Abstract
Purpose
The PI3K/Akt signaling axis contributes to the dysregulation of many dominant features in breast cancer including cell proliferation, survival, metabolism, motility and genomic instability. While multiple studies have demonstrated that basal-like or triple negative breast tumors have uniformly high PI3K/Akt activity, genomic alterations that mediate dysregulation of this pathway in this subset of highly aggressive breast tumors remain to be determined.
Methods
In this study, we present an integrated genomic analysis based on the use of a PI3K gene expression signature as a framework to analyze orthogonal genomic data from human breast tumors, including RNA expression, DNA copy number alterations, and protein expression. In combination with data from a genome-wide RNA-mediated interference screen in human breast cancer cell lines we identified essential genetic drivers of PI3K/Akt signaling.
Results
Our in silico analyses identified SOX4 amplification as a novel modulator of PI3K/Akt signaling in breast cancers and in vitro studies confirmed its role in regulating Akt phosphorylation.
Conclusions
Taken together, these data establish a role for SOX4 mediated PI3K/Akt signaling in breast cancer and suggest that SOX4 may represent a novel therapeutic target and/or biomarker for current PI3K-family therapies.
Keywords: breast cancer, genomics, SOX4, PI3 kinase, Akt
INTRODUCTION
Breast cancer heterogeneity can be observed clinically by widely varying therapeutic responses and at a molecular level by the myriad of genetic alterations driving tumorigenesis [1–6]. Clinically, triple negative breast cancer (TNBC), which is largely synonymous with the basal-like molecular subtype of breast cancer, accounts for 15–20% of breast tumors representing ~20,000 new cases and ~10,000 deaths each year in the United States [3, 7]. Given the lack of drug-able targets expressed by TNBC tumors, including the lack of estrogen receptor (ER) and the HER2 oncogene, few therapeutic options exist beyond currently utilized cytotoxic therapies, and the overall prognosis for these patients remains poor.
Many previous studies, including The Cancer Genome Atlas (TCGA) project, have reported increased phosphatidylinositol-3-OH kinase (PI3K) signaling in basal-like tumors. This pathway mediates, among other processes, cell cycle progression and survival, [3, 8–10] and is highly activated in this subset of tumors despite a low incidence (~7%) of PIK3CA activation mutations. While studies from the TCGA and others have noted copy number alterations or mutations in PTEN (35%), INPP4B (30%) and activation of multiple receptor tyrosine kinases, including EGFR (7%), ERBB2 (4%) and IGFR1 (2%), in addition to other mutations that may affect aberrant PI3K signaling in basal-like tumors, it remains to be determined whether additional mechanisms may contribute to pathway activity and/or the observed lack of response to PI3K family inhibitors [3, 8, 11–14].
To identify genomic alterations mediating PI3K/Akt signaling, specifically those that may represent novel therapeutic targets and/or biomarkers to current therapies, we utilized an integrative genomic strategy based on experimentally-derived gene expression signatures [6]. By analyzing orthogonal proteomic and genomic data from the TCGA in conjunction with data from a genome-wide RNAi proliferation screen in breast cancer cell lines we identified SOX4 as a putative novel regulator of PI3K/Akt signaling and in vitro studies confirmed the role of this gene in mediating Akt phosphorylation.
MATERIALS AND METHODS
Gene expression data
RNA sequencing data (n=1,031) from human tumors (Supplemental Table 1) were acquired from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/) and processed as previously described [15]. PAM50 classification as well as calculation of the PI3K [6, 16], PIK3CA [17], PTEN Deleted and PTEN Wildtype [18] signatures was performed as previously described [1, 6, 19]. Illumina HT-29 v3 expression data for the METABRIC (Molecular Taxonomy of Breast Cancer International Consortium) project (n=1,992) was acquired from the European Genome-phenome Archive at the European Bioinformatics Institute (https://www.ebi.ac.uk/ega/) and data were median centered [4]. Expression data for a panel of 51 breast cancer cell lines was acquired from GEO (GSE12777) [20]. Affymetrix U133+2 data were MAS5.0 normalized using Affymetrix Expression Console (ver1.2.1.20), and log2 transformed. Expression probes were collapsed using the median gene value with the GenePattern [21] module CollapseProbes.
Reverse Phase Protein Array (RPPA) data
RPPA data were acquired (October 24, 2013) from The Cancer Proteome Atlas data portal (http://app1.bioinformatics.mdanderson.org/tcpa/_design/basic/index.html). Replication Based Normalized RPPA data (n=733) containing expression levels for 187 proteins and phosphorylated proteins (Supplementary Table 1) were used to identify differentially expressed proteins. Samples were separated into high (top quartile) and low (all other) subgroups based on pathway score and a t-test used to assess differences in expression for each protein. Additionally, a Spearman-rank correlation was used to compare the overall correlation between pathway activity and protein expression.
Affymetrix SNP 6.0 copy number data
Affymetrix SNP 6.0-derived copy number data (Firehose run April 16, 2014) were acquired from the Firehose data portal (http://gdac.broadinstitute.org/) for the 1,031 samples (Supplementary Table 1) for which both CN and gene expression data were available. Pathway-specific CN alterations were identified as previously described [6]. Briefly, a Spearman rank correlation (both positive and negative), was used to compare gene-level segment scores with pathway activity score. Secondly, the frequency of copy number gains (including high level amplification and gains) or losses (loss of heterozygosity or deletion), as determined by GISTIC 2.0 [22], in samples with high (top quartile) and low (all others) pathway activity were calculated by a Fisher’s exact test.. To identify genes that were significant across both methods, a threshold of q<0.05 was set for validation and q<0.01 for discovery.
Genome-wide RNAi proliferation data
To identify genes required for cell viability in a pathway-dependent manner, data from a genome-wide RNAi screen in a panel of breast cancer cell lines were analyzed [6, 23]. The Gene Activity Ranking Profile (GARP) normalized data were obtained from the DPSC (Donnelly-Princess Margaret Screening Centre) data portal (http://dpsc.ccbr.utoronto.ca/cancer/index.html) and filtered to include those 27 cell lines for which gene expression data (GSE12777) was also available (acquired February 2013). A negative Spearman correlation was used to compare pathway score and GARP score for each sample to identify genes essential for pathway-dependent cell proliferation. A threshold of p<0.05 was considered significant.
Cell culture and siRNA knockdown
HCC38, HCC1143, and MDAMB468 cells were purchased from the American Tissue Culture Collection (Manassas, VA, USA). Cells were cultured in either RPMI-1640 medium with 10% fetal bovine serum and 1% penicillin/streptomycin (HCC38 and HCC1143) or DMEM/F12 medium with 10% fetal bovine serum and 1% penicillin/streptomycin (MDAMB468). Cells were sub-cultured at 60–70% confluence for siRNA transfections. Lipofectamine RNAiMAX (ThermoFisher) was used to transfect cells with 40nm of SMART pool siRNA targeting SOX4 (M011779010005) or scrambled control (D0012061305) according to manufacturer’s instructions (Dharmacon) for 48 hours prior to RNA or protein isolation.
Western blot
HCC38 cells were harvested using Triton lysis buffer containing 25mM HEPES, 100mM NaCl, 1mM EDTA, 10% glycerol and 1% TritonX-100 with 1X protease and phosphatase inhibitor added fresh prior to use. 50 μg of protein was loaded on 4–20% Mini-protean TGX gradient gel (Biorad) at 100V for 2 hours at room temperature and transferred onto nitrocellulose membrane at 100V for 1 hour at 4°C. The membranes were blocked using 5% milk solution, incubated with primary antibody against p-Akt, total Akt and beta-actin (Cell Signaling Technology) overnight at 4°C followed by incubation with HRP-conjugated secondary antibodies (Cell Signaling Technology) for 1 hour at room temperature. The signal was developed using SuperSignal West Pico Chemiluminescent Substrate (ThermoFisher Scientific), digitally imagined using the ChemiDoc Touch Imagining System (BioRad) and signaling intensity quantified by Image J software (https://imagej.nih.gov/ij/).
Immunofluorescence
Cells were grown on coverslips, fixed with 4% paraformaldehyde in PBS and permeabilized with 0.25% Triton X-100 in PBS for 8 minutes at room temperature. Cells were blocked in 1% BSA in PBS with 0.05% Tween20 for 45 minutes at room temperature, incubated with anti-SOX4 (Santa Cruz, sc-17326) and anti-pAKT (Cell Signaling, 9271S) antibodies (1 hour, room temperature) and then incubated with fluorochrome-labeled secondary antibodies for 1 hour (room temperature). The coverslips were counterstained with DAPI and imaged with a Nikon Eclipse TE-2000U fluorescent microscope.
Quantitative real time PCR
Total RNA was isolated using RNeasy plus Mini Kit (Qiagen) and cDNA was synthesized using the QuantiTect Reverse Transcription kit (Qiagen). Quantitative PCR (qPCR) was performed and analyzed using Applied Biosystems 7500 real time thermal cycler system. Human SOX4 primer sequences: Forward: 5′-CTCTCCAGCCTGGGAACTATAA-3′, Reverse: 5′-CGGAGGTGGGTAAAGAGAGAA-3′ and human GAPDH are Forward: 5′-TCTGACTTCAACAGCGACAC -3′, Reverse: 5′-CCAGCCACATACCAGGAAAT -3′.
RESULTS
PI3K gene expression signature corresponds with PI3K/Akt signaling in vivo
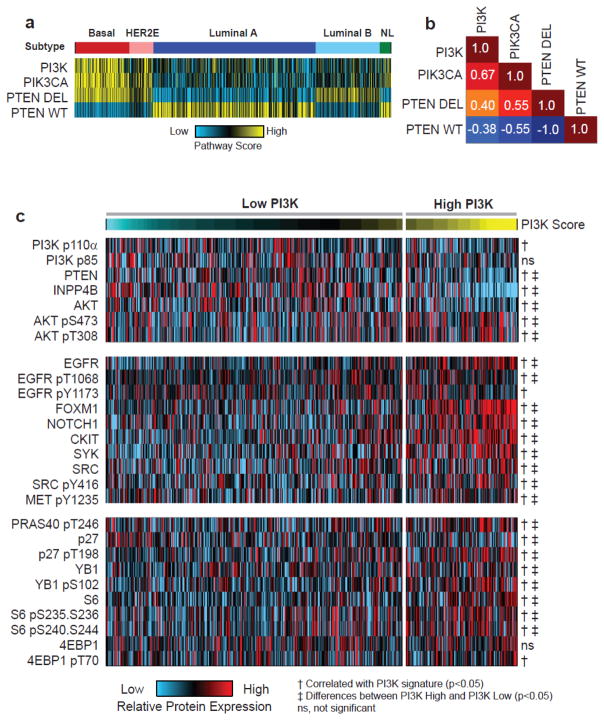
We used four previously published PI3K gene expression signatures, PI3K [6, 16], PIK3CA [17], PTEN Wild-type (PTENWT) and PTEN Deleted (PTENDEL) [18], to calculate PI3K pathway activity in 1,031 human breast tumor samples (Supplementary Table 1) from the TCGA project for which the PAM50 intrinsic subtypes [19, 24] were determined (Figure 1a). Although each signature was independently developed [6, 17, 18], consistent patterns of pathway activity were observed across molecular subtypes with basal-like tumors being characterized by uniformly high levels of PI3K activity; increased signaling was also observed in HER2-Enriched (HER2E) and Luminal B (LumB) tumors and significant differences were noted between each subtype (Supplementary Figure 1). Despite the fact that each signature was developed using different strategies, identifying differentially expressed genes when comparing either GFP expressing cell lines to mutant [17] or wild-type [6, 16] PIK3CA expressing cells or by comparing PTEN wild-type and deleted tumors [18], signature scores were moderately concordant (r=0.4 – 0.67) across the data (Figure 1b and Supplementary Figure 2).
Figure 1. PI3K gene expression signature correlates with protein expression of PI3K/Akt pathway and is up-regulated in basal-like and HER2E tumors.
(A) Patterns of PI3K signaling in breast cancer (n=1,031) correspond with molecular subtype with basal-like (red) and HER2E (pink) having the highest levels, Luminal B (light blue) and Normal-like (NL, green) having intermediate levels, and LumA (dark blue) having the lowest levels. Tumors show consistent patterns of pathway activity across independent gene expression signatures. (B) PI3K pathway gene expression signatures are strongly correlated as calculated by a Pearson correlation (C) PI3K signature (n=733) corresponds with protein and phosphorylated protein expression of PI3K/Akt signaling pathway components. A t- test (‡) was used to assess protein expression levels between the top quartile and all other samples and a Spearman rank correlation (†) was used to assess the overall relationship between pathway score and RPPA expression level.
Focusing on the PI3K signature [6, 16], we confirmed the ability of the mRNA signature to accurately assess functional activation of the pathway by examining relationships between the gene expression signature score and proteins and phospho-protein expression. Analysis of Reverse Phase Protein Array (RPPA) data from breast tumors (n=733), using, both a Spearman correlation (comparing pathway score to protein expression level) and t-test (top quartile against all other samples), confirmed that tumors with a high PI3K score have significantly lower levels of PTEN and INPP4B protein expression, consistent with role of these proteins as negative regulators of PI3K/Akt signaling. Despite lower levels of total Akt and no differences in the expression of the p110-alpha or p85 kinase subunits, tumors with high pathway score expressed significantly higher levels of phosphorylated (p) Akt at S473 and T308 (Figure 1c, Supplementary Table 2). In addition, a number of known drivers of PI3K activity, including EGFR (pY1068 and pY1173), FOXM1, NOTCH1, cKIT, SYK, SRC (pY416), and c-MET (pY1235), had significantly higher levels of protein expression in tumors with a high PI3K score. Importantly, we also observed increased levels (p<0.05) of activated Akt substrates including PRAS40 (pT246), p27 (pT198) and YB1 (pS102) as well as further down-stream targets S6 (pS235-S236 and pS240-S244) and 4EBP1 (pT70) (Figure 1c, Supplementary Table 2) [8, 10].
Identification of copy number alterations associated with PI3K signaling
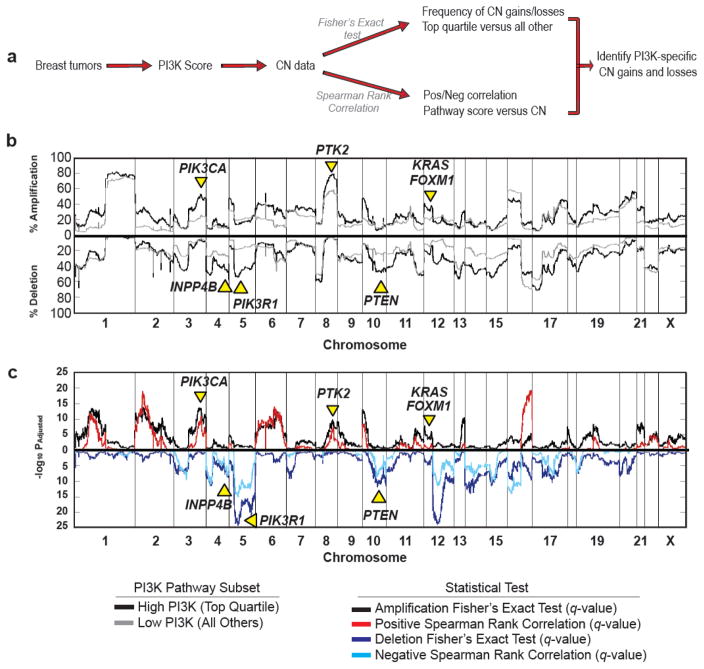
We next sought to identify PI3K-specific copy number (CN) alterations (CNA), including known and potential novel drivers of pathway activity (Figure 2a). Using our previously published strategy [6], breast tumor samples (n=1,031) from the TCGA project were dichotomized into those with high (top quartile) or low (all others) pathway score and the frequency of copy number gains (including high level amplification and gains) or losses (including homozygous deletion and loss of heterozygosity) were calculated for each group (Figure 2b) using a Fisher’s exact test (Figure 2c). Secondly, a Spearman Rank Correlation was used to assess the overall relationship between PI3K pathway score and gene-level segment value (Figure 2c). To reduce potential false-positives associated with either strategy alone, we focused on those alterations that were significant (q<0.05), in both analyses; genes with an increased incidence of CN gains in samples with high PI3K pathway activity (top quartile) and a positive correlation with pathway activity were considered putative drivers of PI3K signaling whereas those genes with an increased frequency of CN losses in tumors with a high PI3K score and a negative correlation were considered potential repressors of PI3K activity. These analyses allow for the identification of chromosomal alterations that are uniquely evident in the context of PI3K signaling while eliminating those regions that, while potentially altered at a high frequency, are not associated with altered signaling. For instance, 50.4% of patients with high PI3K activity but only 25.0% of tumors with low pathway activity have an amplification of chromosome 3q26, which contains PIK3CA (qFisher=1.5 ×10−12; qSpearman=4.0×10−10). Beyond PIK3CA amplification, we also identified CNA (q<0.05) of known drivers of PI3K signaling including amplification of KRAS, PTK2, and FOXM1 as well as losses of pathway inhibitors PTEN, INPP4B, and PIK3R1 (Figure 2b, Supplementary Table 3). Collectively these data suggest that this strategy is able to identify CNA of known PI3K signaling components and may identify CNA of novel regulators of this pathway.
Figure 2. PI3K score identifies PI3K/Akt specific copy number changes.
(A) Overview of method for identifying PI3K-specific CNA. (B) The frequency of copy number gains (including amplification and gains) and losses (both deletion and LOH) were calculated for each gene in the top quartile (black line) and all other samples (gray line); frequencies are plotted according to chromosomal position. (C) A Fisher’s exact test was used to assess differences in the frequency of copy number gains (black line) or losses (dark blue line) between the top quartile and all other samples. A positive (red line) or negative (light blue line) Spearman rank correlation was used to assess the overall relationship between pathway score and gene level copy number expression.
Identification of potential essential, novel regulators of PI3K pathway activity
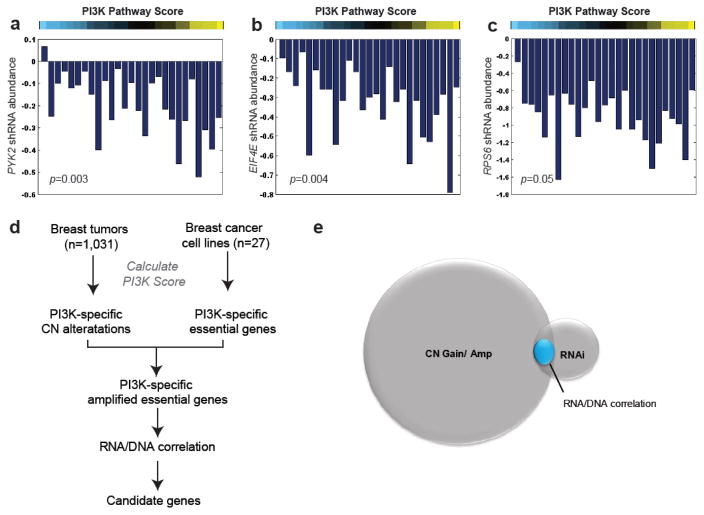
Our previous results identified DNA copy number aberrations that correlate with PI3K activity (Figure 2). By analyzing functional data from a genome-wide RNA-mediated interference (RNAi) screen, we are able to identify specific genes within each amplicon that are essential for cell viability in a pathway-dependent manner. For these analyses, we employed data from a genome-wide RNAi proliferation screen (~16k genes) performed in a panel of 27 breast cancer cell lines [23] for which matching mRNA expression data (GSE12777) was available (Figure 2). For each cell line, the PI3K pathway score was calculated and a negative Spearman rank correlation was used to identify those genes required for cell proliferation in a PI3K dependent manner. Our analyses identified known PI3K/Akt signaling components including PYK2 (Figure 3a, p=0.003) [25] as well as both EIF4E (Figure 3b, p=0.004) and RPS6 (Figure 3c, p=0.05), which are prominent down-stream targets of Akt activity thereby validating this strategy.
Figure 3. Identification of amplified and essential genes associated with PI3K signature.
Identification of genes essential for cell viability in RNAi screen of breast cancer cell lines (n=27) in the context of PI3K signature as calculated by negative Spearman correlation (p<0.05) including A) PKY2 (p=0.003), (B) EIF4E (p=0.004), and (C) RPS6 (p=0.05). The height of each bar indicates shRNA abundance (and cell viability) for each cell line grown in the presence of gene-specific shRNA relative to control shRNA. (D) Schematic outlining the strategy used to identify amplified, essential pathway-specific genes. (E) Venn diagram identifying subset of amplified (q<0.01, n= 5,350) and essential (n=590) candidate genes that have a positive (p<0.05) correlation between mRNA expression and copy number level (n=100).
Next we used these functional data to prioritize candidate genes within each amplicon associated with PI3K activity. To do so we compared the subset of genes that were amplified in a greater percentage (q<0.01) of samples with high PI3K pathway activity, had a positive correlation between PI3K signature score and CN segment score (q<0.01), and which were required for cell viability (p<0.05) in a PI3K-dependent manner (Figure 3d). This subset of genes was further restricted to those that also showed a positive correlation between DNA CN status and mRNA expression (Figure 3e) based on the reasoning that a potential regulator of pathway activity must not only be amplified but also have increased mRNA expression (Supplementary Table 4). Of the identified candidate genes, several, including NEDD9, which has been reported [26] to serve as a scaffolding protein for Akt and required for breast cancer genesis, have been reported to play a role in PI3K/Akt signaling. These data provide evidence that this strategy has the potential to identify both known and potentially novel mediators of PI3K/Akt signaling.
SOX4 is a putative driver of PI3K/AKT signaling
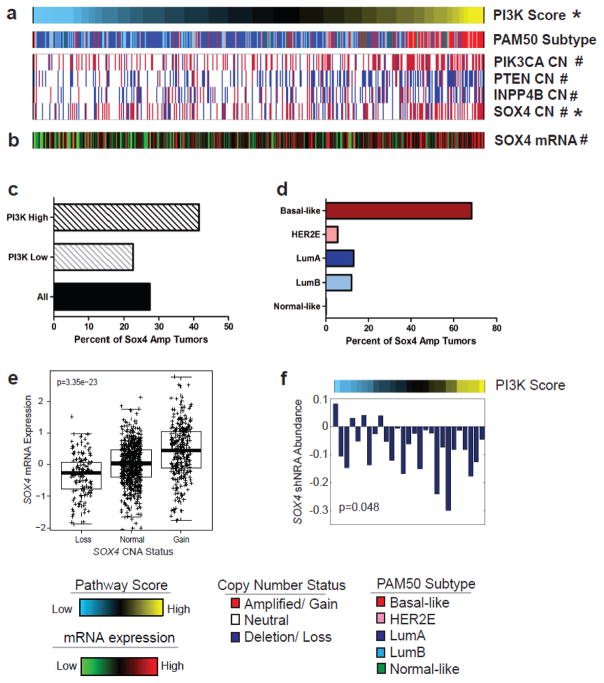
Among the candidate genes identified on chromosome 6p22 was SOX4 (sex-determining region Y-related high-mobility-group box transcription factor 4), which is a previously described oncogenic transcription factor that has been reported to correlate with a poor clinical outcome in human breast cancer and other cancers [27, 28]. We determined that SOX4 CN positively correlated with PI3K score (Figure 4a) and mRNA expression (Figure 4b). In total 41.5% of tumors with high pathway activity (top quartile) were characterized by SOX4 CN gain compared to 22.6% of all other samples (q=2.5×10−08) (Figure 4c). This appears to be largely a subtype-specific event as 68.1% of tumors with high PI3K activity and amplified SOX4 were basal-like (Figure 4d); in total, 57.2% of all basal-like tumors were characterized by SOX4 amplification (Supplementary Figure 3). Furthermore, SOX4 amplified tumors had the highest levels of SOX4 mRNA (Figure 4e, p=4.0×10−23, ANOVA; p<0.0001), and highest levels of PI3K activity (Supplementary Figure 4, p=3.7×10−08, ANOVA; p<0.0001). Analyses of the METABRIC [4] dataset (n=1,992) demonstrated a comparable significant correlation between PI3K pathway score and SOX4 expression (r=0.42, p=2.6×10−87), and SOX4 CN status (r=0.23, p=1.1×10−24), respectively, as well as between SOX4 expression and CN (r=0.27, p=1.5×10−34), thereby validating our findings in an independent cohort (Supplementary Figure 5). Importantly, our analyses of data from a genome-wide RNAi screen demonstrated that breast cancer cell lines with high PI3K activity have a greater dependency (p=0.048) on SOX4 than cell lines with low PI3K activity (Figure 4f).
Figure 4. Identification of SOX4 amplification as a putative driver of PI3K/Akt signaling.
(A) Copy number alterations of modulators of PI3K/Akt signaling PIK3CA, PTEN, INPP4B as well as SOX4 copy number status correlates (q<0.01) with PI3K score (#). (B) SOX4 mRNA expression correlates with PI3K score and SOX4 gene level copy number segment score (*). (C) SOX4 is amplified at a significantly greater frequency of samples with high (top quartile) PI3K pathway activity (41.5%) compared to all other samples (22.6%) (q=2.5×10−08). (D) Samples with an amplification of SOX4 are predominantly (68.1%) basal-like tumors. (E) SOX4 mRNA expression correlates with SOX4 copy number status (p=3.4×10−23). (F) SOX4 is essential for cell viability is breast cancer cell lines with high PI3K pathway activity. Higher expression of the PI3K Score is associated with lower SOX4 shRNA abundance. (Spearman correlation, p=0.048)
Finally, examining the coincidence of SOX4 amplification with copy number alterations of known drivers of PI3K signaling, demonstrated that the frequency of SOX4 amplification in tumors with high (or low) PI3K activity was comparable (q<0.05) to that of PIK3CA (50.4% vs. 25.0%), PTEN (46.9% vs. 25.5%) or INPP4B (37.6% vs. 24.8%) CNA (Figure 4a). These alterations, however, were neither mutually inclusive nor mutually exclusive as SOX4 was solely altered (19.1%) at a comparable frequency to PTEN (23.6%) or INPP4B (18.7%) loss, or PIK3CA gain (21.7%) alone (Supplementary Figure 6).
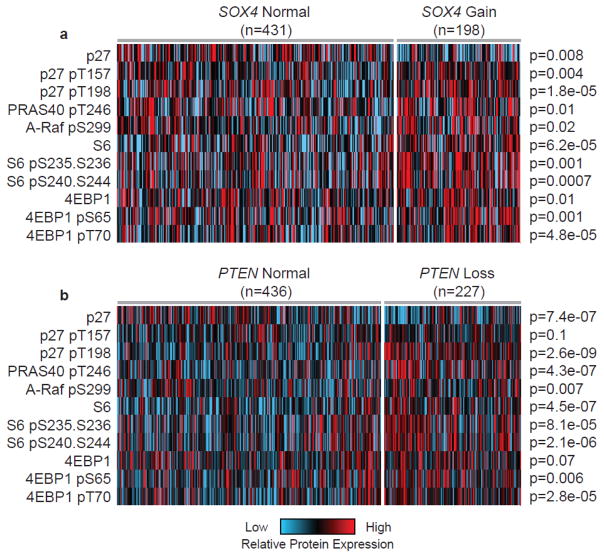
Increased PI3K/AKT signaling in breast tumor samples with amplified SOX4
We next determine whether patients with amplified SOX4 are characterized by increased expression of Akt substrate proteins relative to those tumors that have a normal copy number. Analysis of RPPA-based protein expression in breast tumors with amplified (n=198) and normal (n=431) SOX4 (Figure 5a) demonstrated up-regulation of pAkt substrates in SOX4 amplified tumors including pPRAS40 pT246 (p=0.01); p27 pT157 (p=0.004) and pT198 (p=1.8×10−05), and ARAF pS299 (p=0.02). Furthermore, we determined that down-stream targets p4EBP1 [both pS65 (p=0.001) and pT37 (p=4.8×10−05)] and pS6 at pS235-pS236 (p=0.001) and pS240-S244 (p=0.0007) were also up-regulated in SOX4 amplified tumors. Consistent with these findings, we determined that tumors characterized by loss of PTEN (n=227), relative to samples with normal PTEN (n=436), showed consistent up-regulation of this subset of Akt substrates and down-stream target proteins (Figure 5b). As such, these data support the association of SOX4 with altered PI3K/Akt signaling.
Figure 5. SOX4 amplified tumors are characterized by increased PI3K/Akt protein and phosphoprotein expression.
(A) Breast cancer samples with a SOX4 copy number gain (n=198) had increased levels of expression of active or phosphorylated Akt substrates (both direct and down-stream targets) compared to samples with normal levels of SOX4 (n=431). (B) Comparable patterns (as determined by unpaired t-test) were observed in samples with a loss of PTEN (n=227) compared to tumors with normal PTEN copy number status (n=436).
SOX4 mediates PI3K/Akt signaling in basal-like breast cancer
In order to confirm that SOX4 contributes to altered PI3K signaling, we utilized gene expression data from a panel of 51 breast cancer cell lines (GSE12777) to identify those with the highest PI3K score (Supplementary Figure 7). Of these, basal-like cell lines, HCC38 and HCC1143 both have high PI3K signature score and high SOX4 expression whereas MDAMB468 cells have high PI3K activity but low (below the dataset median) SOX4 expression. In addition to high PI3K activity, each of these cell lines are characterized by either loss of PTEN (HCC38) or EGFR activation (HCC1143) or both PTEN loss and EGFR activation (MDAMB468).
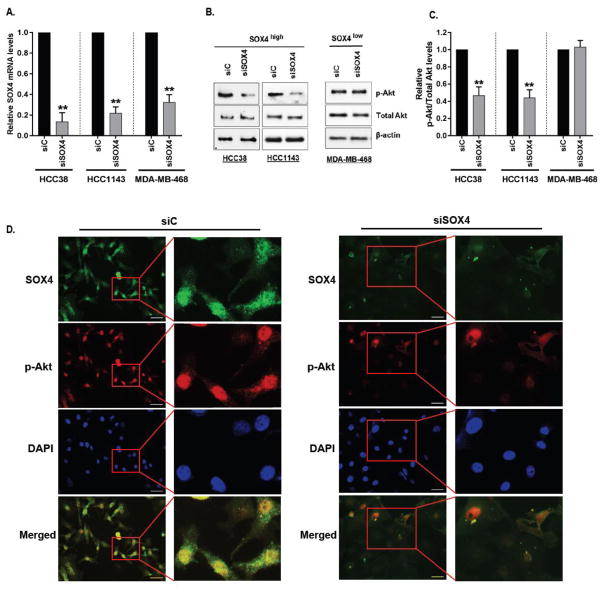
Using these cell lines, we examined the effects of siRNA-mediated silencing of SOX4 on Akt phosphorylation. Quantitative Real-time PCR (qRT-PCR) confirmed that cells transfected with pooled siRNA comprised of a panel of four individual siRNA targeting SOX4 resulted in a significant decrease in SOX4 expression in HCC38 (90%, p=0.002, paired t-test), HCC1143 (79%, p=0.003, paired t-test) and MDAMB468 (70%, p=0.0001, paired t-test) cells, respectively, relative to scrambled control (siC) transfected cells (Figure 6a). Loss of SOX4 resulted in a significant reduction in Akt phosphorylation in both HCC38 and HCC1143 while no change in pAkt levels were observed in MDAMB468 cells; total Akt or β-actin levels were unaffected (Figure 6b). Quantitative analyses of pAkt levels relative to total Akt (Figure 6c) demonstrated that loss of SOX4 resulted in a 54% to 56% reduction in pAkt levels, relative to siC treated cells, in HCC38 (p=0.0008) and HCC1143 (p=0.005), respectively while no quantifiable change was observed in MDAMB468 cells (p=0.53).
Figure 6. Sox4 mediates pAkt expression in vitro.
(A) SOX4 mRNA levels were determined by quantitative RT-PCR in HCC38 (p=0.002), HCC1143 (p=0.0158) and MDA-MB-468 (p=0.0041) cells transfected with either 40nm of control siRNA (siC) or with siRNA against SOX4 (siSOX4) for 48 hours. Fold changes are relative to those of cells transfected with control siRNA and significance was calculated using paired t-test. (B) Immunoblot analyses of p-Akt (S473), total Akt and β-actin expression in cell lines with high (HCC38, HCC1143) and low (MDA-MB-468) SOX4 expression following transfection with either siC or siSOX4. Intensity of the signal was measured for each protein by Image J and p-Akt (S473) expression was analyzed relative to Total AKT (C) for HCC38 (p=0.002), HCC1143 (p=0.005) and MDA-MB-468 (p=0.5310) cell lines. (D) Immunofluorescence microscopy was used to analyze the effects of siC or siSOX4 in HCC38 cells on SOX4 (Cy3) or pAkt (Cy5) protein expression and cell nuclei were visualized by DAPI; scale bar, 100 μm. Images are shown at 20x magnification (left) and digitally enlarged to show detail (right).
Analysis of siC and siSOX4 treated cells by immunofluorescent microscopy (IF) further demonstrates that SOX4 is largely undetectable in siSOX4-treated HCC38 cells and this loss of SOX4 protein expression corresponds with nearly an equal reduction in the levels of pAkt (Figure 6d). Interestingly, the subset of siSOX4 treated HCC38 cells which retain detectable levels of SOX4 also express pAkt suggesting that our western blot analyses may underestimate the total effect of SOX4 on pAkt levels and when compared on a cell to cell basis, SOX4 has a more dramatic effect on the phosphorylation of Akt. As shown in Supplementary Figure 8, similar effects on pAkt levels were observed after SOX4 silencing in HCC1143 cells.
DISCUSSION
PI3K/Akt/mTOR signaling regulates many predominant features of cancer including cellular proliferation, survival, metabolism, motility and genomic instability [8, 9]. This pathway not only contributes to tumor development, but also plays an essential role in regulating therapeutic resistance in breast cancer [29–31]. Not surprisingly, there has been a significant effort to elucidate mechanisms regulating PI3K signaling in order to identify potential therapeutic targets and/or biomarkers to current therapies.
Improper activation of this pathway occurs through various molecular mechanisms including, among others, PIK3CA activating mutations; PTEN silencing mutations; and/or copy number changes in PIK3CA, PTEN, INPP4B or AKT2, depending on the type and/or subtype of cancer [8–10]. In breast cancer, approximately 45% of patients are characterized by PIK3CA activating mutations [1, 3]; however these alterations are largely limited to LumA tumors and the link between these mutations and pathway activation is unclear as there is no correlation with known markers of pathway activity including pAkt, pS6 or p4EBP1 [3, 32–34]. In contrast basal-like or TNBC tumors are characterized by increased PI3K activity and have increased expression of these markers despite the fact that these tumors rarely have PIK3CA activating mutations [3, 12]. While clinical studies have demonstrated that the combination of everolimus plus exemestane resulted in an improved prognosis for patients with advanced stage, ER+ luminal breast cancer [35–37], similar success has not been achieved in TNBC.
In the current study, we sought to identify genomic alterations that contribute to PI3K/Akt signaling and which may represent putative therapeutic opportunities. To address this challenge, we analyzed genomic data from more than 1,000 breast tumor samples from the TCGA [1, 3] within the framework of an experimentally derived PI3K gene expression signature [6]. These in silico analyses identified SOX4 as putative novel regulator of PI3K signaling and this association was validated in an independent cohort of nearly 2,000 breast tumor samples [4]. Since, altered expression of this gene predominantly occurs in basal-like tumors, these data, consistent with previous studies demonstrating increased SOX4 expression in TNBC, suggest that this may be a subtype-specific event [38].
SOX4 is member of the group C family of SOX transcription factors which play an essential role in development but are reported to be over-expressed in many human cancers, including breast cancer where expression correlates with tumor grade, stage and prognosis [27, 28, 39]. Previous studies have reported that SOX4 contributes to tumor development and progression through a number of mechanisms. SOX4 has been shown to be essential in maintenance of stemness in cancer initiating cells, in driving cell proliferation, by promoting cell migration and metastasis and, more recently, through induction of epithelial-to-mesenchymal transition (EMT) in breast cancer [38, 40–43]. While these previous studies have demonstrated a significant role for SOX4 in tumor development, the current study is first to identify SOX4 as a mediator of PI3K/Akt signaling in breast cancer. These results are consistent with recent work that demonstrates a role for SOX4 in regulating PI3K activity in Ph+ ALL (Acute Lymphoblastic Leukemia) [44].
Experimental studies demonstrate that SOX4 regulates pAkt expression in breast cancer cell lines with high SOX4 expression irrespective of whether these cells are characterized by loss of PTEN or activation of EGFR. While the exact mechanism by which SOX4 modulates PI3K/Akt signaling remains to be determined, a number of recent studies offer insight into several potential mechanisms. The role of SOX4 as a transcription factor suggests that its effect on the PI3K pathway is mediated through regulation at the transcript level. However, given that our data demonstrate a clear loss of phosphorylated Akt in the absence of SOX4 expression, but no discernible difference in total Akt, the effect of SOX4 on signaling appears to be upstream of Akt. It was recently reported that SOX4 cooperates with PTEN loss to mediate prostate cancer tumorigenesis [45]; however given that we see a comparable effect on SOX4-mediated Akt phosphorylation in both PTEN normal and PTEN deleted cells, it is unclear whether this is also true in breast cancer. The potential does exist that SOX4 could mediate PTEN activity. In fact, studies in hepatocellular carcinoma recently reported that SOX4 can modulate HUNK kinase (Hormonally-upregulated Neu-associated kinase) expression, which itself, has been shown to regulate Akt signaling in breast cancer by repressing PTEN activity [46, 47]. Unfortunately, we see no difference in HUNK mRNA expression in SOX4 amplified and normal breast tumors.
Alternatively, a number of studies have suggested that SOX4 can mediate activation of potential up-stream drivers of PI3K signaling including transcriptional activation of EGFR [48]. Despite these findings in prostate cancer, we see a SOX4-mediated effect on pAkt levels in both EGFR activated and normal breast cancer cell lines. Moreover, we do not see a difference in pEGFR protein expression in SOX4 amplified or normal breast tumors. Although these data do not eliminate the possibility that SOX4 mediates its effect on pAkt through EGFR, or another receptor tyrosine kinase, it suggests that alternative breast cancer specific mechanisms may exist. Perhaps not surprising, recent analyses have also shown little concordance between genes regulated by SOX4 across tissues type including prostate, lung, hepatocellular carcinoma and adenoid cystic carcinoma [27, 28]. Thus, these results suggest the effects of SOX4 on PI3K signaling, may be affected by tissue specific features. As such, additional studies must be undertaken to delineate these breast cancer specific mechanisms.
Collectively, our cross-platform analyses of orthogonal genomic and proteomic data, along with in vitro studies, have identified and validated SOX4 amplification as a mediator of PI3K/Akt signaling in breast cancer. While the precise mechanisms by which SOX4 modulates PI3K activity in breast cancer remain unknown, our data, in combination with previous studies, suggest that an emphasis should be placed on elucidating these mechanisms but also that SOX4, or its downstream targets, may be a putative therapeutic target and/or biomarker in breast cancer patients whose tumors are characterized by high PI3K activity.
Supplementary Material
Patterns of pathway activity were scored for a collection of 1,031 breast cancer samples. Differences between pathway scores were calculated using a t-test to compare each molecular subtype to all other samples and resulting p-values are reported.
The frequency of copy number gains (including both high-level amplifications and gains) were calculated for each molecular subtype (n=1,031). Basal-like tumors (57.5%) were the only subtype to show a significant enrichment in SOX4 amplification frequency (p<0.0001, Chi-square test).
The relationship between discrete SOX4 copy number status (i.e. gain/amplification, neutral, or LOH/deletion) and PI3K score was calculated (p=3.7×10−08) in the TCGA cohort (n=1,031).
Analysis of the METABRIC dataset (n=1,992) confirmed the positive correlation between SOX4 copy number status, mRNA levels and PI3K signature score. These results confirm the associations identified in the TCGA cohort.
The incidence of the co-occurrence of copy number alterations including copy number gains for PIK3CA and SOX4 or copy number losses for PTEN and INPP4B was calculated in human breast tumors (n=1,031). These results demonstrate that SOX4 CNAs occur in neither a mutually inclusive nor exclusive pattern with each gene.
A panel of 51 breast cancer cell lines (GSE12777) was scored for molecular subtype (basal or luminal), PI3K pathway score and SOX4 mRNA expression. Samples were ranked according to PI3K score and the top three cell lines MDAMB468, HCC1143, and HCC38 (indicated in red text) were selected for in vitro studies.
Immunofluorescence microscopy was used to analyze the effects of siC or siSOX4 in HCC1143 cells on SOX4 (Cy3) or pAkt (Cy5) protein expression and cell nuclei were visualized by DAPI. Images were captured at 40x magnification using Nikon Eclipse TE-2000U fluorescent microscope.
Supplementary Table 1. Summary of Breast Cancer Samples
Supplementary Table 2. Summary of PI3K signature score correlation with PI3K/Akt RPPA data
Supplementary Table 3. Summary of PI3K signature correlation with copy number data
Supplementary Table 4. Candidate gene list
Acknowledgments
We thank members of our laboratory for helpful discussion and suggestions. Research reported in this publication was supported by the National Cancer Institute of the US National Institutes of Health (R00-CA166228), V Foundation for Cancer Research (V2016-013), and from the New Jersey Health Foundation (PC-52-16) to M.L.G. Additional funding for research reported in this study was provided by the National Cancer Institute of the US National Institutes of Health Breast SPORE program grant P50-CA58223-09A1 and R01-CA148761-04, as well as grants from the Susan G. Komen for the Cure and the Breast Cancer Research Foundation to C.M.P.
Footnotes
CONFLICTS OF INTEREST
C.M.P. is an equity stock holder and board of director member of BioClassifier LLC and GeneCentric Diagnostics. C.M.P. is also listed as an inventor on a patent application for the PAM50 molecular assay. J.S.P. is also listed as an inventor on a patent application for the PAM50 molecular assay. All other authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
G.A.M, J.S.P, C.M.P, and M.L.G conceived and designed the study. G.A.M, G.O.S, K.A.H, and M.L.G performed analyses and experiments. G.A.M, C.M.P, and M.L.G wrote the manuscript. All authors have reviewed and approved the final manuscript.
References
- 1.Ciriello G, et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell. 2015;163(2):506–19. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perou CM, et al. Molecular portraits of human breast tumors. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gatza ML, et al. An integrated genomics approach identifies drivers of proliferation in luminal-subtype human breast cancer. Nat Genet. 2014;46(10):1051–1059. doi: 10.1038/ng.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeSantis CE, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64(4):252–71. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 8.Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov. 2014;13(2):140–56. doi: 10.1038/nrd4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodon J, et al. Development of PI3K inhibitors: lessons learned from early clinical trials. Nat Rev Clin Oncol. 2013;10(3):143–53. doi: 10.1038/nrclinonc.2013.10. [DOI] [PubMed] [Google Scholar]
- 10.Wong KK, Engelman JA, Cantley LC. Targeting the PI3K signaling pathway in cancer. Curr Opin Genet Dev. 2010;20(1):87–90. doi: 10.1016/j.gde.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mertins P, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534(7605):55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weigman VJ, et al. Basal-like Breast cancer DNA copy number losses identify genes involved in genomic instability, response to therapy, and patient survival. Breast Cancer Res Treat. 2012;133(3):865–80. doi: 10.1007/s10549-011-1846-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez-Knowles E, et al. PI3K pathway activation in breast cancer is associated with the basal-like phenotype and cancer-specific mortality. Int J Cancer. 2010;126(5):1121–31. doi: 10.1002/ijc.24831. [DOI] [PubMed] [Google Scholar]
- 14.Marty B, et al. Frequent PTEN genomic alterations and activated phosphatidylinositol 3-kinase pathway in basal-like breast cancer cells. Breast Cancer Res. 2008;10(6):R101. doi: 10.1186/bcr2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoadley KA, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929–44. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gatza ML, et al. A pathway-based classification of human breast cancer. Proc Nat’l Acad Sci. 2010;107:6994–6999. doi: 10.1073/pnas.0912708107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutti JE, et al. Oncogenic PI3K mutations lead to NF-kappaB-dependent cytokine expression following growth factor deprivation. Cancer Res. 2012;72(13):3260–9. doi: 10.1158/0008-5472.CAN-11-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saal LH, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci U S A. 2007;104(18):7564–9. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker JS, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoeflich KP, et al. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin Cancer Res. 2009;15(14):4649–64. doi: 10.1158/1078-0432.CCR-09-0317. [DOI] [PubMed] [Google Scholar]
- 21.Reich M, et al. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 22.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marcotte R, et al. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov. 2012;2(2):172–89. doi: 10.1158/2159-8290.CD-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 25.Chapman NM, et al. Proline-rich tyrosine kinase 2 controls PI3-kinase activation downstream of the T cell antigen receptor in human T cells. J Leukoc Biol. 2015;97(2):285–96. doi: 10.1189/jlb.2A1013-568RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izumchenko E, et al. NEDD9 promotes oncogenic signaling in mammary tumor development. Cancer Res. 2009;69(18):7198–206. doi: 10.1158/0008-5472.CAN-09-0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jafarnejad SM, et al. Pleiotropic function of SRY-related HMG box transcription factor 4 in regulation of tumorigenesis. Cell Mol Life Sci. 2013;70(15):2677–96. doi: 10.1007/s00018-012-1187-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vervoort SJ, van Boxtel R, Coffer PJ. The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: friend or foe? Oncogene. 2013;32(29):3397–409. doi: 10.1038/onc.2012.506. [DOI] [PubMed] [Google Scholar]
- 29.Miller TW, et al. Loss of Phosphatase and Tensin homologue deleted on chromosome 10 engages ErbB3 and insulin-like growth factor-I receptor signaling to promote antiestrogen resistance in breast cancer. Cancer Res. 2009;69(10):4192–201. doi: 10.1158/0008-5472.CAN-09-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagata Y, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6(2):117–27. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 31.Perez-Tenorio G, Stal O. Activation of AKT/PKB in breast cancer predicts a worse outcome among endocrine treated patients. Br J Cancer. 2002;86(4):540–5. doi: 10.1038/sj.bjc.6600126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stemke-Hale K, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68(15):6084–91. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bachman KE, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3(8):772–5. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 34.Campbell IG, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64(21):7678–81. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- 35.Baselga J, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366(6):520–9. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piccart M, et al. Everolimus plus exemestane for hormone-receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer: overall survival results from BOLERO-2dagger. Ann Oncol. 2014;25(12):2357–62. doi: 10.1093/annonc/mdu456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zardavas D, Fumagalli D, Loi S. Phosphatidylinositol 3-kinase/AKT/mammalian target of rapamycin pathway inhibition: a breakthrough in the management of luminal (ER+/HER2-) breast cancers? Curr Opin Oncol. 2012;24(6):623–34. doi: 10.1097/CCO.0b013e328358a2b5. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, et al. SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res. 2012;72(17):4597–608. doi: 10.1158/0008-5472.CAN-12-1045. [DOI] [PubMed] [Google Scholar]
- 39.Rhodes DR, et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci U S A. 2004;101(25):9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu P, et al. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res. 2006;66(8):4011–9. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- 41.Tavazoie SF, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451(7175):147–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tiwari N, et al. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell. 2013;23(6):768–83. doi: 10.1016/j.ccr.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 43.Ikushima H, et al. Glioma-initiating cells retain their tumorigenicity through integration of the Sox axis and Oct4 protein. J Biol Chem. 2011;286(48):41434–41. doi: 10.1074/jbc.M111.300863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramezani-Rad P, et al. SOX4 enables oncogenic survival signals in acute lymphoblastic leukemia. Blood. 2013;121(1):148–55. doi: 10.1182/blood-2012-05-428938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bilir B, et al. SOX4 Is Essential for Prostate Tumorigenesis Initiated by PTEN Ablation. Cancer Res. 2016;76(5):1112–21. doi: 10.1158/0008-5472.CAN-15-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao YL, et al. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene. 2008;27(42):5578–89. doi: 10.1038/onc.2008.168. [DOI] [PubMed] [Google Scholar]
- 47.Yeh ES, et al. Hunk negatively regulates c-myc to promote Akt-mediated cell survival and mammary tumorigenesis induced by loss of Pten. Proc Natl Acad Sci U S A. 2013;110(15):6103–8. doi: 10.1073/pnas.1217415110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scharer CD, et al. Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells. Cancer Res. 2009;69(2):709–17. doi: 10.1158/0008-5472.CAN-08-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Patterns of pathway activity were scored for a collection of 1,031 breast cancer samples. Differences between pathway scores were calculated using a t-test to compare each molecular subtype to all other samples and resulting p-values are reported.
The frequency of copy number gains (including both high-level amplifications and gains) were calculated for each molecular subtype (n=1,031). Basal-like tumors (57.5%) were the only subtype to show a significant enrichment in SOX4 amplification frequency (p<0.0001, Chi-square test).
The relationship between discrete SOX4 copy number status (i.e. gain/amplification, neutral, or LOH/deletion) and PI3K score was calculated (p=3.7×10−08) in the TCGA cohort (n=1,031).
Analysis of the METABRIC dataset (n=1,992) confirmed the positive correlation between SOX4 copy number status, mRNA levels and PI3K signature score. These results confirm the associations identified in the TCGA cohort.
The incidence of the co-occurrence of copy number alterations including copy number gains for PIK3CA and SOX4 or copy number losses for PTEN and INPP4B was calculated in human breast tumors (n=1,031). These results demonstrate that SOX4 CNAs occur in neither a mutually inclusive nor exclusive pattern with each gene.
A panel of 51 breast cancer cell lines (GSE12777) was scored for molecular subtype (basal or luminal), PI3K pathway score and SOX4 mRNA expression. Samples were ranked according to PI3K score and the top three cell lines MDAMB468, HCC1143, and HCC38 (indicated in red text) were selected for in vitro studies.
Immunofluorescence microscopy was used to analyze the effects of siC or siSOX4 in HCC1143 cells on SOX4 (Cy3) or pAkt (Cy5) protein expression and cell nuclei were visualized by DAPI. Images were captured at 40x magnification using Nikon Eclipse TE-2000U fluorescent microscope.
Supplementary Table 1. Summary of Breast Cancer Samples
Supplementary Table 2. Summary of PI3K signature score correlation with PI3K/Akt RPPA data
Supplementary Table 3. Summary of PI3K signature correlation with copy number data
Supplementary Table 4. Candidate gene list