Abstract
Genetic or genomic mutation is a major cause of intellectual disability (ID). However, despite the generally anticipated strong genotype/phenotype correlation for ID, there are huge obstacles to gene identification, except perhaps where very distinct syndromic features are observed, because of the high degree of genetic heterogeneity and wide variability of phenotype for different mutations or even with the same mutation within a single gene. A recent review estimates in excess of 2500 genes for ID. Fortunately for researchers and diagnosticians alike, the recent advent of massively parallel sequencing technologies, or next-generation sequencing (NGS) has made an apparently impossible task tractable. Here, we review the ongoing research endeavors to identify new disease genes, as well as strategies and approaches at the clinical level.
The majority of genes that cause intellectual disabilities (perhaps >2500) have remained unidentified. But advances in next-generation sequencing are simplifying this task and may lead to improved diagnostics and clinical care.
About 1% of children worldwide are affected by intellectual disability (ID) (Maulik et al. 2011), often also referred to as mental retardation, which can have a devastating impact on many aspects of the lives of the affected individuals, their families and communities, and is a major challenge at the clinical level. The clinical presentation and etiology of ID is complex with a high degree of heterogeneity, leading to a poor rate of molecular diagnosis resulting in below satisfactory clinical management. ID is characterized by an intelligence quotient (IQ) of 70 or below, and deficits in two or more behaviors related to adaptive functioning with diagnosis by 18 years of age. ID is present in every social class and culture (Leonard and Wen 2002); however, despite its global occurrence, there tends to be a higher prevalence of ID in regions of lower socioeconomic status (using the lack of maternal education as an indicator) and in developing countries, and in particular for milder cases (Drews et al. 1995; Roeleveld et al. 1997; Durkin et al. 1998; Durkin 2002; Emerson 2007). It has been posited that the main reason for this discrepancy is likely caused by environmental factors, in particular, perinatal difficulties, neonatal infections, postnatal brain infections, as well as childhood malnourishment (Roeleveld et al. 1997; Durkin et al. 1998; Emerson 2007). The level of parental consanguinity in specific populations is also considered likely to play a role (Durkin et al. 1998). It is important to note that consanguineous marriages are known to lead to a marked increase in frequency of recessive disorders (Bundey and Alam 1992; Modell and Darr 2002).
Approximately 30% more males than females are diagnosed with ID (McLaren and Bryson 1987; American Psychiatric Association 2000). However, despite the higher prevalence of males than females among milder cases of ID, the ratio actually decreases with decrease in IQ (McLaren and Bryson 1987; American Psychiatric Association 2000). Some studies suggest that severe ID is more prevalent among females (Katusic et al. 1996; Bradley et al. 2002); however, these studies, performed in specific communities, may not necessarily be generalizable to the rest of the world.
Although recent advances in sequencing technology have accelerated the rate of gene discovery for ID, even where family sizes are small, the vast majority of ID genes remain undetected. ID is frequently divided into two groups: nonsyndromic and syndromic ID. A recent review listed 40 known genes for nonsyndromic autosomal recessive ID (NS-ARID), but estimates that there may be in excess of 2500 autosomal ID genes in total, with the majority being recessive (Musante and Ropers 2014). Also, this review estimated that 13%–24% of ID cases in Europe are likely attributed to autosomal recessive causes (Musante and Ropers 2014).
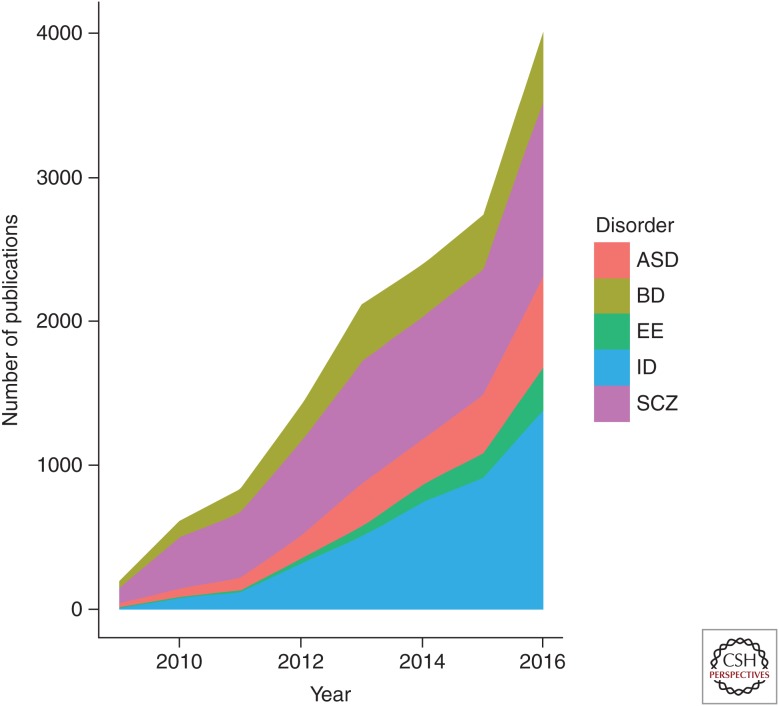
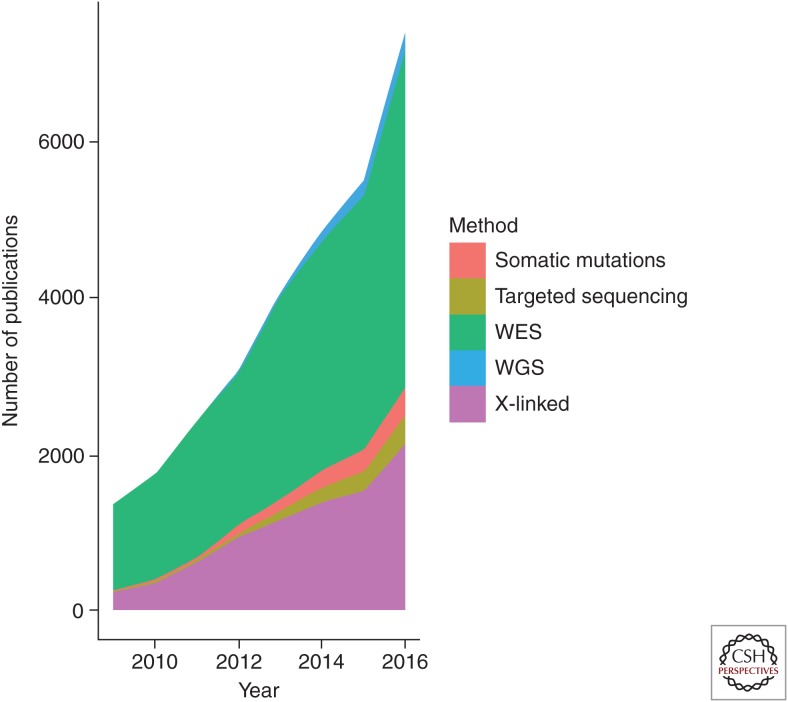
With the advent of massively parallel DNA sequencing, otherwise known as next-generation sequencing (NGS), the identification of genes and mutations causing ID has taken a massive leap forward. Additionally, the potential for using NGS in a clinical setting opens up the possibilities of uncovering the genetic contribution for a large percentage of ID individuals at first onset of symptoms, and hopefully opening up avenues for early therapeutic interventions. There are more than double the number of published studies on NGS and ID than for NGS and epilepsy, NGS and autism, or NGS and other neuropsychiatric disorders (see Fig. 1), most likely reflecting a greater potential usage or uptake of NGS approaches for the identification of new genetic causes of ID. A breakdown of a sequencing strategy in ID is provided in Figure 2.
Figure 1.
Number of publications in PubMed using the search terms “next-generation sequencing,” “autism spectrum disorder” (ASD), “intellectual disability” (ID), “epileptic encephalopathy” (EE), “schizophrenia” (SCZ), and “bipolar disorder” (BD) annually since 2009. For 2016, as PubMed was accessed on August 6, 2016 and thus represents an incomplete year, a factor of 2 was multiplied as an adjustment factor.
Figure 2.
Number of hits or publications per year in Google Scholar broken down into sequencing strategy, using the search terms “intellectual disability” + “whole-exome sequencing” (labeled WES), “intellectual disability” + “whole-genome sequencing” (labeled WGS), “intellectual disability” + “targeted sequencing” (labeled “Targeted sequencing”), “intellectual disability” + “somatic mutations” (labeled “Somatic mutations”), and “intellectual disability” + “X-linked” + “sequencing” (labeled “X-linked”).
PRE-NGS HIGH-THROUGHPUT SEQUENCING RESEARCH FOR GENE IDENTIFICATION
Large-scale sequencing attempts for gene/mutation screening for ID did not begin with NGS, but, for the obvious reasons of cost and labor, were limited in capacity and few and far between. A few examples are described here.
Hamdan et al. (2011) Sanger-sequenced 197 genes involved in glutamate neurotransmission or associated with the glutamatergic synaptic machinery in 95 sporadic cases of nonsyndromic ID (NSID), finding an excess of de novo deleterious mutations, including de novo truncating mutations in SYNGAP1, STXBP1, and SHANK3. Another large-scale Sanger sequencing project involved all coding genes (N = 829) on the X chromosome in 208 families with suspected X-linked ID (XLID), predominantly nonsyndromic (Tarpey et al. 2009). Nine new genes implicated in XLID were reported, including the gene encoding the presynaptic protein CASK. Earlier, smaller-scale sequencing efforts were also performed, for instance, the screening of 47 X-chromosomal genes within a 7.4-Mb region of Xp11 and selected based on brain expression profile in 22 XLID families (Jensen et al. 2007). This study reported four new genes for XLID; however, a microarray hybridization approach was used for mutation screening rather than Sanger sequencing, but still not a massively parallel sequencing approach that would be considered as “next-generation.”
These studies provided a taste of what was to come, but also with some warning as to the issues that would be faced with massively parallel sequencing. In the Tarpey et al. (2009) study, truncating mutations were identified in 19 genes that turned out not to be disease-related, either because they were present in the control population or did not segregate with ID in the families. Large numbers of missense changes were also identified, and the caution in interpretation for these has to be even greater. Currently, we have the advantage of large databases of control exome or genome data with which to compare variants in affected individuals, for example, ExAC, NHLBI ESP, and 1000 Genomes. Data from these can be used to exclude variants if too common (or present as hemizgotes or homozygotes for X and autosomes, respectively) in the controls.
X CHROMOSOME–TARGETED SEQUENCING STUDIES
The X chromosome was for a long time assumed to be associated with ID, in part because males are more frequently affected with ID than females. It is estimated that XLID accounts for 10%–12% of ID in males, which explains why the majority of research for many years focused on the X chromosome, resulting in the identification of >100 ID genes on the X chromosome. There are still many likely X-linked cases in which there is no molecular diagnosis. Hu et al. (2015) performed a study on 405 families, which had clear familial evidence of XLID. These families were termed unresolved because despite microarray-based copy number variation (CNV) analysis and Sanger sequencing of a subset of XLID genes there was no definitive cause of ID. Hu et al. (2015) then proceeded to massively parallel sequence 745 X-chromosomal genes to determine whether there are other genes present in these families that cause ID. They found that 18% (74 of the 405 families) carried variants in previously identified XLID genes that were determined to be causative. The study also identified seven novel ID genes in 2% of their cohort such as CLCN4, CNKSR2, FRMPD4, KLHL15, LAS1L, RLIM, and USP27X. The CLCN4 gene had protein truncating and missense variants in five families. Electrophysiological studies in Xenopus laevis showed that these mutations impaired the function of the protein and resulted in impaired neuronal differentiation. The CNSKR and FRMPD4 proteins were determined to interact with PSD95, which acts as a major scaffolding protein at the postsynaptic density. Their results provided a molecular diagnosis for the families involved; however, approximately half of XLID cases remain unresolved. Another frontier will be to look at the noncoding regions on the X chromosome for regulatory variants, which may cause disease. This study also shows that even in a well-characterized chromosome that has been long associated with ID, there are still many loci/genes to be found related to ID, emphasizing the inherent genetic heterogeneity in ID. X-chromosomal inheritance, however, does not explain a large portion of the ID cases and, thus, the autosomes need further study in ID research.
TARGETED SEQUENCING STUDIES
XLID has been a major focus in ID research but XLID only represents 10%–12% of patients with ID (Ropers and Hamel 2005). It has become apparent that autosomal forms of ID are more common than previously appreciated. Autosomal recessive intellectual disability (ARID) has not been studied as extensively because Western countries tend to have smaller families and fewer consanguineous marriages. This has led to a gap in knowledge regarding the role recessive variants play in ID.
Najmabadi et al. (2011) sought to identify ARID genes by mapping 272 consanguineous Iranian families and determining the single linkage interval regions, to design a custom sequence capture system for these regions. Subsequently, 136 consanguineous families were sequenced using these custom-generated gene panels, which covered the genes in the single interval regions mapped in the consanguineous families.
The study identified 50 novel genes for ARID as well as 26 families that harbor known mostly syndromic forms of ARID. The study also noted that only two of the novel genes were found in more than one family, which highlights the heterogeneity of ARID genes. As with XLID, identification of multiple ID families with mutations in a specific gene is important for validation as an ARID gene, and many of these genes are still awaiting supporting evidence from additional families. The variants detected for this cohort ranged from housekeeping genes such as LARP7 (a negative transcriptional regulator of polymerase II) to variants in genes with no obvious link with ID, including ADK, which causes adenosine kinase deficiency. Several variants detected in the Najmabadi et al. (2011) study also had associations with brain-specific functions including CACNA1G (a T-type calcium channel protein) and ZBTB40, which has a role in glia differentiation.
This targeted approach has proven useful in identifying novel and rare variants associated with ID, and implicating new ID genes. It also has the benefit of reducing the cost of the massively parallel sequencing approach while increasing the number of samples screened. This approach, however, is biased to the targeted capture regions, and relies on having consanguineous families, preferably large and with microarray data, and the good fortune to identify clear autozygous (homozygous-by-descent) regions. This bias may result in ID genes or unexpected variants being overlooked in some instances.
A recent study focused on targeted sequencing of 107 XLID genes in 50 patients with a family history suggestive of XLID, and in 100 apparently sporadic ID cases (Tzschach et al. 2015). In 13 (26%) of the familial cases, likely pathogenic mutations were identified. However, only five pathogenic mutations were identified in the 100 sporadic patients. Targeted sequencing approaches, however, are biased toward the selected regions, whereas an unbiased approach may be required for more exploratory studies such as for the identification of new disease genes.
WHOLE-EXOME SEQUENCING FOR GENE IDENTIFICATION
Whole-exome sequencing (WES) targets only the 2% of the genome that codes for proteins. Most disease-causing variants identified thus far occur within the protein-coding regions of the genome, in which variants are more likely to alter the protein’s structure, thus results in dysfunction. This approach is also popular owing to the difficulty in interpreting noncoding variants. WES has become an excellent research and diagnostic tool that balances cost with disease-related information. Alongside pedigree and familial data, WES of mother/father/proband trios can be used to detect variants from different modes of inheritance as well as sporadic de novo instances of disease mutations in an unbiased fashion.
De novo mutations have been found to be more enriched in patients with ID than in control populations. In Western countries, where environmental and familial causes of ID are not obvious, de novo mutations may pose a plausible genetic model in many cases. Because of the lack of diagnostic, familial, and environmental clues, de Ligt et al. (2012) focused primarily on de novo mutations in their study. After an assessment by clinical geneticists, 100 patients who lacked any diagnosis by CNV analysis were enrolled in this study. Both parents and probands were sequenced and the data was filtered based on mode of inheritance and predicted pathogenicity of variants (based on algorithms such as SIFT and PolyPhen, as well as gene ontology). De novo mutations were identified in 53% of the patients sequenced and offered conclusive diagnosis for 13% of the patients sequenced. An additional 3% were found to be X-linked, and de novo mutations were found in 22 patients, with three being confirmed as having characteristic phenotypes among the confirmation series of 765 ID patients. All screening was performed with high-throughput sequencing, and variant prioritization as outlined by de Ligt et al. (2012). Some variants discovered in this study can lead to treatments such as antiepileptic and avoidance of sodium channel blockers for a patient with SCNA2 de novo mutations or dietary considerations for a patient with a PDHA1 mutation. Clinical evaluation and counseling was offered to families once the study was completed.
This study showcased the usefulness of unbiased sequencing as a potential diagnostic tool for ID and other complex heterogeneous disorders. A robust, scalable, and integrated method for identifying causal variants, managing the data, and disseminating the results as a national health care initiative is needed. The logistics of this type of undertaking are daunting especially when considering the scalable application of ethics, counseling, and follow-up of patients afflicted with ID.
The use of WES in clinical practice has raised many questions concerning the appropriate methods to sift through genomic data for relevant pathogenic variants and the ethical delivery of results to patients and families in light of incidental findings. The Deciphering Developmental Disorders (DDD) group is a collaborative group in the United Kingdom that aims to translate high-resolution genomic and phenotypic information into the National Health Service (NHS) (Wright et al. 2015). In conjunction with the UK NHS and the Irish Regional Genetics Service, Wright et al. (2015) recruited ∼2000 families in the first year. Another 8000 families were recruited within 3 years. This study made use of the first 1133 family trios collected and integrated recruitment, sample tracking, data generation, analysis, variant filtration, and clinical feedback to patients with the approval of the UK Research Ethics Committee. A team of research coordinators was essential in procuring informed consent, and clinical geneticists entered phenotypic data after detailed assessments. After sample collection, whole-genome microarrays were used for CNV detection and whole-exome sequencing (WES) was performed on both parents and the proband. Variants detected in exome sequencing were disregarded if they were common (minor allele frequency >1%) and unlikely to disrupt protein function (e.g., synonymous changes filtered out). The pathogenic variants are then compared with the Developmental Disorders Genotype to Phenotype (DDG2P) in-house database. This database contains >1000 genes that are continually being updated as patients are recruited and scientific literature advances.
Clinical results are evaluated by a multidisciplinary team including clinical geneticists and genetic scientists in a weekly meeting. Results are provided to families and then are deposited on the DECIPHER database. In total, from ∼80,000 genomic variants identified, 400 rare variants were predicted to be causal protein-altering variants. The use of trios reduced the number of putative causal variants by 10-fold and allowed a diagnostic yield of 27% by focusing on segregating and de novo variants.
Despite these advances, the majority of cases of ID remain with no identified genetic cause. WES represents 2% of the genome, but extending the unbiased approach of high-throughput sequencing has proven informative. With advances in whole-genome sequencing (WGS), ID research may benefit from interrogating the noncoding regions of the genome.
WHOLE-GENOME SEQUENCING FOR GENE IDENTIFICATION
WGS is the most comprehensive diagnostic test to date because it sequences the entire genome of an organism. Gilissen et al. (2014) recently sequenced 50 patients with severe ID and their parents, who have already undergone a comprehensive diagnostic workup including targeted sequencing, genomic microarray, and WES with no molecular diagnosis yet established.
Gilissen et al. (2014) chose to focus on de novo CNVs and single-nucleotide variants (SNVs) because of their known importance in ID, and discovered on average 84 de novo CNVs and 82 de novo SNVs per genome. This accords well with previous studies. The investigators found that there was a significant enrichment for loss-of-function (LoF) mutations, and mutations in known ID genes. WGS also allowed for the detection of structural variants and indels that may be outside the exons of ID genes. This was illustrated in a patient with a partial duplication of TENM3 within the known ID gene IQSEC2. This duplication led to a gene fusion product that disrupted the function of IQSEC2, and was confirmed with RNA studies. Interestingly, three of the 10 de novo SNVs were identified in a mosaic state in patients. WGS, as long as depth of coverage is sufficient, allows for the detection of mosaic variations as seen in a fraction of the reads from sequencing.
This study conclusively diagnosed 21 out of the 50 patients, resulting in a diagnostic yield of 42%. Gilissen et al. (2014) attempted to systematically characterize variants by selecting high confidence variants located in promoter, introns, and untranslated regions, such as those described by the ENCODE project (ENCODE Project Consortium 2012), and validated 43 variants by Sanger sequencing. The ENCODE project provided a rich set of resources for transcription factor-binding sites and chromatin state segments in nine different human cell types. However, data mining using these resources still yielded no pathogenic information about the noncoding variants identified in this study. Scientific understanding of noncoding regulatory variation is still in its infancy, and rigorous functional work will be needed to understand the role of noncoding genetic variation in disease.
There has been recent success in understanding regulatory variation in the human genome through computational techniques such as deep neural network learning algorithms and experimental functional work. Huang et al. (2012) used targeted massively paralleled sequencing to identify a noncoding, regulatory variant in the known ID gene HCFC1. Because no coding mutations had been identified despite the solid XLID mapping evidence, the investigators hypothesized that the variant may reside outside of the coding region on the X chromosome. After extensive functional work, it was determined that this variant disrupted the YY1 transcription factor-binding site. These investigators also experimentally validated their claims to show that this variant reduced neurite growth and increased astrocyte production. Two further families with ID were also found to harbor this mutation.
WGS is the most exhaustive and complete diagnostic test available but it is hampered by the high cost of sequencing and difficulties in data analysis, management, and storage. The real strengths of WGS are the sequencing of the noncoding regulatory regions of the genome, also the much greater ability to identify CNVs in comparison to WES data. WGS is still a cutting-edge technology and techniques to analyze WGS data are continually being developed, but careful validation is required for clinical diagnostic implementation.
COPY NUMBER VARIATION DETECTION THROUGH NGS FOR GENE IDENTIFICATION
One of the major areas of development in NGS analysis is the detection of CNV. CNV analysis is still an active area of research in NGS variant analysis and has long been important in ID research.
Iqbal et al. (2012) encountered a case involving a patient with what they believed was a novel form of NSID from a consanguineous family in Pakistan. The family had microarray single nucleotide polymorphism (SNP) genotyping performed and there was a homozygous ∼100-kb deletion on 2q37.1 found that did not segregate with the rest of the family. After homozygosity mapping, four autozygous regions were identified, but no obvious candidate genes within these regions. A targeted panel, designed for these homozygous regions was developed and the entire family was sequenced. There was a five-exon deletion in the TPO gene present, which did segregate with the family. Reanalysis of the SNP microarray data (Affymetrix 250K) revealed that there was only a single SNP within this deletion region, and thus beyond the limits of CNV-detection algorithms. This deletion could potentially have been identified using higher-density SNP arrays, but in this instance targeted exonic NGS provided the resolution required for detection. Subsequent validation was confirmed with qPCR. Iqbal et al. (2012) proved that the unbiased approach of NGS can be used to detect causal CNV variants for ID.
CNV analysis is routinely performed with CGH microarrays in diagnostic laboratories because of its low cost, robustness, and reliability. The major drawback of this approach is that CGH microarrays can only detect unbalanced structural variants. “Apparently balanced chromosomal rearrangements” (ABCRs) occur in 1.54% of live births and contribute to 6% of abnormal phenotypes including multiple congenital abnormalities and ID (MCA/ID). Previous methods such as break point cloning using FISH and BAC, Southern Blot, inverse PCR, and long-range PCR are time-consuming, labor-intensive, and may not provide the desired resolution required for clinical diagnostics. Because of the above reasons, this area remains under-investigated.
Schluth-Bolard et al. (2013) used WGS to characterize ABCR in four patients where CGH arrays had provided no diagnosis. NGS identified ABCR in three of the four patients including all breakpoints, inversions, and balanced translocations. In three of the four patients, disruption of genes (SHANK2, TCF4, PPFIA1, RAB19, and KCNQ1) was the probable cause of MCA/ID. The method was also validated with FISH and PCR followed by Sanger sequencing. The investigators concluded that WGS is a reliable method for breakpoint mapping, but capture methods may have lower coverage around ABCR because they often occur near repetitive sequences, which are difficult to map back to the reference sequence.
Recently, many software programs have been developed to accurately detect CNVs and other structural variations such as CONIFER (Krumm et al. 2012; O’Roak et al. 2012) and XHMM (Fromer et al. 2012; Poultney et al. 2013; Fromer and Purcell 2014). It is an active area of research and large cohort studies need to be performed to determine the clinical relevance of NGS structural variation identification for ID.
NGS STRATEGIES FOR CLINICAL DIAGNOSTICS
Because of a higher diagnostic yield and reduction in costs, NGS is increasingly being deployed in clinical laboratories for the diagnosis of genetic syndromes and cancer (Yang et al. 2013; Omoyinmi et al. 2014). For individuals with ID, chromosomal microarray analysis (CMA) is recommended as the first-line diagnostic test as it offers much higher diagnostic yield (15%–20%) compared with G-banded karyotype analysis (3%) (Miller et al. 2010). However, the genetic etiology of 80% to 85% of patients still remains unknown. For these cases, NGS-based testing (targeted multigene panels, WES or WGS) has a great potential to achieve diagnosis. A recent study has shown a diagnostic yield of ∼28% in patients affected with developmental delay using clinical WES (Lee et al. 2014). In another study, using clinical WES, molecular diagnosis was archived in ∼25% cases with neurological phenotypes (Yang et al. 2014). Using the NGS-based, targeted gene panel approach, Tan et al. has identified the disease-causing variants in 21% of the ID cases (Tan et al. 2015). These studies show the clinical use of NGS-based tests. Although targeted gene panels and WES are the commonly used NGS tests in clinical laboratories, WGS offers additional benefits as it has the potential to uncover all forms of genetic variation in one test (Pang et al. 2014) and, consequently, it potentially offers a higher diagnostic yield. In a recent study of patients with severe ID, a diagnostic yield of 42% was observed, which is a significant improvement over diagnostic yield obtained by microarray, gene panels, or WES (Gilissen et al. 2014).
These observations indicate that NGS will undoubtedly have a remarkable impact for patients with ID and other genetic disorders. However, the widespread adoption of NGS technologies in diagnostic settings is hindered by several factors such as cost, processing time, clinical interpretation, and storage of an enormous amount of data. The American College of Medical Genetics and Genomics has issued the professional standards and guidelines for implementation of NGS in clinical laboratories (Rehm et al. 2013). In the light of these guidelines, Patwardhan et al. (2015) have evaluated the performance of the commercially available exome-capture methods and sequencing technologies. Insufficient reads coverage was observed across several disease-associated genes using each of the conventional exome capture and whole-genome platforms. These findings highlight the need for further improvements in the exome capture methods, sequencing technologies, and computational algorithms to improve the accuracy and sensitivity of NGS testing.
Finally, ethical concerns need also be considered for the application of NGS to ID in both clinical and research settings, in particular, the possibility of incidental findings as had been discussed in many articles (e.g., see Knoppers et al. 2014; Howard et al. 2015). For gene-panel approaches, issues of incidental findings are obviously much less of a concern. Also, for trio-based NGS efforts aimed strictly at identifying de novo mutations, the concerns would possibly also be lower.
SUMMARY
To maximize the usage of NGS for ID, it is of the utmost importance to make variant data available for WES/WGS of ID cases, as well as for sufficient numbers of controls of all ethnicities. Without cooperation and corroboration across studies, evidence for the accumulation of variants that may be necessary to implicate genes as being associated with disease may be missed, and potentially etiological variants may remain as variants of unknown significance. The identification of compound heterozygous mutations is particularly problematic, as analytic pipelines are not currently set up to determine whether genes containing variants are likely to be recessive or dominant; also without haplotype information (unless haploid DNA NGS is performed, haplotype is determined independently by genotyping or from sequencing parents, or if the two variants occur within the same read) it is impossible to know whether two mutations in the same gene occur on the same (cis) or different (trans) alleles. To help address this, first, we need greater effort in identifying autosomal recessive ID genes using consanguineous families, and, second, we need WGS/WES data from ID cohorts to be made available to researchers to enable cross-referencing between family studies and studies of unrelated individuals. A number of efforts are underway to share NGS data for ID. For instance, data from DECIPHER and UK10K studies of ID patients are available to some degree. Matchmaker Exchange (matchmakerexchange.org/) provides a mechanism for matching studies that have identified potential etiological variants in the same gene. In addition, groups using consanguineous families to identify autosomal recessive ID genes have come together to share data under the umbrella of the Consortium for Autosomal Recessive ID (CARID), hosted by Rami Abou Jamra at Erlangen University, Germany.
Possible overlap of genetic etiology between ID and other neuropsychiatric disorders also needs to be considered. This is particularly relevant for autism spectrum disorders (ASDs), as estimates of comorbidity are high. ∼28% of ID individuals may meet criteria for ASD (Bryson et al. 2008). Estimates of ASD individuals with some degree of ID are frequently in the range of 40%–70% (71% [Chakrabarti and Fombonne 2001], 63% [Bertrand et al. 2001], 40% [Baird et al. 2006], and 50% [Charman et al. 2011]). Many genes have already been shown to cause both ID and/or ASD, including PTCHD1, SHANK3, NLGN4, NRXN1, CNTNAP2, UBE3A, NF1, TSC1, TSC2, FMR1, MECP2, and others. There is also increasing evidence that this may be true for other disorders such as schizophrenia and bipolar disorder. For example, McCarthy et al. (2014) reported a de novo missense mutation, Arg190Cys, in MECP2. This mutation, while outside the methyl CpG–binding domain, is located within the so-called intervening domain but at a highly conserved residue within an AT-hook motif. This motif is a cationic peptide with affinity for AT-rich DNA. Although AT-hooks have weak affinity individually, the cooperative action of multiple AT-hooks can achieve high DNA-binding affinity (Geierstanger et al. 1994). Arg190Cys disrupts one of the canonical motif residues, replacing a positively charged amino acid with a neutral polar residue. Thus, this de novo mutation in a known ID gene is almost certainly disrupting the protein function, and reaffirms the link between this ID gene and psychosis previously reported for Ala140Val (Cohen et al. 2002) and many others. McCarthy et al. (2014) also reported de novo mutations in schizophrenia patients in other ID genes such as TRAPPC9 and HUWE1. Additionally, Heidari et al. (2015) reporting the use of WES to identify missense mutations in the histamine N-methyltransferase gene HNMT as a cause of nonsyndromic autosomal recessive ID, also made the point that rare heterozygous loss CNVs or LoF mutations disrupting HNMT have also been reported for bipolar disorder (Zhang et al. 2009), autism spectrum disorder (DECIPHER), and for schizophrenia (Genebook). The issue of pleiotropy across ID and neuropsychiatric disorders was raised in a recent publication by Li et al. (2015). In this research article, trio sequence data was analyzed across ID, ASD, schizophrenia (SCZ) and epileptic encephalopathy (EE) cohorts and genes harboring de novo mutations were compared. Significant numbers of genes harboring de novo mutations were shared between ASD and SCZ, also between ASD and ID, between ID and EE, but more surprisingly between ID and SCZ (e.g., genes such as POGZ, MYH9, LRP1, STAG1, SYNGAP1, GRIN2A, [the aforementioned] MECP2, and TANC2). Interestingly, de novo mutations in the EE gene SCN2A (OMIM 182390) were present in all four cohorts. The investigators discuss the possible overlap in genetic architecture and biological pathways, despite the apparent distinct pathogenesis for these disorders. In another review article, LoF mutations shared across different neurodevelopmental or neuropsychiatric disorders are discussed (Vissers et al. 2015). In addition to LoF mutations in many genes shared between ASD and ID, the review lists LoF mutations in AUTS2 in both ID and SCZ, and in POGZ, SCN2A, and SYNGAP1 in ID, SCZ, and ASD.
The vast majority of individuals with ID currently receive no molecular diagnosis—a shortcoming that significantly impacts health and life span. Research suggests that individuals with ID die on average 15 years younger than people without ID (Glover and Ayub 2010). There is also a strongly negative correlation of survival with severity of ID (Bittles et al. 2002). Knowing which genes carry mutations that cause ID can have huge benefits for diagnosis in clinics, can lead to better understanding of each patient’s health issues, more appropriate care and treatment, improved overall health and life span, and appropriate counseling and planning for families. As a stark example of this, the average life span for individuals with the most recognized and most common known form of ID (1/650–1000 births), Down syndrome, has increased from 12 to 60 years over the last two generations (reviewed in Bittles et al. 2002, 2007). For many other genetic forms of ID, there is insufficient data on life expectancy to determine the improvement since the discovery of the specific genetic cause; however, it has been estimated that the mean age of death for ID in general has increased from 19 years in the 1930s to 66 years in the 1990s (reviewed in Coppus 2013). A significant proportion of this increase is likely attributed to improved health management resulting from knowledge of the underlying genetic etiology. Increases in the rate of gene and mutation identification for ID through NGS should add significantly to this improvement. Also, along with gene identification, we hope to achieve a better understanding of the pathophysiology, which would, one would hope, ultimately lead to treatments and cures. Finally, to quote J.B.S. Haldane’s preface to The Biology of Mental Defect by Penrose (1949), “We cannot do so much about mental defect as had been hoped in the recent past. Nor could we do as much about flying four hundred years ago as Leonardo da Vinci had hoped. But there is no reason to hope that one problem is more insoluble than the other… . We do not know what data will be required. But among them is certainly a knowledge of a great deal of normal human genetics.” Penrose himself wished to see each ID individual “as an integral part of the human race in its struggle for evolution and survival, unwittingly yielding up information of the greatest value in the progressive understanding of the biological structure of the whole group” (from Berg 1998). Given our 21st century access to NGS, transgenic animal models, gene-editing technology, stem-cell technology, improved (and personalized) drug design strategies, etc., surely we can and should be able to do so much more than Penrose had ever dared to wish for.
Footnotes
Editors: W. Richard McCombie, Elaine R. Mardis, James A. Knowles, and John D. McPherson
Additional Perspectives on Next-Generation Sequencing in Medicine available at www.perspectivesinmedicine.org
REFERENCES
- American Psychiatric Association. 2000. Diagnostic criteria from DSM-IV-TR, diagnostic and statistical manual of mental disorders, 4th ed American Psychiatric Association, Arlington, VA. [Google Scholar]
- Baird G, Simonoff E, Pickles A, Chandler S, Loucas T, Meldrum D, Charman T. 2006. Prevalence of disorders of the autism spectrum in a population cohort of children in South Thames: The Special Needs and Autism Project (SNAP). Lancet 368: 210–215. [DOI] [PubMed] [Google Scholar]
- Berg J. 1998. Lionel Sharples Penrose (1898–1972): Aspects of the man and his works, with particular reference to his undertakings in the fields of intellectual disability and mental disorder. J Intellect Disabil Res 42: 104–111. [DOI] [PubMed] [Google Scholar]
- Bertrand J, Mars A, Boyle C, Bove F, Yeargin-Allsopp M, Decoufle P. 2001. Prevalence of autism in a United States population: The Brick Township, New Jersey, investigation. Pediatrics 108: 1155–1161. [DOI] [PubMed] [Google Scholar]
- Bittles A, Petterson B, Sullivan S, Hussain R, Glasson E, Montgomery P. 2002. The influence of intellectual disability on life expectancy. J Gerontol A Biol Sci Med Sci 57: M470–M472. [DOI] [PubMed] [Google Scholar]
- Bittles AH, Bower C, Hussain R, Glasson EJ. 2007. The four ages of Down syndrome. Eur J Public Health 17: 221–225. [DOI] [PubMed] [Google Scholar]
- Bradley EA, Thompson A, Bryson SE. 2002. Mental retardation in teenagers: Prevalence data from the Niagara region, Ontario. Can J Psychiatry 47: 652–659. [DOI] [PubMed] [Google Scholar]
- Bryson SE, Bradley EA, Thompson A, Wainwright A. 2008. Prevalence of autism among adolescents with intellectual disabilities. Can J Psychiatry 53: 449–459. [DOI] [PubMed] [Google Scholar]
- Bundey S, Alam H. 1992. A five-year prospective study of the health of children in different ethnic groups, with particular reference to the effect of inbreeding. Eur J Hum Genet 1: 206–219. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S, Fombonne E. 2001. Pervasive developmental disorders in preschool children. JAMA 285: 3093–3099. [DOI] [PubMed] [Google Scholar]
- Charman T, Jones CR, Pickles A, Simonoff E, Baird G, Happé F. 2011. Defining the cognitive phenotype of autism. Brain Res 1380: 10–21. [DOI] [PubMed] [Google Scholar]
- Cohen D, Lazar G, Couvert P, Desportes V, Lippe D, Mazet P, Heron D. 2002. MECP2 mutation in a boy with language disorder and schizophrenia. Am J Psychiatry 159: 148–149. [DOI] [PubMed] [Google Scholar]
- Coppus A. 2013. People with intellectual disability: What do we know about adulthood and life expectancy? Dev Disabil Res Rev 18: 6–16. [DOI] [PubMed] [Google Scholar]
- de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, Vulto-van Silfhout AT, Koolen DA, de Vries P, Gilissen C, et al. 2012. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 367: 1921–1929. [DOI] [PubMed] [Google Scholar]
- Drews CD, Yeargin-Allsopp M, Decoufle P, Murphy CC. 1995. Variation in the influence of selected sociodemographic risk factors for mental retardation. Am J Public Health 85: 329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin M. 2002. The epidemiology of developmental disabilities in low-income countries. Ment Retard Dev Disabil Res Rev 8: 206–211. [DOI] [PubMed] [Google Scholar]
- Durkin MS, Hasan Z, Hasan K. 1998. Prevalence and correlates of mental retardation among children in Karachi, Pakistan. Am J Epidemiol 147: 281–288. [DOI] [PubMed] [Google Scholar]
- Emerson E. 2007. Poverty and people with intellectual disabilities. Ment Retard Dev Disabil Res Rev 13: 107–113. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Purcell SM. 2014. Using XHMM software to detect copy number variation in whole-exome sequencing data. Curr Protoc Hum Genet 81: 7.23.1–7.23.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Moran JL, Chambert K, Banks E, Bergen SE, Ruderfer DM, Handsaker RE, McCarroll SA, O’Donovan MC, Owen MJ, et al. 2012. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet 91: 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geierstanger BH, Mrksich M, Dervan PB, Wemmer DE. 1994. Design of a GC-specific DNA minor groove-binding peptide. Science 266: 646–650. [DOI] [PubMed] [Google Scholar]
- Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BW, Willemsen MH, Kwint M, Janssen IM, Hoischen A, Schenck A, et al. 2014. Genome sequencing identifies major causes of severe intellectual disability. Nature 511: 344–347. [DOI] [PubMed] [Google Scholar]
- Glover G, Ayub M. 2010. How people with learning disabilities die—Improving health and lives. Learning Disabilities Observatory, Durham, UK. [Google Scholar]
- Hamdan FF, Gauthier J, Araki Y, Lin DT, Yoshizawa Y, Higashi K, Park AR, Spiegelman D, Dobrzeniecka S, Piton A, et al. 2011. Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am J Hum Genet 88: 306–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidari A, Tongsook C, Najafipour R, Musante L, Vasli N, Garshasbi M, Hu H, Mittal K, McNaughton AJ, Sritharan K, et al. 2015. Mutations in the histamine N-methyltransferase gene, HNMT, are associated with nonsyndromic autosomal recessive intellectual disability. Hum Mol Genet 24: 5697–5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard HC, Knoppers BM, Cornel MC, Clayton EW, Sénécal K, Borry P. 2015. Whole-genome sequencing in newborn screening? A statement on the continued importance of targeted approaches in newborn screening programmes. Eur J Hum Genet 23: 1593–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Haas SA, Chelly J, Van Esch H, Raynaud M, de Brouwer AP, Weinert S, Froyen G, Frints SG, Laumonnier F, et al. 2015. X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol Psychiatry 21: 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Jolly LA, Willis-Owen S, Gardner A, Kumar R, Douglas E, Shoubridge C, Wieczorek D, Tzschach A, Cohen M, et al. 2012. A noncoding, regulatory mutation implicates HCFC1 in nonsyndromic intellectual disability. Am J Hum Genet 91: 694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal Z, Neveling K, Razzaq A, Shahzad M, Zahoor MY, Qasim M, Gilissen C, Wieskamp N, Kwint MP, Gijsen S, et al. 2012. Targeted next generation sequencing reveals a novel intragenic deletion of the TPO gene in a family with intellectual disability. Arch Med Res 43: 312–316. [DOI] [PubMed] [Google Scholar]
- Jensen LR, Lenzner S, Moser B, Freude K, Tzschach A, Wei C, Fryns JP, Chelly J, Turner G, Moraine C, et al. 2007. X-linked mental retardation: A comprehensive molecular screen of 47 candidate genes from a 7.4 Mb interval in Xp11. Eur J Hum Genet 15: 68–75. [DOI] [PubMed] [Google Scholar]
- Katusic SK, Colligan RC, Beard CM, O’Fallon WM, Bergstralh EJ, Jacobsen SJ, Kurland LT. 1996. Mental retardation in a birth cohort, 1976–1980, Rochester, Minnesota. Am J Ment Retard 100: 335–344. [PubMed] [Google Scholar]
- Knoppers BM, Avard D, Sénécal K, Zawati MnH. 2014. Return of whole-genome sequencing results in paediatric research: A statement of the P3G international paediatrics platform. Eur J Hum Genet 22: 3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm N, Sudmant PH, Ko A, O’Roak BJ, Malig M, Coe BP; NHLBI Exome Sequencing Project, Quinlan AR, Nickerson DA, Eichler EE. 2012. Copy number variation detection and genotyping from exome sequence data. Genome Res 22: 1525–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F, Das K, Toy T, Harry B, Yourshaw M, et al. 2014. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA 312: 1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard H, Wen X. 2002. The epidemiology of mental retardation: Challenges and opportunities in the new millennium. Ment Retard Dev Disabil Res Rev 8: 117–134. [DOI] [PubMed] [Google Scholar]
- Li J, Cai T, Jiang Y, Chen H, He X, Chen C, Li X, Shao Q, Ran X, Li Z, et al. 2015. Genes with de novo mutations are shared by four neuropsychiatric disorders discovered from NPdenovo database. Mol Psychiatry 21: 298. [DOI] [PubMed] [Google Scholar]
- Maulik PK, Mascarenhas MN, Mathers CD, Dua T, Saxena S. 2011. Prevalence of intellectual disability: A meta-analysis of population-based studies. Res Dev Disabil 32: 419–436. [DOI] [PubMed] [Google Scholar]
- McCarthy SE, Gillis J, Kramer M, Lihm J, Yoon S, Berstein Y, Mistry M, Pavlidis P, Solomon R, Ghiban E, et al. 2014. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry 19: 652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren J, Bryson SE. 1987. Review of recent epidemiological studies of mental retardation: Prevalence, associated disorders, and etiology. Am J Ment Retard 92: 243–254. [PubMed] [Google Scholar]
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ, et al. 2010. Consensus statement: Chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 86: 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modell B, Darr A. 2002. Genetic counselling and customary consanguineous marriage. Nat Rev Genet 3: 225–229. [DOI] [PubMed] [Google Scholar]
- Musante L, Ropers HH. 2014. Genetics of recessive cognitive disorders. Trends Genet 30: 32–39. [DOI] [PubMed] [Google Scholar]
- Najmabadi H, Hu H, Garshasbi M, Zemojtel T, Abedini SS, Chen W, Hosseini M, Behjati F, Haas S, Jamali P, et al. 2011. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 478: 57–63. [DOI] [PubMed] [Google Scholar]
- Omoyinmi E, Melo Gomes S, Standing A, Rowczenio DM, Eleftheriou D, Klein N, Aróstegui JI, Lachmann HJ, Hawkins PN, Brogan PA. 2014. Brief report: Whole-exome sequencing revealing somatic NLRP3 mosaicism in a patient with chronic infantile neurologic, cutaneous, articular syndrome. Arthritis Rheumatol 66: 197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. 2012. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485: 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang AW, MacDonald JR, Yuen RK, Hayes VM, Scherer SW. 2014. Performance of high-throughput sequencing for the discovery of genetic variation across the complete size spectrum. G3 (Bethesda) 4: 63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan A, Harris J, Leng N, Bartha G, Church DM, Luo S, Haudenschild C, Pratt M, Zook J, Salit M, et al. 2015. Achieving high-sensitivity for clinical applications using augmented exome sequencing. Genome Med 7: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penrose L. 1949. The biology of mental defect, pp. 109–112. Grune & Stratton, New York. [Google Scholar]
- Poultney CS, Goldberg AP, Drapeau E, Kou Y, Harony-Nicolas H, Kajiwara Y, De Rubeis S, Durand S, Stevens C, Rehnström K, et al. 2013. Identification of small exonic CNV from whole-exome sequence data and application to autism spectrum disorder. Am J Hum Genet 93: 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, Brown KK, Deignan JL, Friez MJ, Funke BH, Hegde MR, Lyon E, et al. 2013. ACMG clinical laboratory standards for next-generation sequencing. Genet Med 15: 733–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeleveld N, Zielhuis GA, Gabreëls F. 1997. The prevalence of mental retardation: A critical review of recent literature. Dev Med Child Neurol 39: 125–132. [DOI] [PubMed] [Google Scholar]
- Ropers HH, Hamel BC. 2005. X-linked mental retardation. Nat Rev Genet 6: 46–57. [DOI] [PubMed] [Google Scholar]
- Schluth-Bolard C, Labalme A, Cordier MP, Till M, Nadeau G, Tevissen H, Lesca G, Boutry-Kryza N, Rossignol S, Rocas D, et al. 2013. Breakpoint mapping by next generation sequencing reveals causative gene disruption in patients carrying apparently balanced chromosome rearrangements with intellectual deficiency and/or congenital malformations. J Med Genet 50: 144–150. [DOI] [PubMed] [Google Scholar]
- Tan CA, Topper S, Del Gaudio D, Nelakuditi V, Shchelochkov O, Nowaczyk MJ, Zeesman S, Brady L, Russell L, Meeks N, et al. 2015. Characterization of patients referred for non-specific intellectual disability testing: The importance of autosomal genes for diagnosis. Clin Genet 10.111/cge.12575. [DOI] [PubMed] [Google Scholar]
- Tarpey PS, Smith R, Pleasance E, Whibley A, Edkins S, Hardy C, O’Meara S, Latimer C, Dicks E, Menzies A, et al. 2009. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet 41: 535–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzschach A, Grasshoff U, Beck-Woedl S, Dufke C, Bauer C, Kehrer M, Evers C, Moog U, Oehl-Jaschkowitz B, Di Donato N, et al. 2015. Next-generation sequencing in X-linked intellectual disability. Eur J Hum Genet 23: 1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LE, Gilissen C, Veltman JA. 2015. Genetic studies in intellectual disability and related disorders. Nat Rev Genet 17: 9–18. [DOI] [PubMed] [Google Scholar]
- Wright CF, Fitzgerald TW, Jones WD, Clayton S, McRae JF, van Kogelenberg M, King DA, Ambridge K, Barrett DM, Bayzetinova T, et al. 2015. Genetic diagnosis of developmental disorders in the DDD study: A scalable analysis of genome-wide research data. Lancet 385: 1305–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, Braxton A, Beuten J, Xia F, Niu Z, et al. 2013. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med 369: 1502–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, Ward P, Braxton A, Wang M, Buhay C, et al. 2014. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 312: 1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Cheng L, Qian Y, Alliey-Rodriguez N, Kelsoe JR, Greenwood T, Nievergelt C, Barrett TB, McKinney R, Schork N, et al. 2009. Singleton deletions throughout the genome increase risk of bipolar disorder. Mol Psychiatry 14: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]