Abstract
In this review, we will discuss how the cell of origin may modulate breast cancer intratumoral heterogeneity (ITH) as well as the role of ITH in the evolution of cancer. The clonal evolution and the cancer stem cell (CSC) models, as well as a model that integrates clonal evolution with a CSC hierarchy, have all been proposed to explain the development of ITH. The extent of ITH correlates with clinical outcome and reflects the cellular complexity and dynamics within a tumor. A unique subtype of breast cancer, the claudin-low subtype that is highly resistant to chemotherapy and most closely resembles mammary epithelial stem cells, will be discussed. Furthermore, we will review how the interactions among various tumor cells, some with distinct mutations, may impact breast cancer treatment. Finally, novel technologies that may help advance our understanding of ITH and lead to improvements in the design of new treatments also will be discussed.
Intratumoral heterogeneity (ITH) reflects tumor complexity and has implications for clinical outcome and treatment. The cell of origin may modulate breast cancer ITH and evolution—as observed in the claudin-low subtype.
The subclonal diversity of primary breast cancers has been revealed by multiregion sequencing (Yates et al. 2015). Whole-genome and targeted genome sequencing were used to analyze 303 primary tumors from 50 breast cancer patients. Mutations in genes such as AKT, FGFR, PIK3CA, and TP53 were identified as subclonal in 13 out of 50 cancers. Clinical features of disease progression, such as chemoresistance, invasiveness, and metastatic capability, also were detected in subclones of the original lesions. These results highlight the importance of analyzing the complex subclonal structure of breast cancers. However, to decipher the evolutionary relationships between different clones, it will be necessary to compare these results from primary tumors with those of matched metastases similar to the pioneering studies reported for clear cell renal cell carcinoma (Gerlinger et al. 2012).
In 1977, Hamburger and Salmon (1977) first suggested that a subset of cancer cells named human tumor stem cells, now referred to as cancer stem cells (CSCs), may drive tumorigenesis. CSCs, a limited subpopulation of tumor-initiating cells (TICs), are functionally defined as cancer cells that retain extensive self-renewal potential in xenotransplantation assays through a series of generations and have the ability to recreate the heterogeneity of the original tumor through asymmetric division. Following the pioneering studies of Bonnet and Dick (1997) defining leukemia-initiating cells in acute myeloid leukemia (AML), a similar strategy was applied by Michael Clarke and his colleagues to solid cancers. The first of these studies was reported in 2003 by Al-Hajj et al. (2003), who identified a subset of tumorigenic breast cancer cells, isolated either from patient-derived mouse xenografts or directly from patients’ samples, by fluorescence-activated cell sorting (FACS) isolation using cell-surface markers CD44 and CD24. These cells were able to form tumors after transplantation into the mammary fat pad in immunocompromised recipient mice. Thereafter, a small subpopulation of tumor-initiating cells bearing distinct cell-surface markers has been identified from a variety of solid tumors using a similar strategy as described above (see review by Clarke and colleagues [Lobo et al. 2007]).
In normal human mammary epithelium, CD44high(H)/+/CD24low(L) cells within the basal EpCAM−/L/CD49f+ population also have the highest repopulating ability using in vitro functional colony- and mammosphere-forming assays, both of which are surrogate assays measuring single-cell survival and stem cell self-renewal (Ghebeh et al. 2013). By comparing the gene expression profiles of human breast cancer lineage (Lin)−CD44+CD24−/L cells with those of normal breast epithelial and myoepithelial cells, a 186 gene “signature” was identified. This signature was able to predict the recurrence risk in patients with cancers of the breast, lung, and prostate and medulloblastoma, and also showed a strong correlation with overall and metastasis-free survival in breast cancer patients (Liu et al. 2007b).
Subsets of tumor-initiating cells have been identified from multiple mouse mammary tumor models dependent on their genetic backgrounds, indicating that the malignant transformation events may occur in different cell types in different tumors (Liu et al. 2007a; Cho et al. 2008; Vaillant et al. 2008). Using limiting dilution transplantation and in vitro mammosphere assays, we have identified a Lin−CD29HCD24H subpopulation of TICs, or CSCs, from a genetically engineered mouse (GEM) syngeneic p53-null mammary tumor model that closely mimics human breast cancer (Jerry et al. 2000; Zhang et al. 2008). The resulting tumors derived from the tumorigenic subpopulation contained cells of all lineages and displayed properties similar to the primary tumor. Analysis of biomarkers indicates the tumorigenic subpopulation may have arisen from a bipotent mammary progenitor. In addition, gene expression microarrays identified a number of epigenetic regulators critical for stem cell self-renewal as well as those involved in DNA damage response and repair processes, which were differentially expressed in the tumor-initiating cell population. Studies from the Clarke laboratory showed a low level of reactive oxygen species (ROS) in the normal mammary epithelial stem cells and CSCs of both mouse and human tumors relative to mature progeny cells and non-CSCs, respectively (Diehn et al. 2009), also supporting the stem-cell origin of cancer hypothesis.
WHAT DRIVES BREAST TUMORIGENESIS?
Deciphering the transforming events in the normal mammary stem cells or more committed progenitors is critical for understanding what drives breast tumorigenesis. Breast cancers in BRCA1 germline mutation carriers show basal-like phenotypes (Foulkes et al. 2003; Sorlie et al. 2003), suggesting that they might originate from normal mammary stem-cell/basal-cell populations. However, BRCA1-mutant pre-neoplastic tissues displayed an increased luminal progenitor population as compared to normal breast tissues (Lim et al. 2009). Further analysis of gene expression profiles showed that breast tissue from heterozygous BRCA1 mutation carriers and basal breast tumors were more similar to normal luminal progenitor cells than other stem- and differentiated cell subpopulations. Consistently, targeted Brca1 loss in stem cells did not generate tumors that reproduce the features of human BRCA1 tumors, whereas homozygous deletion of BRCA1 in luminal progenitors produced tumors resembling human BRCA1-associated cancers, implying that luminal progenitors are most likely the cell of origin of human BRCA1-associated breast cancers (Molyneux et al. 2010). Recently, more elegant studies using lineage tracing performed by Bentires-Alj (Meyer et al. 2011) and Blanpain (Van Keymeulen et al. 2011, 2015) have provided more evidence that oncogenic events in different cell types lead to distinct tumor types and that these differences correlate with clinical outcomes. PIK3CA-activating mutations occur in approximately 30% of breast cancers. In a mouse model conditionally expressing PIK3CAH1047R, expression of the mutant allele in luminal mammary epithelium induces heterogeneous tumors that express both luminal and basal markers and are positive for the estrogen receptor (ER) (Meyer et al. 2011), suggesting that the PIK3CAH1047R oncogene targets a multipotent progenitor cell. In addition, PIK3CAH1047R expression in unipotent progenitor cells has been shown to reprogram these cells. Expression of PIK3CAH1047R in unipotent basal cells gave rise to luminal-like cells, whereas its expression in unipotent luminal cells gave rise to basal-like cells before progressing into invasive tumors displaying intratumoral heterogeneity (Van Keymeulen et al. 2011, 2015).
Although CSC may be crucial during tumorigenesis and the CSC hierarchical model may account, at least in part, for the observed intratumoral heterogeneity (ITH) that appears to be a property of many cancers, tumor progression appears to result from the evolution of a large population of genetically and epigenetically distinct cells (Merlo et al. 2006; Polyak 2014). Neoplastic cells with different orders and/or numbers of mutations may compete for space and resources, and cooperate to disperse and colonize new organs. The evolution of neoplastic cells provides new insights into neoplastic progression, intratumoral heterogeneity, and the clinical treatment of cancer (Beca and Polyak 2016). The clonal evolution and CSC models are not mutually exclusive (Kreso and Dick 2014). Malignant transformation may occur in both normal multipotent stem cells as well as more differentiated progenitors through clonal evolution, which then results in the existence of multiple cell lineages (Polyak and Weinberg 2009; Greaves and Maley 2012; Shibata and Shen 2013).
SIMILAR REGULATORY NETWORKS DRIVING ORGANOGENESIS AND TUMORIGENESIS
The identification and characterization of tumor-initiating cells and the molecular pathways responsible for their self-renewal and survival are critical to design therapies that preferentially target these cells and sensitize them to conventional therapies such as radiation and chemotherapy. Given the apparent similarities between the normal stem cells and cancer (stem) cells, it is not surprising that a number of key developmental pathways play a common role in the regulation of both normal tissue and malignant stem cells (Reya et al. 2001).
The canonical Wnt/β-catenin signaling pathway is known to regulate stem cell self-renewal, and its abnormal activation has been associated with the development of various cancers (Reya and Clevers 2005). Aberrant Wnt signaling leads to perturbed mammary gland development and results in mammary tumor development in mice (Nusse et al. 1984; Li et al. 2000). When Wnt-1 is induced in the mammary glands of mice heterozygous for Pten, the tumors formed contain both myoepithelial and luminal epithelial cells sharing a common Pten loss mutation. However, when Neu, H-Ras, or polyoma middle-T antigen are induced, the resulting tumors lose the myoepithelial cell population, indicating that a common stem cell and/or progenitor may be the target for Wnt1-induced oncogenesis (Li et al. 2003). In addition, interference with the Wnt signaling pathway in cancers of the skin, prostate, and intestine resulted in the loss of self-renewal ability and increased differentiation of the CSCs, suggesting that Wnt signaling plays a role in maintaining the stem cell function of CSCs in many cancers (Malanchi et al. 2008; Bisson and Prowse 2009; Zheng et al. 2010).
Using limiting dilution transplantation performed on p53-null tumor cells transduced with Wnt reporter lentivirus, we showed that FACS sorting of cells expressing the TOP-eGFP canonical Wnt reporter resulted in a marked enrichment for CSCs characterized previously using cell-surface markers. Pten/Akt/Wnt signaling was shown to confer the enhanced radiation resistance observed in these cells, and pharmacological inhibition of the signaling pathways was able to inhibit canonical Wnt signaling as well as DNA damage repair selectively in CSCs, sensitizing them to ionizing radiation treatment (Zhang et al. 2010).
Hedgehog (Hh) signaling is a key regulator in organogenesis and tissue repair through regulation of adult stem cells (Lewis 2001; Liu et al. 2006). Hh signaling also has been implicated in the development of cancers through regulation of CSCs (Liu et al. 2006; Clement et al. 2007; Coni et al. 2013). Activation of Hh signaling by overexpressing Hh transcriptional effectors, GLI1/GLI2, results in increased size and formation number of mammospheres, whereas inhibition of Hh signaling with a primary inhibitor, cyclopamine, results in a reduced mammosphere-forming potential (Liu et al. 2006). In addition, an expansion of the cytokeratin 6-positive cells, a putative progenitor cell population, has been reported in GLI1-induced tumors, suggesting the importance of Hh signaling in stem/progenitor function and breast cancer development (Fiaschi et al. 2009).
The Notch signaling pathway also has been implicated in the regulation of asymmetric cell-fate decisions in human mammary stem cells (Dontu et al. 2004a,b, 2005; Dontu and Wicha 2005; Liu et al. 2005). Expression of the activated form of the Notch1 receptor, Notch1 receptor intracellular domain (N1IC), in mammary cells of mouse mammary tumor virus (MMTV)/N1IC transgenic mice increased the survival potential of the CD29HCD24+ progenitor cells and led to the formation of basal-like ductal tumors through a cyclin D1-dependent pathway (Ling et al. 2010).
These shared regulatory mechanisms create an opportunity for targeting tumorigenic cancer cells, but may also cause the pleotropic side effects on normal cells. Therefore, identification of the unique features of CSCs may provide a more efficient and less toxic course of treatment in cancer. For example, unique mutations within the CSCs may cause an increase in the frequency of symmetric versus asymmetric divisions resulting in a change in their population dynamics. Therefore, systemic therapies targeting symmetric cell division mechanisms may provide a promising approach to eradicating CSCs while sparing the normal stem cells (Boman et al. 2007).
Whether tumors are developed from normal stem cells, progenitor cells, or differentiated cells undergoing de-differentiation to regain self-renewal potential, similar regulatory mechanisms seem to drive organogenesis and tumorigenesis. Leukemia stem cells (LSCs) are the most well-studied CSCs. They maintain features of normal stem cells with their ability to self-renew and differentiate to generate the heterogeneity of blood cancers, providing the strongest support that the LSCs are derived from the normal hematopoietic stem cells (Hope et al. 2004), and are the driving force for tumorigenesis. Additional studies need to be performed in breast tissue to better understand the cellular origin of different breast cancer subtypes.
CLAUDIN-LOW, EPITHELIAL-MESENCHYMAL-TRANSITION (EMT), AND CHEMOTHERAPY RESISTANCE
As mentioned previously, breast cancer is a disease that displays both inter- and intratumoral heterogeneity. Uncovering relationships between tumor subtypes and their potential normal cellular counterparts is critical for understanding the cellular origin of breast cancer. Using a meta-analysis approach, Pfefferle et al. (2015) derived consensus gene signatures for both mouse and human species and used these to relate tumors to normal mammary epithelial cell phenotypes. They found most human and murine tumor subtypes shared some, but not all, features with a specific FACS-purified normal cell type; thus for most tumors a potential distinct cell type of “origin” could be assigned. We will focus on discussing the claudin-low subtype, which most closely resembles mammary epithelial stem cells.
In collaboration with our clinical colleague, Dr. Jenny Chang, we asked whether a similar CSC subpopulation of breast cancer cells might be resistant to chemotherapy and responsible for relapse. Specifically, we wanted to determine whether these markers could be used in paired breast cancer core biopsies from patients with primary breast cancer before and after neoadjuvant chemotherapy. We also were able to compare the potential of cells taken before and after treatment to form mammospheres. These studies were the first to provide clinical evidence for a subpopulation of chemotherapy-resistant breast cancer–initiating cells (Li et al. 2008). Parallel studies performed in a series of patients with human epidermal growth factor receptor 2 (HER2)-positive tumors treated with lapatinib also suggested that specific pathway inhibitors might provide a therapeutic strategy for eliminating these cells to decrease recurrence and improve long-term survival. Taking advantage of these clinical samples, it was possible to perform gene expression profiling on the CD44+/CD24− subpopulation compared to the other FACS populations, as well as mammosphere-forming cells compared to the bulk tumors. Somewhat disappointingly, the gene expression signature derived from these comparisons was not prognostic for the majority of human breast cancers, and only overlapped with an identified “claudin-low” molecular subtype (Creighton et al. 2009). The claudin-low breast cancer subtype is characterized by the low to absent expression of luminal differentiation markers, high enrichment for many EMT-associated genes, and immune-response genes (Prat et al. 2010). The claudin-low subtype also most closely resembles mammary epithelial stem cells, leading to the hypothesis that the most primitive mammary stem cells may be the cell of origin for the claudin-low subtype of breast cancer (Prat and Perou 2009). The majority of the claudin-low tumors are triple-negative breast cancers lacking the estrogen and progesterone receptors and HER2 receptors. Despite the apparent lack of prognostic significance for bulk tumors, the claudin-low signature was enriched in residual tumor tissue remaining after either endocrine therapy using an aromatase inhibitor, letrozole, or chemotherapy treatment with docetaxel (Creighton et al. 2009). Strikingly, using quantitative immunofluorescent analysis double-positive cells expressing the mesenchymal marker, vimentin, and the pan-keratin, epithelial marker were enriched following letrozole treatment. This suggests the presence of an intermediate or “partial EMT,” which has been proposed to be a hybrid E/M phenotype between the epithelial to mesenchymal transition (Jolly et al. 2014, 2015).
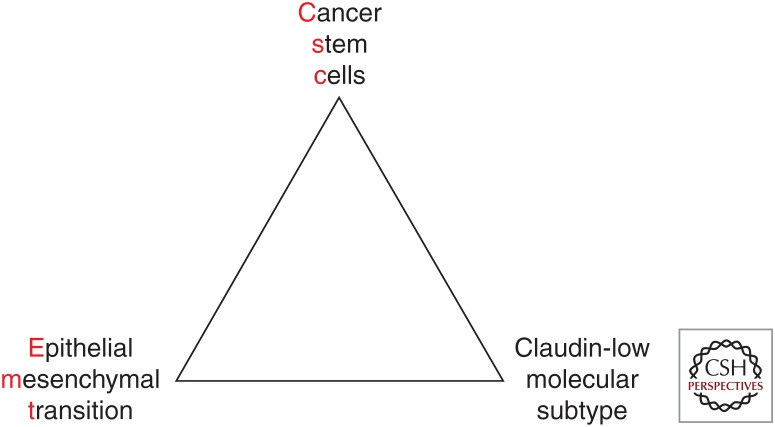
While these studies using clinical samples were in progress, parallel experiments with transcription factors, such as SNAIL and TWIST, conditionally activated in immortalized human mammary epithelial cells (HMLEs) were being conducted in the Weinberg laboratory (Mani et al. 2008). These transcription factors induced the expression of mesenchymal markers and more importantly led to an increase in stem-like behavior. More recently, a core EMT interactome gene expression signature obtained by overexpressing several different EMT-inducing transcription factors or by treating HMLEs with transforming growth factor β (TGF-β) was shown to associate closely with the claudin-low and metaplastic breast cancer subtypes and correlated negatively with pathological complete response (Taube et al. 2010). The last piece of this puzzle came about during the characterization of tumors derived in the p53-null mammary tumor model described previously. A rare subset of these tumors was discovered that had similar gene expression signatures as the human claudin-low tumors (Herschkowitz et al. 2012). These murine claudin-low tumors showed high expression of EMT inducers, low expression of members of the miR-200 family, and several other microRNAs thought to be absent in normal mammary stem cells consistent with the earlier observations of a TGF-β-miR-200 double-negative feedback loop important for the EMT transition (Gregory et al. 2008). More importantly, it was possible to isolate cells from these tumors to perform functional limiting dilution transplantation assays into the cleared fat pad of syngeneic mice. These studies showed that CSCs were highly enriched in the claudin-low tumors as compared to other basal-like and luminal-like tumors developed in the same model (Herschkowitz et al. 2012). In fact, the high frequency of CSCs in this model was analogous to earlier studies in melanoma, suggesting that CSCs were not a rare population in these tumors (Quintana et al. 2008). Gene expression profiling of the mouse melanoma and claudin-low GEM models showed considerable overlap as well as similarities in their response to targeted therapies with MEK and PI3K inhibitors (Roberts et al. 2012). Thus, there is an intimate association of EMT and CSCs and claudin-low subtype of breast cancer as shown in Figure 1. However, this should not be interpreted to mean that these three states are all equivalent.
Figure 1.
The claudin-low molecular subtype, epithelial-mesenchymal-transition (EMT), and cancer stem cell (CSC) phenotypes display many similarities but are not necessarily identical.
Why is this important in breast cancer evolution? In a seminal opinion piece in Nature Reviews Cancer, Brabletz et al. (2005) first proposed the concept of “migrating cancer stem cells—an integrated concept of malignant tumor progression.” These investigators suggested that “a loss of epithelial differentiation and the acquisition of a migratory phenotype is a hallmark of tumor progression.” The foundation for this concept comes from basic studies of normal development (Thiery 2003). In the context of breast cancer, the CD44+/CD24− CSCs isolated from patient-derived cells have been shown in mouse xenografts to not only be tumor-initiating cells, but also to give rise to lung metastasis (Liu et al. 2010). The observation that cells can undergo a transient EMT suggests that EMT/MET plasticity may play a critical role in tumor progression. The “conventional” model suggests that tumor cells first have to undergo EMT to migrate and intravasate into the vasculature, but at distant sites after extravasation they may have to then revert to an MET state to support their proliferation into macrometastases. Support for this model comes from elegant studies using a doxycycline-inducible Twist skin carcinogenesis model (Tsai et al. 2012). Furthermore, recent studies that used a similar inducible Twist model showed that a transient EMT must occur for CSC formation, followed by a subsequent MET that still can retain CSC characteristics even after Twist is deactivated, suggesting an epigenetic reprogramming mechanism (Schmidt et al. 2015). In contrast, two recent studies have suggested that EMT is not required for metastasis, highlighting the controversy surrounding the role of EMT in metastasis (Fischer et al. 2015; Zheng et al. 2015). However, both of these latter studies also reported that EMT still endowed cells with chemotherapy resistance in primary and metastatic tumors (Fischer et al. 2015; Zheng et al. 2015). This is probably the most consistent observation in many solid cancers where the EMT state is often the “default” phenotype observed during therapeutic resistance. A discussion of the strengths and weaknesses of these studies has been published recently (Li and Kang 2016). EMT may be accompanied by a significant reduction in proliferation (Vega et al. 2004; Tsai et al. 2012), suggesting that cell quiescence may endow a cell with resistance to therapies that target proliferating cells. However, this is most likely not the only mechanism of resistance, as mesenchymal-like, claudin-low breast cancers, which are proliferative, also display characteristics associated with therapy resistance, suggesting that alternative resistance mechanisms likely exist (Creighton et al. 2009, 2010). Perhaps EMT/CSCs are better able to repair their DNA in response to damage as well as induce stress pathways and inhibit apoptosis, as has recently been shown for CSCs in the p53-null breast cancer model (Chang et al. 2015). The ZEB1/miR-200 axis has additional, non-cell-autonomous roles in cancer pathogenesis. For example, it has recently been shown to affect immune recognition of cancer cells, whereby ZEB1 suppression of the miR-200 family leads to up-regulation of PD-L1, a direct miR-200 family target. This leads to subsequent evasion of immune cells by the tumor cells (Chen et al. 2015). Therefore, the EMT transition also may play a role in immunosuppression, thus exacerbating treatment response and potentially increasing metastasis (Chen et al. 2015; Mak et al. 2016).
Because metastases are responsible for >90% of cancer-associated mortality, and cells with EMT properties display resistance to “standard of care” treatments of radiation and chemotherapy, efforts have been made to develop small molecule inhibitors to reverse EMT and sensitize CSCs to treatment (Gupta et al. 2009; Pattabiraman et al. 2016). Unfortunately, these approaches have been unsuccessful because none of these drugs to date have been moved forward to the clinic for use in patients because of their toxicity. A proof of concept, however, has been obtained by several studies in preclinical GEM models in which reexpression of miR-200 family members has been able to reverse EMT, decrease the CSC population, and sensitize cells to both chemotherapy and targeted therapies (Adam et al. 2009; Knezevic et al. 2015). One potential approach to reactivate the endogenous expression of miR-200 family members, which are silenced by both DNA methylation and chromatin modification, may be to use epigenetic therapies (Azad et al. 2013; Lim et al. 2013).
ROLE OF INTRATUMORAL HETEROGENEITY IN EVOLUTION OF CANCER
Next, we discuss changes in intratumoral heterogeneity and the role it may play in tumor development. Evaluating changes in ITH during cancer progression is complicated by the paucity of longitudinal samples. Although many studies have characterized the ITH of primary cancers, such as The Cancer Genome Atlas (TCGA) pan-cancer analysis (Andor et al. 2016), relatively few have studied metastatic breast cancer and changes that occur between primary and metastatic lesions. Two genome-wide studies of a single case of paired primary-metastatic breast cancer provided an initial view of ITH in primary breast cancer and its evolution in metastatic disease (Shah et al. 2009; Ellis et al. 2012). A recent genome-wide analysis of primary and metastatic samples from a single patient showed that each metastatic site harbored distinct mutational events, with independent inactivating mutations in PTEN indicating convergent and parallel evolution (Juric et al. 2015). Despite these significant advances, fundamental questions remain in the understanding of the evolution of breast cancer metastasis. For example, a large study of paired primary-metastatic samples using a small hotspot-sequencing panel found 85.7% of pairs had identical mutations (Goswami et al. 2015). This is in contrast to a recent study of exome sequencing 21 paired primary and brain metastases, which indicated extensive evolution of metastatic disease consistent with branched evolution (Brastianos et al. 2015).
Importantly, the majority of these studies have focused on single nucleotide mutations/variants. This is in part because massively parallel sequencing is adept at identifying single nucleotide variants (SNVs), but detection of structural variants such as copy number changes and translocations has, until recently, remained challenging. However, breast cancer has on average one of the lowest levels of SNVs in solid tumors (Kandoth et al. 2013), and a panomic analysis by TCGA categorized breast cancer into a copy number variant (CNV)-driven disease (Ciriello et al. 2013). Early studies using CNV arrays revealed genome-wide changes in DNA copy number between primary tumors and metastases (Nishizaki et al. 1997). Studies analyzing CNVs in single disseminated cancer cells confirmed that CNVs are both gained and lost during progression, but interestingly, the lower number of CNVs found in disseminated tumor cells compared with the primary tumor suggested that tumor cells disseminate very early in a less progressed evolutionary state (Schmidt-Kittler et al. 2003). Focused analysis of CNV changes in breast cancer has recently given insight into tumor evolution and the timing of different genetic changes (Janiszewska et al. 2015). A focus on the use of CNVs for tumor deconvolution and evolution (Chowdhury et al. 2013, 2014, 2015) found extensive ITH in paired ductal carcinoma in situ (DCIS) and invasive ductal carcinomas (IDCs) (Heselmeyer-Haddad et al. 2012) and was found to be far more diverse than is apparent from analysis of SNVs. This conclusion is supported by the prevalence of chromosome instability phenotypes in breast cancers (Heng et al. 2006, 2013) and suggests that SNVs may be the dominant mechanism of evolution in tumor progression, such as they are in normal evolution of speciation (Zack et al. 2013).
Although regional (Gerlinger et al. 2012) and single-cell sequencing (Navin et al. 2011) have expanded the understanding of ITH (Burrell et al. 2013), the use of solid tumor biopsies to study cancer evolution is ultimately hindered by the impractical nature of the biopsy, which comes at both cost and potential clinical complications. In contrast, accessing DNA from bodily fluids such as blood or urine has the potential to allow real-time monitoring of tumor burden (Dawson et al. 2013) and the evolution of SNVs in response to therapy (Murtaza et al. 2013). Recent studies in breast cancer highlight the use of SNVs in circulating cell-free DNA (cfDNA) for monitoring tumor relapse (Garcia-Murillas et al. 2015) and selecting and monitoring response to targeted therapies (Frenel et al. 2015). A recent single case report measured SNVs over a 3-year tumor period (eight biopsies and nine cfDNA samples) to study tumor evolution (Murtaza et al. 2015). cfDNA SNVs reflected the clonal hierarchy of the solid tumor biopsies, and low allele frequency cfDNA mutations were found to change in response to therapy (Murtaza et al. 2015). This report highlights the potential for cfDNA to refine phylogenetic studies of evolution in solid tumor biopsies.
COOPERATION OF CANCER CELLS IN TUMOR HETEROGENEITY AND EVOLUTION
ITH has been shown to drive neoplastic progression and therapeutic resistance because many treatments may only target certain tumor cell subpopulations. Using the bioinformatics tools “expanding ploidy and allele frequency on nested subpopulations” (EXPANDS) and PyClone, clones that are present at a >/=10% frequency in 1165 exome sequences from tumors in TCGA, were investigated. 86% of tumors from 12 cancer types had at least two clones (Andor et al. 2016). In addition, the observation that a high concentration of cells was required for successful growth of tumor-derived cell lines suggested the cooperation between various tumor cells under in vitro growth conditions (Von Hoff et al. 1986). To date, only a few studies have been reported that show the importance of functional ITH. Using an MMTV-driven Wnt1 transgenic mouse model, Gunther and colleagues showed that both the basal and luminal cells were required for efficient tumor formation. This appears similar to the development of normal mammary tissues in which the cross talk between basal/myoepithelial and luminal cells depends on Wnt expression in luminal cells (Cleary et al. 2014). Polyak and colleagues (Marusyk et al. 2014) also showed the competition-triggered clonal expansion within tumors derived from transplanting various artificially engineered clones of weakly tumorigenic MDA-MB-468 cells. A small population of cells was able to promote tumor growth and progression by overcoming environmental constraints to allow the fast proliferation of all tumors cells in a non-cell-autonomous manner. Heterogeneous clones of tumor cells are highly dynamic and competitive during tumor development. Cross talk between various subpopulations of tumor cells has also been reported in metastasis of small-cell lung tumor (Calbo et al. 2011). Similarly, a minor subclone within glioblastoma was also shown to drive tumor growth and maintain tumor heterogeneity (Inda et al. 2010).
Cross talk between various tumor cells in the p53-null mouse model of breast cancer provides another example showing the cooperation of tumor cells during tumor initiation (Zhang et al. 2015). Perturbation of gene expression by shRNA knockdown of ligands up-regulated in mesenchymal-like cells and their corresponding receptors in the CSCs led to reduced tumorigenicity and increased tumor latency, illustrating the non-cell-autonomous properties and importance of cooperativity between tumor subpopulations during tumor initiation.
CHALLENGES TO CANCER THERAPIES AND TECHNIQUES THAT MAY HELP ADVANCE OUR UNDERSTANDING OF ITH AND IMPROVE CANCER TREATMENT
Both intrinsic and acquired resistance to radiation/chemotherapeutics are the major reasons for failure in cancer treatment. Increasing evidence has been accumulated showing the existence of the low-frequency resistant clones within a tumor that expand under selective pressure after initial therapies (Turner and Reis-Filho 2012). Genetic changes, environmental differences, and dynamic conversion among tumor cells within a tumor may all lead to phenotypic and functional heterogeneity (Meacham and Morrison 2013). In addition, using somatic cell fusions and integrated genetic and epigenetic analyses, Polyak and colleagues (Su et al. 2015) found that the basal features of basal-like breast cancers, which maintain a high degree of ITH usually correlating with poor clinical outcome, are largely defined by epigenetic transcription repression of the luminal factors. These results revealed a remarkable degree of epigenetic plasticity between different breast cell types. Technically, it remains a challenge in cancer genomics to detect minor and genetically distinct subpopulations within tumors. It also remains a challenge to determine individual cell fate after drug treatment or environmental changes without applications of lineage tracing and deep sequencing, which may help to determine the extent to which ITH accounts for therapy resistance and disease progression (Meacham and Morrison 2013).
Using a novel in situ single-cell-based method, STAR-FISH (specific-to-allele PCR-FISH), to detect both single-nucleotide and copy number alterations in single cells in intact archived tissues, the Polyak laboratory has assessed the clinical impact of changes in the frequency and topology of PIK3CA mutations and HER2 (ERBB2) amplification in HER2-positive breast cancer during neoadjuvant therapy. Their results suggested that the two independent genetic events do not always occur simultaneously within the same cells. In nearly all treatment-naïve samples, a minor subpopulation of cells with a preexisting PIK3CA mutation was able to regulate genetic diversity within tumors under the pressure of chemotherapy selection, implying that the resistance to HER2-targeted drugs may arise as a result of chemotherapy (Janiszewska et al. 2015). Most recently, Caldas and colleagues (Pereira et al. 2016) sequenced 173 genes in more than 2000 primary breast tumors, and identified 40 mutation-driver genes, including mutations in PIK3CA that were highly associated with reduced survival in three subgroups of ER-positive cancers. Although high levels of ITH are in general associated with a worse outcome, highly aggressive tumors with amplification of alleles at 11q13-14 locus showed low levels of ITH. This indicates the importance of genome-based stratification for the treatment of breast cancer, which will be an important prerequisite for personalized genomic medicine (Aparicio and Caldas 2013; Baird and Caldas 2013). In addition, using deep-genome and single-cell sequencing methods, Eirew and colleagues (2015) analyzed DNAs from patient-derived xenograft (PDX) lines as well as their matched patient samples to identify any clonal variations between primary samples, and early as well as subsequent engraftments at the single-cell level. Varying degrees of clonal selection, from rare clone (<5% of starting population) to moderate, polyclonal engraftment, were observed in all ten primary and five metastatic breast tumors. PDX models were shown to recapitulate the clonal heterogeneity, with some drift. Thus, these technologies and models hold promise to study ITH and clonal evolution both before and after therapy, and may impact the design of future combinatorial therapies (Tabassum and Polyak 2015).
CONCLUDING REMARKS
The key to understanding tumor initiation and progression, and to designing more efficient therapeutic drugs, is to detect which genes, when mutated, initiate tumorigenesis, and how they, under selection pressure after initial therapies, determine individual cell fate. Thus, one of the main fields of genome research includes identifying the existence of SNVs that may be the driving force during breast tumor evolution. In addition, understanding evolutionary dynamics among various cancer cells will also help lead to the development of novel therapeutic approaches. Furthermore, understanding the importance of inter- and intratumoral heterogeneity combined with the rapid development of technologies, including deep sequencing and lineage tracing, will improve our understanding of various breast tumor subtypes, and should help determine the extent to which ITH accounts for therapy resistance and disease progression.
ACKNOWLEDGMENTS
The authors apologize to those investigators whose work is not cited because of space limitations. M.Z. is supported by a National Institutes of Health/National Cancer Institute (NIH/NCI) Pathway to Independence (PI) Award CA14289, A.V.L. is funded as a Komen Scholar and by the Breast Cancer Research Foundation, and J.M.R. is also funded as a Komen Scholar and is supported, in part, by NCI CA16303.
Footnotes
Editors: Charles Swanton, Alberto Bardelli, Kornelia Polyak, Sohrab Shah, and Trevor A. Graham
Additional Perspectives on Cancer Evolution available at www.perspectivesinmedicine.org
REFERENCES
- Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, Calin G, Wang H, Siefker-Radtke A, McConkey D, et al. 2009. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res 15: 5060–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. 2003. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci 100: 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C, Ji HP, Maley CC. 2016. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med 22: 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio S, Caldas C. 2013. The implications of clonal genome evolution for cancer medicine. N Engl J Med 368: 842–851. [DOI] [PubMed] [Google Scholar]
- Azad N, Zahnow CA, Rudin CM, Baylin SB. 2013. The future of epigenetic therapy in solid tumours—Lessons from the past. Nat Rev Clin Oncol 10: 256–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird RD, Caldas C. 2013. Genetic heterogeneity in breast cancer: The road to personalized medicine? BMC Med 11: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beca F, Polyak K. 2016. Intratumor heterogeneity in breast cancer. Adv Exp Med Biol 882: 169–189. [DOI] [PubMed] [Google Scholar]
- Bisson I, Prowse DM. 2009. WNT signaling regulates self-renewal and differentiation of prostate cancer cells with stem cell characteristics. Cell Res 19: 683–697. [DOI] [PubMed] [Google Scholar]
- Boman BM, Wicha MS, Fields JZ, Runquist OA. 2007. Symmetric division of cancer stem cells—A key mechanism in tumor growth that should be targeted in future therapeutic approaches. Clin Pharmacol Ther 81: 893–898. [DOI] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. 1997. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 3: 730–737. [DOI] [PubMed] [Google Scholar]
- Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. 2005. Opinion: Migrating cancer stem cells—An integrated concept of malignant tumour progression. Nat Rev Cancer 5: 744–749. [DOI] [PubMed] [Google Scholar]
- Brastianos PK, Carter SL, Santagata S, Cahill DP, Taylor-Weiner A, Jones RT, Van Allen EM, Lawrence MS, Horowitz PM, Cibulskis K, et al. 2015. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov 5: 1164–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell RA, McGranahan N, Bartek J, Swanton C. 2013. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501: 338–345. [DOI] [PubMed] [Google Scholar]
- Calbo J, van Montfort E, Proost N, van Drunen E, Beverloo HB, Meuwissen R, Berns A. 2011. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell 19: 244–256. [DOI] [PubMed] [Google Scholar]
- Chang CH, Zhang M, Rajapakshe K, Coarfa C, Edwards D, Huang S, Rosen JM. 2015. Mammary stem cells and tumor-initiating cells are more resistant to apoptosis and exhibit increased DNA repair activity in response to DNA damage. Stem Cell Rep 5: 378–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Heymach JV, Qin FX, Gibbons DL. 2015. The mutually regulatory loop of epithelial-mesenchymal transition and immunosuppression in cancer progression. Oncoimmunology 4: e1002731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RW, Wang X, Diehn M, Shedden K, Chen GY, Sherlock G, Gurney A, Lewicki J, Clarke MF. 2008. Isolation and molecular characterization of cancer stem cells in MMTV-Wnt-1 murine breast tumors. Stem Cells 26: 364–371. [DOI] [PubMed] [Google Scholar]
- Chowdhury SA, Shackney SE, Heselmeyer-Haddad K, Ried T, Schaffer AA, Schwartz R. 2013. Phylogenetic analysis of multiprobe fluorescence in situ hybridization data from tumor cell populations. Bioinformatics 29: 189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury SA, Shackney SE, Heselmeyer-Haddad K, Ried T, Schaffer AA, Schwartz R. 2014. Algorithms to model single gene, single chromosome, and whole genome copy number changes jointly in tumor phylogenetics. PLoS Comput Biol 10: e1003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury SA, Gertz EM, Wangsa D, Heselmeyer-Haddad K, Ried T, Schaffer AA, Schwartz R. 2015. Inferring models of multiscale copy number evolution for single-tumor phylogenetics. Bioinformatics 31: 258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. 2013. Emerging landscape of oncogenic signatures across human cancers. Nat Genet 45: 1127–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary AS, Leonard TL, Gestl SA, Gunther EJ. 2014. Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature 508: 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. 2007. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol 17: 165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coni S, Infante P, Gulino A. 2013. Control of stem cells and cancer stem cells by Hedgehog signaling: Pharmacologic clues from pathway dissection. Biochem Pharmacol 85: 623–628. [DOI] [PubMed] [Google Scholar]
- Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, Rimm DL, Wong H, Rodriguez A, Herschkowitz JI, et al. 2009. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci 106: 13820–13825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Chang JC, Rosen JM. 2010. Epithelial-mesenchymal transition (EMT) in tumor-initiating cells and its clinical implications in breast cancer. J Mammary Gland Biol Neoplasia 15: 253–260. [DOI] [PubMed] [Google Scholar]
- Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, et al. 2013. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 368: 1199–1209. [DOI] [PubMed] [Google Scholar]
- Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M, et al. 2009. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 458: 780–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G, Wicha MS. 2005. Survival of mammary stem cells in suspension culture: Implications for stem cell biology and neoplasia. J Mammary Gland Biol Neoplasia 10: 75–86. [DOI] [PubMed] [Google Scholar]
- Dontu G, El-Ashry D, Wicha MS. 2004a. Breast cancer, stem/progenitor cells and the estrogen receptor. Trends Endocrinol Metab 15: 193–197. [DOI] [PubMed] [Google Scholar]
- Dontu G, Jackson KW, McNicholas E, Kawamura MJ, Abdallah WM, Wicha MS. 2004b. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res 6: R605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G, Liu S, Wicha MS. 2005. Stem cells in mammary development and carcinogenesis: Implications for prevention and treatment. Stem Cell Rev 1: 207–213. [DOI] [PubMed] [Google Scholar]
- Eirew P, Steif A, Khattra J, Ha G, Yap D, Farahani H, Gelmon K, Chia S, Mar C, Wan A, et al. 2015. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 518: 422–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, et al. 2012. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486: 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiaschi M, Rozell B, Bergstrom A, Toftgard R. 2009. Development of mammary tumors by conditional expression of GLI1. Cancer Res 69: 4810–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J, et al. 2015. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527: 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes WD, Stefansson IM, Chappuis PO, Begin LR, Goffin JR, Wong N, Trudel M, Akslen LA. 2003. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Ins 95: 1482–1485. [DOI] [PubMed] [Google Scholar]
- Frenel JS, Carreira S, Goodall J, Roda D, Perez-Lopez R, Tunariu N, Riisnaes R, Miranda S, Figueiredo I, Nava-Rodrigues D, et al. 2015. Serial next-generation sequencing of circulating cell-free DNA evaluating tumor clone response to molecularly targeted drug administration. Clin Cancer Res 21: 4586–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, Cheang M, Osin P, Nerurkar A, Kozarewa I, et al. 2015. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 7: 302ra133. [DOI] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. 2012. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366: 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghebeh H, Sleiman GM, Manogaran PS, Al-Mazrou A, Barhoush E, Al-Mohanna FH, Tulbah A, Al-Faqeeh K, Adra CN. 2013. Profiling of normal and malignant breast tissue show CD44high/CD24low phenotype as a predominant stem/progenitor marker when used in combination with Ep-CAM/CD49f markers. BMC Cancer 13: 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami RS, Patel KP, Singh RR, Meric-Bernstam F, Kopetz ES, Subbiah V, Alvarez RH, Davies MA, Jabbar KJ, Roy-Chowdhuri S, et al. 2015. Hotspot mutation panel testing reveals clonal evolution in a study of 265 paired primary and metastatic tumors. Clin Cancer Res 21: 2644–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves M, Maley CC. 2012. Clonal evolution in cancer. Nature 481: 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. 2008. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10: 593–601. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. 2009. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 138: 645–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger AW, Salmon SE. 1977. Primary bioassay of human tumor stem cells. Science 197: 461–463. [DOI] [PubMed] [Google Scholar]
- Heng HH, Liu G, Bremer S, Ye KJ, Stevens J, Ye CJ. 2006. Clonal and non-clonal chromosome aberrations and genome variation and aberration. Genome 49: 195–204. [DOI] [PubMed] [Google Scholar]
- Heng HH, Bremer SW, Stevens JB, Horne SD, Liu G, Abdallah BY, Ye KJ, Ye CJ. 2013. Chromosomal instability (CIN): What it is and why it is crucial to cancer evolution. Cancer Metastasis Rev 32: 325–340. [DOI] [PubMed] [Google Scholar]
- Herschkowitz JI, Zhao W, Zhang M, Usary J, Murrow G, Edwards D, Knezevic J, Greene SB, Darr D, Troester MA, et al. 2012. Comparative oncogenomics identifies breast tumors enriched in functional tumor-initiating cells. Proc Natl Acad Sci 109: 2778–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heselmeyer-Haddad K, Berroa Garcia LY, Bradley A, Ortiz-Melendez C, Lee WJ, Christensen R, Prindiville SA, Calzone KA, Soballe PW, Hu Y, et al. 2012. Single-cell genetic analysis of ductal carcinoma in situ and invasive breast cancer reveals enormous tumor heterogeneity yet conserved genomic imbalances and gain of MYC during progression. Am J Pathol 181: 1807–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope KJ, Jin L, Dick JE. 2004. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nat Immunol 5: 738–743. [DOI] [PubMed] [Google Scholar]
- Inda MM, Bonavia R, Mukasa A, Narita Y, Sah DW, Vandenberg S, Brennan C, Johns TG, Bachoo R, Hadwiger P, et al. 2010. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev 24: 1731–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiszewska M, Liu L, Almendro V, Kuang Y, Paweletz C, Sakr RA, Weigelt B, Hanker AB, Chandarlapaty S, King TA, et al. 2015. In situ single-cell analysis identifies heterogeneity for PIK3CA mutation and HER2 amplification in HER2-positive breast cancer. Nat Genet 47: 1212–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerry DJ, Kittrell FS, Kuperwasser C, Laucirica R, Dickinson ES, Bonilla PJ, Butel JS, Medina D. 2000. A mammary-specific model demonstrates the role of the p53 tumor suppressor gene in tumor development. Oncogene 19: 1052–1058. [DOI] [PubMed] [Google Scholar]
- Jolly MK, Huang B, Lu M, Mani SA, Levine H, Ben-Jacob E. 2014. Towards elucidating the connection between epithelial-mesenchymal transitions and stemness. J R Soc Interface 11: 20140962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly MK, Jia D, Boareto M, Mani SA, Pienta KJ, Ben-Jacob E, Levine H. 2015. Coupling the modules of EMT and stemness: A tunable “stemness window” model. Oncotarget 6: 25161–25174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juric D, Castel P, Griffith M, Griffith OL, Won HH, Ellis H, Ebbesen SH, Ainscough BJ, Ramu A, Iyer G, et al. 2015. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kα inhibitor. Nature 518: 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. 2013. Mutational landscape and significance across 12 major cancer types. Nature 502: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezevic J, Pfefferle AD, Petrovic I, Greene SB, Perou CM, Rosen JM. 2015. Expression of miR-200c in claudin-low breast cancer alters stem cell functionality, enhances chemosensitivity and reduces metastatic potential. Oncogene 34: 5997–6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreso A, Dick JE. 2014. Evolution of the cancer stem cell model. Cell Stem Cell 14: 275–291. [DOI] [PubMed] [Google Scholar]
- Lewis MT. 2001. Hedgehog signaling in mouse mammary gland development and neoplasia. J Mammary Gland Biol Neoplasia 6: 53–66. [DOI] [PubMed] [Google Scholar]
- Li W, Kang Y. 2016. Probing the fifty shades of EMT in metastasis. Trends Cancer 2: 65–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hively WP, Varmus HE. 2000. Use of MMTV-Wnt-1 transgenic mice for studying the genetic basis of breast cancer. Oncogene 19: 1002–1009. [DOI] [PubMed] [Google Scholar]
- Li Y, Welm B, Podsypanina K, Huang S, Chamorro M, Zhang X, Rowlands T, Egeblad M, Cowin P, Werb Z, et al. 2003. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci 100: 15853–15858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, Hilsenbeck SG, Pavlick A, Zhang X, Chamness GC, et al. 2008. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst 100: 672–679. [DOI] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, et al. 2009. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med 15: 907–913. [DOI] [PubMed] [Google Scholar]
- Lim YY, Wright JA, Attema JL, Gregory PA, Bert AG, Smith E, Thomas D, Lopez AF, Drew PA, Khew-Goodall Y, et al. 2013. Epigenetic modulation of the miR-200 family is associated with transition to a breast cancer stem-cell-like state. J Cell Sci 126: 2256–2266. [DOI] [PubMed] [Google Scholar]
- Ling H, Sylvestre JR, Jolicoeur P. 2010. Notch1-induced mammary tumor development is cyclin D1-dependent and correlates with expansion of pre-malignant multipotent duct-limited progenitors. Oncogene 29: 4543–4554. [DOI] [PubMed] [Google Scholar]
- Liu S, Dontu G, Wicha MS. 2005. Mammary stem cells, self-renewal pathways, and carcinogenesis. Breast Cancer Res 7: 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW, Suri P, Wicha MS. 2006. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res 66: 6063–6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JC, Deng T, Lehal RS, Kim J, Zacksenhaus E. 2007a. Identification of tumorsphere- and tumor-initiating cells in HER2/Neu-induced mammary tumors. Cancer Res 67: 8671–8681. [DOI] [PubMed] [Google Scholar]
- Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, Sherlock G, Lewicki J, Shedden K, Clarke MF. 2007b. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med 356: 217–226. [DOI] [PubMed] [Google Scholar]
- Liu H, Patel MR, Prescher JA, Patsialou A, Qian D, Lin J, Wen S, Chang YF, Bachmann MH, Shimono Y, et al. 2010. Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc Natl Acad Sci 107: 18115–18120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo NA, Shimono Y, Qian D, Clarke MF. 2007. The biology of cancer stem cells. Annu Rev Cell Dev Biol 23: 675–699. [DOI] [PubMed] [Google Scholar]
- Mak MP, Tong P, Diao L, Cardnell RJ, Gibbons DL, William WN, Skoulidis F, Parra ER, Rodriguez-Canales J, Wistuba II, et al. 2016. A patient-derived, pan-cancer EMT signature identifies global molecular alterations and immune target enrichment following epithelial-to-mesenchymal transition. Clin Cancer Res 22: 609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malanchi I, Peinado H, Kassen D, Hussenet T, Metzger D, Chambon P, Huber M, Hohl D, Cano A, Birchmeier W, et al. 2008. Cutaneous cancer stem cell maintenance is dependent on β-catenin signalling. Nature 452: 650–653. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. 2008. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell 133: 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusyk A, Tabassum DP, Altrock PM, Almendro V, Michor F, Polyak K. 2014. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature 514: 54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham CE, Morrison SJ. 2013. Tumour heterogeneity and cancer cell plasticity. Nature 501: 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo LM, Pepper JW, Reid BJ, Maley CC. 2006. Cancer as an evolutionary and ecological process. Nature Rev Cancer 6: 924–935. [DOI] [PubMed] [Google Scholar]
- Meyer DS, Brinkhaus H, Muller U, Muller M, Cardiff RD, Bentires-Alj M. 2011. Luminal expression of PIK3CA mutant H1047R in the mammary gland induces heterogeneous tumors. Cancer Res 71: 4344–4351. [DOI] [PubMed] [Google Scholar]
- Molyneux G, Geyer FC, Magnay FA, McCarthy A, Kendrick H, Natrajan R, Mackay A, Grigoriadis A, Tutt A, Ashworth A, et al. 2010. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell 7: 403–417. [DOI] [PubMed] [Google Scholar]
- Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong AS, et al. 2013. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497: 108–112. [DOI] [PubMed] [Google Scholar]
- Murtaza M, Dawson SJ, Pogrebniak K, Rueda OM, Provenzano E, Grant J, Chin SF, Tsui DW, Marass F, Gale D, et al. 2015. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun 6: 8760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. 2011. Tumour evolution inferred by single-cell sequencing. Nature 472: 90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizaki T, DeVries S, Chew K, Goodson WH III, Ljung BM, Thor A, Waldman FM. 1997. Genetic alterations in primary breast cancers and their metastases: Direct comparison using modified comparative genomic hybridization. Genes Chromosomes Cancer 19: 267–272. [DOI] [PubMed] [Google Scholar]
- Nusse R, van Ooyen A, Cox D, Fung YK, Varmus H. 1984. Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. Nature 307: 131–136. [DOI] [PubMed] [Google Scholar]
- Pattabiraman DR, Bierie B, Kober KI, Thiru P, Krall JA, Zill C, Reinhardt F, Tam WL, Weinberg RA. 2016. Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science 351: aad3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ, et al. 2016. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun 7: 11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferle AD, Spike BT, Wahl GM, Perou CM. 2015. Luminal progenitor and fetal mammary stem cell expression features predict breast tumor response to neoadjuvant chemotherapy. Breast Cancer Res Treat 149: 425–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K. 2014. Tumor heterogeneity confounds and illuminates: A case for Darwinian tumor evolution. Nat Med 20: 344–346. [DOI] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. 2009. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat Rev Cancer 9: 265–273. [DOI] [PubMed] [Google Scholar]
- Prat A, Perou CM. 2009. Mammary development meets cancer genomics. Nat Med 15: 842–844. [DOI] [PubMed] [Google Scholar]
- Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. 2010. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 12: R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. 2008. Efficient tumour formation by single human melanoma cells. Nature 456: 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Clevers H. 2005. Wnt signalling in stem cells and cancer. Nature 434: 843–850. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. 2001. Stem cells, cancer, and cancer stem cells. Nature 414: 105–111. [DOI] [PubMed] [Google Scholar]
- Roberts PJ, Usary JE, Darr DB, Dillon PM, Pfefferle AD, Whittle MC, Duncan JS, Johnson SM, Combest AJ, Jin J, et al. 2012. Combined PI3K/mTOR and MEK inhibition provides broad antitumor activity in faithful murine cancer models. Clin Cancer Res 18: 5290–5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JM, Panzilius E, Bartsch HS, Irmler M, Beckers J, Kari V, Linnemann JR, Dragoi D, Hirschi B, Kloos UJ, et al. 2015. Stem-cell-like properties and epithelial plasticity arise as stable traits after transient Twist1 activation. Cell Rep 10: 131–139. [DOI] [PubMed] [Google Scholar]
- Schmidt-Kittler O, Ragg T, Daskalakis A, Granzow M, Ahr A, Blankenstein TJ, Kaufmann M, Diebold J, Arnholdt H, Muller P, et al. 2003. From latent disseminated cells to overt metastasis: Genetic analysis of systemic breast cancer progression. Proc Natl Acad Sci 100: 7737–7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, et al. 2009. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461: 809–813. [DOI] [PubMed] [Google Scholar]
- Shibata M, Shen MM. 2013. The roots of cancer: Stem cells and the basis for tumor heterogeneity. BioEssays 35: 253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. 2003. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci 100: 8418–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Subedee A, Bloushtain-Qimron N, Savova V, Krzystanek M, Li L, Marusyk A, Tabassum DP, Zak A, Flacker MJ, et al. 2015. Somatic cell fusions reveal extensive heterogeneity in basal-like breast cancer. Cell Rep 11: 1549–1563. [DOI] [PubMed] [Google Scholar]
- Tabassum DP, Polyak K. 2015. Tumorigenesis: It takes a village. Nat Rev Cancer 15: 473–483. [DOI] [PubMed] [Google Scholar]
- Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, Hartwell K, Onder TT, Gupta PB, Evans KW, et al. 2010. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci 107: 15449–15454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP. 2003. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol 15: 740–746. [DOI] [PubMed] [Google Scholar]
- Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. 2012. Spatiotemporal regulation of epithelial–mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 22: 725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner NC, Reis-Filho JS. 2012. Genetic heterogeneity and cancer drug resistance. Lancet Oncol 13: e178–185. [DOI] [PubMed] [Google Scholar]
- Vaillant F, Asselin-Labat ML, Shackleton M, Forrest NC, Lindeman GJ, Visvader JE. 2008. The mammary progenitor marker CD61/β3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res 68: 7711–7717. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, Sharma N, Dekoninck S, Blanpain C. 2011. Distinct stem cells contribute to mammary gland development and maintenance. Nature 479: 189–193. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Lee MY, Ousset M, Brohee S, Rorive S, Giraddi RR, Wuidart A, Bouvencourt G, Dubois C, Salmon I, et al. 2015. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature 525: 119–123. [DOI] [PubMed] [Google Scholar]
- Vega S, Morales AV, Ocana OH, Valdes F, Fabregat I, Nieto MA. 2004. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev 18: 1131–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Hoff DD, Forseth BJ, Huong M, Buchok JB, Lathan B. 1986. Improved plating efficiencies for human tumors cloned in capillary tubes versus Petri dishes. Cancer Res 46: 4012–4017. [PubMed] [Google Scholar]
- Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P, Aas T, Alexandrov LB, Larsimont D, Davies H, et al. 2015. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 21: 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang CZ, Wala J, Mermel CH, et al. 2013. Pan-cancer patterns of somatic copy number alteration. Nat Genet 45: 1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Behbod F, Atkinson RL, Landis MD, Kittrell F, Edwards D, Medina D, Tsimelzon A, Hilsenbeck S, Green JE, et al. 2008. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res 68: 4674–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Atkinson RL, Rosen JM. 2010. Selective targeting of radiation-resistant tumor-initiating cells. Proc Natl Acad Sci 107: 3522–3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Tsimelzon A, Chang CH, Fan C, Wolff A, Perou CM, Hilsenbeck SG, Rosen JM. 2015. Intratumoral heterogeneity in a Trp53-null mouse model of human breast cancer. Cancer Discov 5: 520–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Ying H, Wiedemeyer R, Yan H, Quayle SN, Ivanova EV, Paik JH, Zhang H, Xiao Y, Perry SR, et al. 2010. PLAGL2 regulates Wnt signaling to impede differentiation in neural stem cells and gliomas. Cancer Cell 17: 497–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, Kalluri R. 2015. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527: 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]