35) and (40)]. MCL1 transcript levels correlated with the ABT-263 IC50 in ERα+ and HER2-amplified cells, but not in triple negative breast cancer (TNBC) cells (Figure 5A). Interestingly, BCL2 and BCL2L1 transcript levels did not correlate directly or inversely with the ABT-263 IC50 in any cell type. These findings support the notion that Mcl-1 may be for a marker of de novo resistance of breast cancers to ABT-263. To test this idea directly, we overexpressed Mcl-1 in HCC1428, MCF7, and T47D cells (Figure 5B). Although Bim interactions with Mcl-1 were upregulated by 20-45% in cells transduced with the his-Mcl-1 adenovirus, we found ABT-263 more potently upregulated Mcl-1/Bim interactions in transduced cells (Figure 5C), demonstrating that Mcl-1 acts as a sink for Bim upon ABT-263 treatment and that ABT-263 promotes maximal Mcl-1/Bim interactions. Consistent with the idea that Mcl-1 levels contribute to de novo ABT-263 resistance, we found that Mcl-1 overexpression substantially decreased caspase-3/7 activation in ABT-263-treated cells (Figure 5D), further suggesting that ERα+ breast cancer cells use upregulation of Mcl-1 to evade cell death when challenged with ABT-263 (Figure 5E).
Figure 5. Mcl-1 expression correlates with reduced sensitivity to ABT-263.
A. MCL1, BCL2, and BCL2L1 (Bcl-xL) mRNA expression in luminal breast cancer cell lines (CCLE) was compared to the ABT-263 IC50 (Sanger Institute). Data was fit to a linear regression. R2 and P-values shown for MCL1. R2 and P-values for BCL2 are 0.05, 0.37 (ERα+), 0.05, 0.55 (HER2), and 0.07, 0.28 (TNBC), respectively; and for BCL2L1 are 0.00, 0.97 (ERα+), 0.03, 0.52 (HER2), and 0.05, 0.58 (TNBC), respectively.
- B. Whole cell lysates after 48h treatment with adenovirus.
- C. PLA in HCC1428 and T47D cells treated with an his-Mcl1 adenovirus for 48h, then treated with ABT-263 (1.0μM) for 4h N=3 (assessed in triplicate), Student's unpaired 2-tailed T-test.
- D. Relative caspase activity of cells treated ABT-263 (1.0μM) for 4h. Average (±S.E.) relative luminescence is shown, N=3, Bonfer roni post hoc test followed by Student's unpaired 2-tailed T-test.
E. Model for Mcl-1-mediated resistance to ABT-263.
Discussion
Nearly 40,000 breast cancer deaths occur annually in the United States, often in the context of recurrent and/or therapeutically resistant disease (15). Additional molecularly targeted treatment strategies that effectively induce tumor cell killing could potentially decrease tumor recurrences, and would increase therapeutic options for patients with resistant disease. Tumors often rely on anti-apoptotic Bcl-2 family proteins to evade tumor cell death. Given the clinical success of ABT-199 in CLL (27), and ongoing clinical investigations of ABT-263 and ABT-199 in breast cancers (28), we were motivated to understand which breast cancers express high levels of Bcl-2 family members, as this might indicate which breast cancers would benefit from Bcl-2 and/or Bcl-xL inhibition. We also investigated the Bcl-2 family member Mcl-1, given recently developed compounds that target Mcl-1 activity that have transitioned into early phase clinical trials for hematological malignancies (41). Data shown herein suggest that Mcl-1 may be a key survival factor across all breast cancer subtypes.
In previous studies, a dependency screening approach identified Mcl-1 as a key survival factor in TNBCs (42), consistent with our observations presented herein that among Bcl-2 family members, MCL1 mRNA expression was frequently higher than BCL2 and BCL2L1 mRNA in breast cancers, including TNBCs (Figure 1C). At the protein level, immunohistochemical Mcl-1 staining of breast cancer tissue microarrays demonstrated profoundly elevated Mcl-1 in breast cancers as compared to normal breast epithelium (Figure 1F). Further, Mcl-1 levels were elevated in breast tissue containing ERα expression, suggesting that Mcl-1 may confer a selective survival advantage to breast cancer cells, particularly luminal breast cancer cells, but may also represent a vulnerability that can be exploited therapeutically. Mcl-1 knockdown in luminal breast cancer cells decreased Mcl-1 activity, as measured in situ by Mcl-1/Bim interactions (Figure 2B). Importantly, Mcl-1 knockdown decreased tumor cell survival and tumor cell growth (Figure 2C-E). These studies complement earlier studies from other groups indicating that Mcl-1 knockdown in some triple negative (20, 43) and/or HER2-amplified breast cancer cells (44, 45) increases caspase activity. In contrast, combined Bcl-2 and Bcl-xL inhibition using ABT-263 had only an acute impact on caspase activation (Figure 3A) and did not affect tumor cell growth (Figure 3B&C). These findings are in agreement with studies from other groups demonstrating that ABT-263 was insufficient as a single agent to induce tumor cell killing in luminal breast cancer cells, but only once cells were primed with tamoxifen did Bcl-2/Bcl-xL or selective Bcl-2 inhibition with ABT-199 induce cell death (33).
Several studies show Mcl-1 production and stability is responsive to cellular cues, including inhibition of other Bcl-2 family members (46-50), making Mcl-1 ideally suited for rapidly evading therapeutically-induced tumor cell death [reviewed in (2)], and emphasizing the value of Mcl-1 as a therapeutic target. Interestingly, we found that rapid and sustained Mcl-1 induction occurred in response to ABT-263 in luminal breast cancer cells (Figure 4), although Mcl-1 depletion did not result uniformly in Bcl-2 or Bcl-xL upregulation in luminal breast cancer cells (Figure 2A). This suggests that Mcl-1 may be a dominant anti-apoptotic signal in luminal breast cancers. Previous studies in hematological malignancies, colorectal cancers, and small cell lung cancers show that Mcl-1 drives innate resistance to ABT-263, while Mcl-1 suppression could restore sensitivity to ABT-263 (46, 50, 51). Although this idea needs to be tested further in luminal breast cancers, it is possible that maximal tumor cell killing may only be achieved when Bcl-2, Bcl-xL and Mcl-1 are inhibited, suggesting that the toxicities associated with targeting all three family members need to be explored. Additionally, we show herein that MCL1 gene expression levels correlated inversely with sensitivity to ABT-263 (Figure 5A), suggesting that MCL1 might be used as predictor of patient response to ABT-263 or ABT-199, a hypothesis that could be tested in ongoing and future clinical trials.
In summary, we find that MCL1 gene expression and amplification are frequent occurrences in breast cancers, and that genetic Mcl-1 inhibition increased apoptosis in luminal breast cancer cells, resulting in increased growth inhibition. In contrast, Bcl-2/Bcl-xL inhibition using ABT-263 did not sustain tumor cell killing or growth inhibition. Cells rapidly responded to ABT-263 by increasing Mcl-1 expression, but Mcl-1 depletion did not induce Bcl-2 or Bcl-xL upregulation. Together, these findings support a role for Mcl-1 in survival of breast cancer cells, warranting consideration of Mcl-1 in the design and interpretation of clinical trials investigating Bcl-2 family inhibitors.
Supplementary Material
Implications.
Mcl-1 levels predict breast cancer response to inhibitors targeting other Bcl-2 family members, and demonstrate the key role played by Mcl-1 in resistance to this drug class.
Acknowledgments
R.S. Cook received NIH award R01CA143126 and Susan G. Komen for the Cure grant KG100677. M. Morrison-Joly received NRSA F31 predoctoral award CA186329-01. M. Williams received NRSA F31 predoctoral award CA195989-01 and an award from the Vanderbilt Institute for Clinical and Translational Research [CTSA UL1TR000445 from the National Center for Advancing Translational Sciences]. We thank Dr. Stephen Fesik for critical review of this manuscript. We are grateful to Dr. Carlos Arteaga for thoughtful discussion of this manuscript. We also acknowledge Joshua Bauer from the Vanderbilt High Throughput Screening Facility for his assistance with imaging and analysis of the Proximity Ligation Assay.
Funding. This work was supported by Specialized Program of Research Excellence (SPORE) grant NIH P50 CA098131 (VICC), Cancer Center Support grant NIH P30 CA68485 (VICC), NIH F31 CA195989-01 (MMW) and CTSA UL1TR000445 from the National Center for Advancing Translational Sciences.
Footnotes
The authors declare no conflicts of interest with the execution or presentation of this research. This manuscript and all information herein is not under consideration for publication elsewhere.
References
- 1.Williams MaCR. Bcl-2 family proteins in breast development and cancer: could Mcl-1 targeting overcome therapeutic resistance? Oncotarget. 2015;6:3519–3530. doi: 10.18632/oncotarget.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merino D, Lok SW, Visvader JE, Lindeman GJ. Targeting BCL-2 to enhance vulnerability to therapy in estrogen receptor-positive breast cancer. Oncogene. 2016;35:1877–1887. doi: 10.1038/onc.2015.287. [DOI] [PubMed] [Google Scholar]
- 3.Sarosiek KA, Letai A. Directly targeting the mitochondrial pathway of apoptosis for cancer therapy with BH3 mimetics: recent successes, current challenges and future promise. FEBS J. 2016 doi: 10.1111/febs.13714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tait SW, Green DR. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol. 2010;11:621–632. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- 5.Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 6.Chipuk JE, Bouchier-Hayes L, Green DR. Mitochondrial outer membrane permeabilization during apoptosis: the innocent bystander scenario. Cell Death Differ. 2006;13:1396–1402. doi: 10.1038/sj.cdd.4401963. [DOI] [PubMed] [Google Scholar]
- 7.Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, Adams JM, Huang DCS. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-x(L), but not Bcl-2, until displaced by BH3-only proteins. Genes & Development. 2005;19:1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng EH, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T, Korsmeyer SJ. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol Cell. 2001;8:705–711. doi: 10.1016/s1097-2765(01)00320-3. [DOI] [PubMed] [Google Scholar]
- 9.Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, Taniguchi T, Tanaka N. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 10.She QB, Solit DB, Ye Q, O'Reilly KE, Lobo J, Rosen N. The BAD protein integrates survival signaling by EGFR/MAPK and PI3K/Akt kinase pathways in PTEN-deficient tumor cells. Cancer Cell. 2005;8:287–297. doi: 10.1016/j.ccr.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ploner C, Kofler R, Villunger A. Noxa: at the tip of the balance between life and death. Oncogene. 2008;27(1):S84–92. doi: 10.1038/onc.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craig RW. MCL1 provides a window on the role of the BCL2 family in cell proliferation, differentiation and tumorigenesis. Leukemia. 2002;16:444–454. doi: 10.1038/sj.leu.2402416. [DOI] [PubMed] [Google Scholar]
- 13.Perciavalle RM, Opferman JT. Delving deeper: MCL-1's contributions to normal and cancer biology. Trends in Cell Biology. 2013;23:22–29. doi: 10.1016/j.tcb.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cory S, Huang DC, Adams JM. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene. 2003;22:8590–8607. doi: 10.1038/sj.onc.1207102. [DOI] [PubMed] [Google Scholar]
- 15.A.C. Society, editor. American Cancer Society. Atlanta: 2016. Cancer Facts & Figures 2016. [Google Scholar]
- 16.Gee JM, Robertson JF, Ellis IO, Willsher P, McClelland RA, Hoyle HB, Kyme SR, Finlay P, Blamey RW, Nicholson RI. Immunocytochemical localization of BCL-2 protein in human breast cancers and its relationship to a series of prognostic markers and response to endocrine therapy. Int J Cancer. 1994;59:619–628. doi: 10.1002/ijc.2910590508. [DOI] [PubMed] [Google Scholar]
- 17.Keitel U, Scheel A, Thomale J, Halpape R, Kaulfuss S, Scheel C, Dobbelstein M. Bcl-xL mediates therapeutic resistance of a mesenchymal breast cancer cell subpopulation. Oncotarget. 2014;5:11778–11791. doi: 10.18632/oncotarget.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams CC, Basu A, El-Gharbawy A, Carrier LM, Smith CL, Rowan BG. Identification of four novel phosphorylation sites in estrogen receptor alpha: impact on receptor-dependent gene expression and phosphorylation by protein kinase CK2. BMC Biochem. 2009;10:36. doi: 10.1186/1471-2091-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thrane S, Pedersen AM, Thomsen MB, Kirkegaard T, Rasmussen BB, Duun-Henriksen AK, Laenkholm AV, Bak M, Lykkesfeldt AE, Yde CW. A kinase inhibitor screen identifies Mcl-1 and Aurora kinase A as novel treatment targets in antiestrogen-resistant breast cancer cells. Oncogene. 2014 doi: 10.1038/onc.2014.351. [DOI] [PubMed] [Google Scholar]
- 20.Balko JM, Giltnane JM, Wang K, Schwarz LJ, Young CD, Cook RS, Owens P, Sanders ME, Kuba MG, Sanchez V, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov. 2014;4:232–245. doi: 10.1158/2159-8290.CD-13-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Real PJ, Sierra A, De Juan A, Segovia JC, Lopez-Vega JM, Fernandez-Luna JL. Resistance to chemotherapy via Stat3-dependent overexpression of Bcl-2 in metastatic breast cancer cells. Oncogene. 2002;21:7611–7618. doi: 10.1038/sj.onc.1206004. [DOI] [PubMed] [Google Scholar]
- 22.Teixeira C, Reed JC, Pratt MA. Estrogen promotes chemotherapeutic drug resistance by a mechanism involving Bcl-2 proto-oncogene expression in human breast cancer cells. Cancer Research. 1995;55:3902–3907. [PubMed] [Google Scholar]
- 23.Crawford A, Nahata R. Targeting Bcl-2 in Herceptin-Resistant Breast Cancer Cell Lines. Current Pharmacogenomics. 2011;9:184–190. doi: 10.2174/187569211796957584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9:631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 25.Zhou H, Zhang Y, Fu Y, Chan L, Lee AS. Novel mechanism of anti-apoptotic function of 78-kDa glucose-regulated protein (GRP78): endocrine resistance factor in breast cancer, through release of B-cell lymphoma 2 (BCL-2) from BCL-2-interacting killer (BIK) Journal of Biological Chemistry. 2011;286:25687–25696. doi: 10.1074/jbc.M110.212944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 27.Cang S, Iragavarapu C, Savooji J, Song Y, Liu D. ABT-199 (venetoclax) and BCL-2 inhibitors in clinical development. J Hematol Oncol. 2015;8:129. doi: 10.1186/s13045-015-0224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindeman GS, K, Lok Wen S. A Phase 1b Study of Bcl-2 inhibition with ABT-199 in combination with Tamoxifen in Metastatic ER-Positive Breast Cancer Peter MacCallum Cancer Centre. Royal Melbourne Hospital: Austrialian New Zealand Clinical Trials Registry [Google Scholar]
- 29.Roberts AW, Advani RH, Kahl BS, Persky D, Sweetenham JW, Carney DA, Yang J, Busman TB, Enschede SH, Humerickhouse RA, et al. Phase 1 study of the safety, pharmacokinetics, and antitumour activity of the BCL2 inhibitor navitoclax in combination with rituximab in patients with relapsed or refractory CD20+ lymphoid malignancies. Br J Haematol. 2015;170:669–678. doi: 10.1111/bjh.13487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts AW, Seymour JF, Brown JR, Wierda WG, Kipps TJ, Khaw SL, Carney DA, He SZ, Huang DC, Xiong H, et al. Substantial susceptibility of chronic lymphocytic leukemia to BCL2 inhibition: results of a phase I study of navitoclax in patients with relapsed or refractory disease. J Clin Oncol. 2012;30:488–496. doi: 10.1200/JCO.2011.34.7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson WH, O'Connor OA, Czuczman MS, LaCasce AS, Gerecitano JF, Leonard JP, Tulpule A, Dunleavy K, Xiong H, Chiu YL, et al. Navitoclax, a targeted high-affinity inhibitor of BCL-2, in lymphoid malignancies: a phase 1 dose-escalation study of safety, pharmacokinetics, pharmacodynamics, and antitumour activity. Lancet Oncol. 2010;11:1149–1159. doi: 10.1016/S1470-2045(10)70261-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oakes SR, Vaillant F, Lim E, Lee L, Breslin K, Feleppa F, Deb S, Ritchie ME, Takano E, Ward T, Fox SB, Generali D, Smyth GK, Strasser A, Huang DC, Visvader JE, Lindeman GJ. Sensitization of BCL-2-expressing breast tumors to chemotherapy by the BH3 mimetic ABT-737. Proc Natl Acad Sci U S A. 2012 Feb 21;109(8):2766–71. doi: 10.1073/pnas.1104778108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaillant F, Merino D, Lee L, Breslin K, Pal B, Ritchie ME, Smyth GK, Christie M, Phillipson LJ, Burns CJ, et al. Targeting BCL-2 with the BH3 mimetic ABT-199 in estrogen receptor-positive breast cancer. Cancer Cell. 2013;24:120–129. doi: 10.1016/j.ccr.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 34.Chen J, Jin S, Abraham V, Huang X, Liu B, Mitten MJ, Nimmer P, Lin X, Smith M, Shen Y, et al. The Bcl-2/Bcl-X(L)/Bcl-w inhibitor, navitoclax, enhances the activity of chemotherapeutic agents in vitro and in vivo. Mol Cancer Ther. 2011;10:2340–2349. doi: 10.1158/1535-7163.MCT-11-0415. [DOI] [PubMed] [Google Scholar]
- 35.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gullberg M, Anderson AC. Visualization and quantification of protein-protein interactions in cells and tissues. Nature Methods. 2010;7:5–6. [Google Scholar]
- 40.Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, Bindal N, Beare D, Smith JA, Thompson IR, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.ClinicalTrials.gov, editor. AMG 176 First in HUman Trial in Subjects with Relapsed or Refractory Multiple Myeloma. 2016. [Google Scholar]
- 42.Petrocca F, Altschuler G, Tan SM, Mendillo ML, Yan H, Jerry DJ, Kung AL, Hide W, Ince TA, Lieberman J. A genome-wide siRNA screen identifies proteasome addiction as a vulnerability of basal-like triple-negative breast cancer cells. Cancer Cell. 2013;24:182–196. doi: 10.1016/j.ccr.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goodwin CM, Rossanese OW, Olejniczak ET, Fesik SW. Myeloid cell leukemia-1 is an important apoptotic survival factor in triple-negative breast cancer. Cell Death Differ. 2015;22:2098–2106. doi: 10.1038/cdd.2015.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bashari MH, Fan F, Vallet S, Sattler M, Arn M, Luckner-Minden C, Schulze-Bergkamen H, Zornig I, Marme F, Schneeweiss A, et al. Mcl-1 confers protection of Her2-positive breast cancer cells to hypoxia: therapeutic implications. Breast Cancer Res. 2016;18:26. doi: 10.1186/s13058-016-0686-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao Y, Nimmer P, Sheppard GS, Bruncko M, Hessler P, Lu X, Roberts-Rapp L, Pappano WN, Elmore SW, Souers AJ, et al. MCL-1 Is a Key Determinant of Breast Cancer Cell Survival: Validation of MCL-1 Dependency Utilizing a Highly Selective Small Molecule Inhibitor. Mol Cancer Ther. 2015;14:1837–1847. doi: 10.1158/1535-7163.MCT-14-0928. [DOI] [PubMed] [Google Scholar]
- 46.Faber AC, Coffee EM, Costa C, Dastur A, Ebi H, Hata AN, Yeo AT, Edelman EJ, Song Y, Tam AT, et al. mTOR Inhibition Specifically Sensitizes Colorectal Cancers with KRAS or BRAF Mutations to BCL-2/BCL-XL Inhibition by Suppressing MCL-1. Cancer Discovery. 2014;4:42–52. doi: 10.1158/2159-8290.CD-13-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leverson JD, Zhang H, Chen J, Tahir SK, Phillips DC, Xue J, Nimmer P, Jin S, Smith M, Xiao Y, et al. Potent and selective small-molecule MCL-1 inhibitors demonstrate on-target cancer cell killing activity as single agents and in combination with ABT-263 (navitoclax) Cell Death Dis. 2015;6:e1590. doi: 10.1038/cddis.2014.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choudhary GS, Al-Harbi S, Mazumder S, Hill BT, Smith MR, Bodo J, Hsi ED, Almasan A. MCL-1 and BCL-xL-dependent resistance to the BCL-2 inhibitor ABT-199 can be overcome by preventing PI3K/AKT/mTOR activation in lymphoid malignancies. Cell Death Dis. 2015;6:e1593. doi: 10.1038/cddis.2014.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faber AC, Farago AF, Costa C, Dastur A, Gomez-Caraballo M, Robbins R, Wagner BL, Rideout WM, 3rd, Jakubik CT, Ham J, et al. Assessment of ABT-263 activity across a cancer cell line collection leads to a potent combination therapy for small-cell lung cancer. Proc Natl Acad Sci U S A. 2015;112:E1288–1296. doi: 10.1073/pnas.1411848112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yecies D, Carlson NE, Deng J, Letai A. Acquired resistance to ABT-737 in lymphoma cells that up-regulate MCL-1 and BFL-1. Blood. 2010;115:3304–3313. doi: 10.1182/blood-2009-07-233304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mazumder S, Choudhary GS, Al-harbi S, Almasan A. Mcl-1 Phosphorylation Defines ABT-737 Resistance That Can Be Overcome by Increased NOXA Expression in Leukemic B cells. Cancer Research. 2012;72:3069–3079. doi: 10.1158/0008-5472.CAN-11-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.