Abstract
Cilia and flagella are microtubule-based organelles whose assembly requires a motile process, known as intraflagellar transport (IFT), to bring tubulin and other components to the distal tip of the growing structure. The IFT system uses a multiprotein complex with components that appear to be specialized for the transport of different sets of cargo proteins. The mechanisms by which cargo is selected for ciliary import and transport by IFT remain an area of active research. The complex dynamics of cilia and flagella are under constant regulation to ensure proper length control, and this regulation appears to involve regulation at the stage of IFT injection into the flagellum, as well as regulation of flagellar disassembly and, possibly, of cargo binding. Cilia and flagella thus represent a convenient model system to study how multiple motile and signaling pathways cooperate to control the assembly and dynamics of a complex cellular structure.
A bidirectional protein transport system inside cilia carries components (e.g., tubulin) from the cell body to the tip and returns the products of turnover back to the cell body. It requires the coordination of many pathways.
1. INTRODUCTION
Cilia are hair-like organelles that protrude from the cell surface in organisms as diverse as single-celled eukaryotes and humans. Depending on the cell type, cilia are sometimes also referred to as “flagella.” These organelles play important roles in the motility of single cells, generating fluid flow and sensing chemical and mechanical cues. Within the human body, cilia are present in almost all cell types and are involved in many areas of physiology and development. It is, therefore, not surprising that defects in cilia can lead to a wide spectrum of diseases, known collectively as “ciliopathies” (Badano et al. 2006). From a more fundamental perspective, the complex structure of the cilium poses exciting challenges for our understanding of molecular assembly processes, and indeed the cilium has long served as a paradigm for understanding the regulation of macromolecular assembly (Randall 1969; Johnson and Rosenbaum 1993; Dutcher 1995). A central feature of these organelles is that they are assembled and maintained by an active motility known as intraflagellar transport (IFT), which appears to select components to assemble, move them to the correct place to assemble, and regulate the rate of assembly to control the size of the final structure.
2. STRUCTURE AND FUNCTION OF CILIA
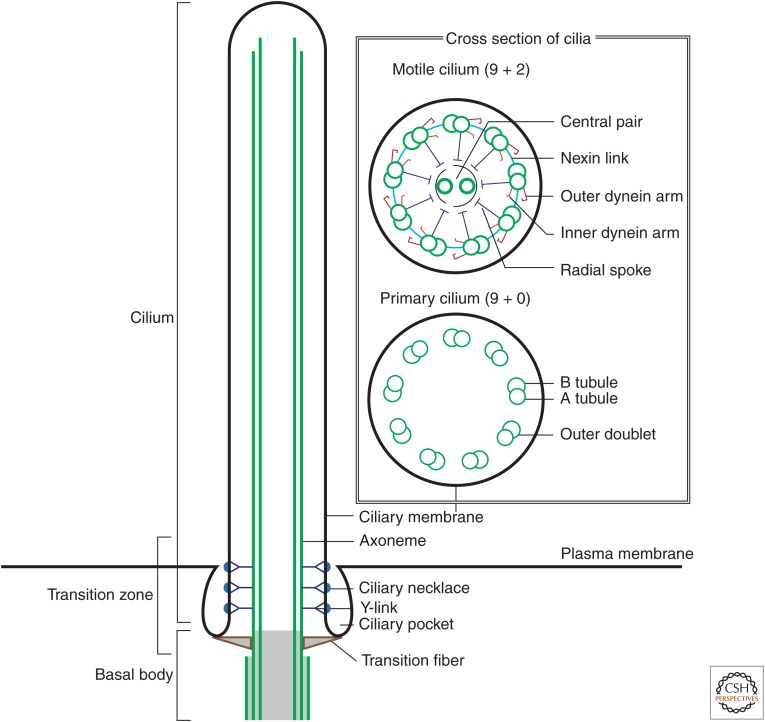
The cilium consists of a microtubule-based core structure—the axoneme—surrounded by the ciliary membrane (Fig. 1). All axonemes comprise nine parallel doublet microtubules called outer doublets. Motile axonemes have a central pair of microtubules. The outer doublets extend from the basal body, which is derived from the mother centriole (the older of the two centrioles in the centrosome). The doublet microtubules consist of one complete microtubule (A tubule) connected to an incomplete second microtubule (B tubule). These microtubules are oriented with their plus ends at the tip of the cilium. The tubulin of the outer doublet is usually subject to posttranslational modifications, including acetylation, glutamylation, and glycylation. Those tubulin modifications are known to be important for the stability, assembly, and motility of cilia (Gaertig and Wloga 2008; Ikegami and Setou 2010). The axoneme is not only the core of the cilium but also works as “rails” for transport of ciliary proteins.
Figure 1.
The architecture of cilia. Schematic, foreshortened drawing of a longitudinal section of the primary cilium. The inset shows cross sections of motile and primary cilia.
The region between the cilium and basal body is called the transition zone. Transition fibers are thought to be the distal appendages of the mother centriole (reviewed by Vertii et al. 2016) and are required for anchoring the mother centriole to the plasma membrane (Tateishi et al. 2013). More distally in the transition zone, bridges, called Y-links, connect the axoneme microtubules to the overlying ciliary membrane, as originally observed by transmission electron microscopy (Gilula and Satir 1972). In some types of cilia, the plasma membrane in the transition zone has unique patterns of particles called the ciliary necklace (Gilula and Satir 1972). Some types of mammalian cells and trypanosomatids have the “ciliary pocket,” an invagination of the plasma membrane at the root of the cilium (Molla-Herman et al. 2010).
Cilia are conventionally categorized into two types—primary and motile cilia. Primary cilia are defined as the cilia that form from the mother centriole that a cell was born with. Primary cilia are almost always nonmotile (with the notable exception of the primary cilia in the embryonic node) and occur at a frequency of one per cell in a variety of vertebrate cells. Although primary cilia were discovered more than 100 years ago (Zimmermann 1898), they were ignored as vestigial structures until the 1990s. Subsequent studies revealed that the primary cilium acts as an “antenna” to sense the extracellular signals and environment (Goetz and Anderson 2010; Drummond 2012). Primary cilia also work as a signaling center for processing various signaling pathways, such as Hedgehog, Notch, Wnt, and growth factor signaling (Singla and Reiter 2006; Berbari et al. 2009). The axonemes of primary cilia have nine outer doublet microtubules and no central pair, and so they are called “9 + 0” axonemes (Fig. 1, insets).
Some sensory organs have specialized primary cilia, which differ in appearance from generic primary cilia but share the same fundamental structure of a microtubule axoneme surrounded by a membrane. These specialized primary cilia include the connecting cilium of photoreceptors, which detects light, and olfactory cilia, which detect odorants.
Motile cilia are usually present in large numbers on epithelial cells in the trachea and oviducts, and ependymal cells in the ventricles of the brain. Sperm also has the motile cilium as its tail. Motile cilia play important roles for developing a force for locomotion and making fluid flow (Ostrowski et al. 2011; Vincensini et al. 2011). Motile cilia take a “9 + 2” configuration, with two extra singlet microtubules in the center of the axoneme (called the central pair) and have additional components for their motility, such as dynein arms, radial spokes, and nexin links (reviewed by Viswanadha et al. 2016). In the unicellular green alga Chlamydomonas reinhardtii, the motile cilia are known as flagella, and because there are two per cell, the cells are referred to as “biflagellate.”
These conventionally classified categories have some exceptions. The cilia in the node during early vertebrate development (also called nodal cilia) are similar in appearance to generic primary cilia, but they are motile. Moreover, motile cilia in the trachea are known to sense chemical stimuli for regulating their motility.
3. IFT
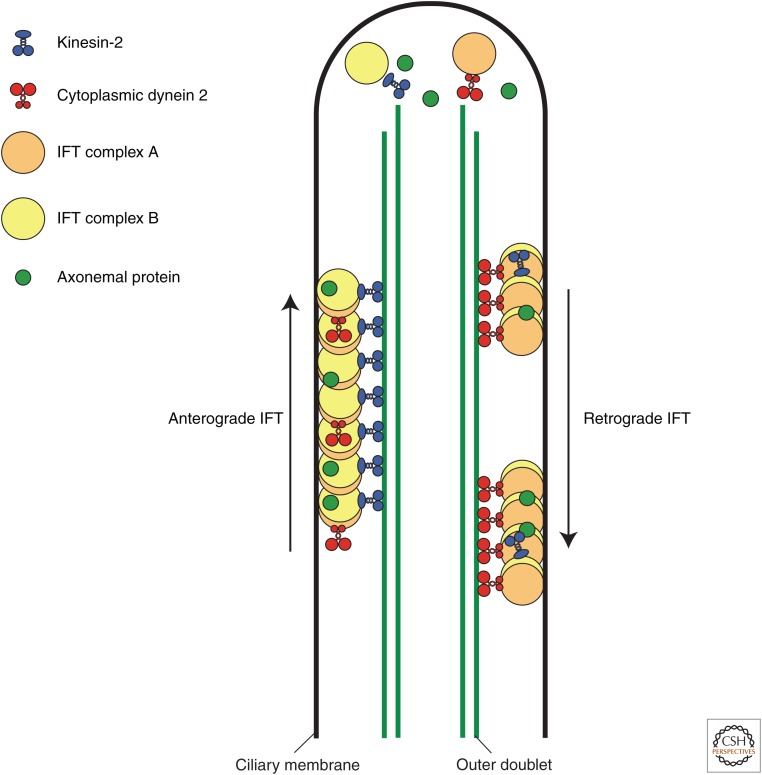
Proteins cannot be synthesized within the cilium, and so all materials required to assemble cilia are transported from the cell body. In addition, assembly of the outer doublets occurs exclusively at the distal end of the cilium (Johnson and Rosenbaum 1992). Therefore, an efficient transport system is necessary for protein transport from the base to the tip of the cilium. IFT—the bidirectional protein transport system inside cilia—carries ciliary components from the cell body to the tip of cilia and sends the products of turnover back to the cell body from cilia (Fig. 2) (Rosenbaum and Witman 2002; Scholey 2003; Ishikawa and Marshall 2011). IFT was first observed as the bidirectional movement of granule-like particles (so-called “IFT trains”) along the cilium of the biflagellate green alga Chlamydomonas by using differential interference contrast (DIC) microscopy (Kozminski et al. 1993). The IFT system has since been shown to be present in most ciliated organisms, including human. IFT trains were observed by transmission electron microscopy to consist of a varying number of electron-dense particles assembled into linear arrays between the outer doublets and the overlying ciliary membrane (Kozminski et al. 1993, 1995; Pigino et al. 2009). IFT particles move continuously along the length of cilia in both directions. The speed of IFT varies with conditions and the types of organisms. For example, the anterograde transport rate is ∼2.0 µm/sec and retrograde transport is ∼3.5 µm/sec in Chlamydomonas (Kozminski et al. 1993; Dentler 2005), whereas the velocities are slower in mammalian cells (∼1.2 µm/sec anterograde and ∼0.9 µm/sec retrograde) (He et al. 2014; Ishikawa et al. 2014).
Figure 2.
Intraflagellar transport. The anterograde intraflagellar transport (IFT) motor kinesin-2 transports IFT complexes A and B, axonemal proteins, and cytoplasmic dynein 2 to the tip of a cilium. At the tip of the cilium, anterograde IFT “trains” release axonemal proteins and rearrange their conformation for retrograde IFT. Cytoplasmic dynein 2 transports retrograde IFT trains to the cell body.
Two types of microtubule motors drive IFT. Kinesin-2 moves IFT trains toward the tip of the axoneme, and cytoplasmic dynein 2 moves IFT trains toward the minus ends of the microtubules at the base of the axoneme (Fig. 2). Two kinesin-2 motors—heterotrimeric and homodimeric—have been implicated in anterograde IFT. The canonical anterograde IFT motor is heterotrimeric kinesin-2, which consists of two heterodimerized kinesin-2 motor subunits and an accessory subunit, kinesin-associated protein (KAP). Heterotrimeric kinesin-2 was first purified from sea urchin eggs as a plus end–directed microtubule motor (Cole et al. 1992, 1993) and is necessary for the assembly and maintenance of cilia in most ciliated organisms. For example, deleting the gene for a subunit of heterotrimeric kinesin-2 prevents the assembly of cilia (Nonaka et al. 1998). In Chlamydomonas, a temperature-sensitive mutant of the motor subunit of heterotrimeric kinesin-2 displays gradual shortening of its cilia at restrictive temperatures (Kozminski et al. 1995). Homodimeric kinesin-2 also contributes to assembly of some types of cilia, such as sensory cilia of the nematode Caenorhabditis elegans and the connecting cilium of photoreceptors in the vertebrate retina (Snow et al. 2004; Insinna et al. 2009). In C. elegans, homodimeric kinesin-2 works cooperatively with heterotrimeric kinesin-2 for anterograde IFT and can substitute the function of heterotrimeric kinesin-2 in sensory cilia. Homodimeric kinesin-2 is also localized to cilia in mammalian somatic cells, but its function remains unclear.
The retrograde IFT motor cytoplasmic dynein 2 is a multiprotein complex comprising at least four different subunits: a heavy chain, a light intermediate chain, an intermediate chain, and a light chain (Pazour et al. 1998, 1999; Porter et al. 1999; Grissom et al. 2002; Perrone et al. 2003; Schafer et al. 2003; Rompolas et al. 2007; Patel-King et al. 2013). Cytoplasmic dynein 2 was originally identified from sea urchin embryos by cDNA analysis of dynein heavy chains (Gibbons et al. 1994), and subsequent genetic and motility studies showed that cytoplasmic dynein 2 is essential for retrograde IFT (Pazour et al. 1998, 1999). The function of cytoplasmic dynein 2 is necessary for the assembly of cilia but is dispensable for their maintenance (Engel et al. 2012).
IFT trains were first isolated from Chlamydomonas cilia and were found to consist of two large complexes, termed IFT complex A and complex B (Piperno and Mead 1997; Cole et al. 1998). IFT complexes A and B are loosely associated and seem to move together within the cilium, but they have distinct functions. IFT complex B contributes to anterograde transport with kinesin and is necessary for the assembly and maintenance of cilia (Fig. 2). In most cases, mutation or knockdown of IFT complex B proteins results in absent or very short cilia (Pazour et al. 2000; Brazelton et al. 2001; Haycraft et al. 2003; Sun et al. 2004; Follit et al. 2006). In contrast, IFT complex A is required for the retrograde transport that returns proteins to the cell body for turnover, but it does not appear to be necessary for ciliary assembly. For example, cilia can assemble with a mutation in an IFT complex A protein, but have abnormal bulges containing accumulated IFT complex B proteins (Iomini et al. 2001, 2009; Efimenko et al. 2006; Tsao and Gorovsky 2008).
Immunoprecipitation of IFT complexes from Chlamydomonas cilia coprecipitates ciliary proteins, such as components of dynein arms and radial spokes (Qin et al. 2004). This result suggests that the IFT complex binds ciliary proteins as cargo. For example, IFT46, an IFT complex B protein, transports components of outer arm dynein into cilia by a cargo-specific adaptor (Hou et al. 2004; Ahmed et al. 2008). Recent structural analysis has revealed that two core IFT complex B proteins, IFT74 and IFT81, form a tubulin-binding module and transport tubulin into cilia (Bhogaraju et al. 2013). IFT is also required to localize signal-transduction proteins to cilia (Keady et al. 2012). These proteins include receptors for Hedgehog and Wnt, important signaling pathways for organizing the body plan and organogenesis (Singla and Reiter 2006; Veland et al. 2009).
The behavior of the cargo proteins has been visualized in Chlamydomonas flagella and C. elegans cilia (Hao et al. 2011; Wren et al. 2013; Craft et al. 2015), where the IFT conveys cargo proteins, such as tubulin and other axonemal components, to the tip of the cilium, followed by release and diffusion in the ciliary lumen before docking at their axonemal assembly site (Wren et al. 2013). However, the mechanisms of loading and unloading IFT components with cargo are still unclear.
The complexity of the IFT system raises the question of how such a complicated molecular machine could have evolved. The same question has also been raised about the cilium itself, given that normal bending motility seems to involve the interaction of many different protein complexes. One possible answer to both questions is that cilia, and IFT, might have first evolved to mediate gliding motility rather than swimming motility. Glycoproteins of the ciliary membrane undergo anterograde and retrograde motions that can move attached beads back and forth and can cause cells to move over flat surfaces by gliding (Bloodgood 1977, 1988; Laib et al. 2009). Dynein generates the motile force for gliding, which is mediated by retrograde IFT motion of membrane glycoproteins (Shih et al. 2013). Gliding and bead movement would be extremely useful for a cell that lacked other forms of motility and presumably conferred a fitness advantage by allowing it to move from one place to another, and to capture and move food particles toward the cell body (Mitchell 2007). Gliding seems to involve the motion of a single membrane protein and thus would not require a complex of many IFT proteins to select different cargos. Moreover, the entire machinery of flagellar motility (inner and outer dynein arms, radial spokes, etc.) is not required for gliding. It thus seems possible that a much simpler cilium-like structure could have evolved first to allow gliding, and then subsequently been modified to perform other motile functions. Given that both cilia- and actin-based amoeboid motility were apparently present in the last eukaryotic common ancestor (Cavalier-Smith 2002; Fritz-Laylin et al. 2010), it is possible that cilium-based gliding evolved before actin-mediated amoeboid motility.
4. INTRACELLULAR TRANSPORT AND CILIARY IMPORT
For assembly and maintenance of cilia, the component materials must be transported from the cytoplasm or Golgi apparatus in the cell body and be accumulated in the vicinity of the basal body. In addition to their role in intraciliary transport, IFT proteins are also implicated in vesicle transport from the Golgi apparatus to cilia. IFT20—an IFT complex B protein—is not only localized to the basal body and cilia along with most other IFT proteins but is also associated with the Golgi apparatus and is required for the vesicle trafficking of membrane proteins to the cilium (Follit et al. 2006). Rab small GTPases mediate membrane vesicle trafficking to cilia. For example, ciliogenesis depends on interaction of Rab8 with the IFT machinery through an endocytosis regulator (Rabaptin5) to mediate vesicle trafficking to cilia (Yoshimura et al. 2007; Omori et al. 2008).
Bardet–Biedl syndrome (BBS; reviewed by Vertii et al. 2016) is a genetic disease characterized by a collection of symptoms, including polydactyly, retinitis pigmentosa, and obesity, which are characteristic of ciliary defects (Badano et al. 2006). BBS is caused by mutations in a set of genes encoding proteins that play important roles in ciliary vesicle trafficking through interactions with Rab8 (Nachury et al. 2007). BBS proteins are required for the localization of some membrane proteins to cilia (Berbari et al. 2008b). Some BBS proteins interact with the small GTPase Arl6 to form a coat complex that sorts membrane proteins to cilia (Jin et al. 2010).
Some ciliary proteins contain a targeting sequence that is necessary to transport these proteins into cilia (Geng et al. 2006; Berbari et al. 2008a; Dishinger et al. 2010; Follit et al. 2010; Humbert et al. 2012; Loktev and Jackson 2013). Although several ciliary targeting sequences have been reported, no unique consensus sequence has been identified for transport of ciliary proteins (Malicki and Avidor-Reiss 2014). This suggests that multiple systems are involved in the ciliary transport of these proteins.
Moreover, cytosolic axonemal proteins have been found associated with cytoplasmic membrane vesicles bearing IFT proteins, as well as ciliary membrane proteins during growth of cilia in Chlamydomonas (Wood and Rosenbaum 2014). This finding suggested that cytosolic ciliary proteins are also transported to cilia as membrane vesicles. It is hypothesized that fusion of membrane vesicles, either with the plasma membrane near the base of the cilium or with the ciliary membrane near the transition zone, would orient any proteins associated with the outer surface of the vesicles toward the interior of the cilium.
To sense the extracellular environment and process signaling pathways, cilia accumulate specific membrane receptors, channels, and cytosolic signaling molecules. For example, type III adenylyl cyclase, the somatostatin receptor 3, and the melanin-concentrating hormone receptor 1 are exclusively localized to cilia (Händel et al. 1999; Bishop et al. 2007; Berbari et al. 2008a). To retain these molecules, cilia must make a cellular compartment—like other organelles do. However, the ciliary membrane is continuous with the plasma membrane and the base of the cilium is open to the cytoplasm of the cell body. The cilium uses several systems to maintain its compartment.
Two mechanisms segregate transmembrane proteins between the plasma membranes of the cilium and the cell body. The ciliary membrane has a diffusion barrier at its base. This barrier is established in part by septin2 and septin7, members of the septin family of guanosine triphosphatases. These peripheral membrane proteins form a ring at the base of cilia and restrict lateral diffusion of transmembrane proteins into the cilium (Hu et al. 2010; Kim et al. 2010). The transition zone at the base of cilia forms a second diffusion barrier for transmembrane proteins (Dowdle et al. 2011; Garcia-Gonzalo et al. 2011; Williams et al. 2011; Chih et al. 2012). The mechanism by which such membrane proteins are able to form a selective diffusion barrier remains a mystery. Most proteins localized at the transition zone, including both axonemal and membrane-associated proteins, contribute to the diffusion barrier. Some proteins might be components of Y-links, which connect the axonemal microtubules to the ciliary membrane (Craige et al. 2010; Williams et al. 2011). Mutations of these proteins cause human ciliopathies. Other plasma membrane proteins of the cell body are excluded from the vicinity of cilia by tethering to cortical actin filaments (Vieira et al. 2006; Francis et al. 2011).
Because the ciliary lumen is also connected with the cytoplasm of the cell body, maintaining the ciliary compartment depends on controlling the traffic of cytosolic molecules. However, the pores that exclude diffusion of soluble molecules into cilia have a diameter of ∼8 nm (Breslow et al. 2013; Lin et al. 2013). Electron microscopy showed much larger pores of ∼53 nm adjacent to the outer doublets at the base of cilia in Tetrahymena pyriformis (Ounjai et al. 2013). This is slightly larger than the size of the ∼37.5-nm IFT particle itself (Pigino et al. 2009). Thus, the physical pore that prevents larger molecules from freely passing must be restricted by molecules that are not clearly visible by electron microscopy.
How do larger protein complexes move through the pore and enter the cilium? The motile force of the IFT kinesin might help the IFT particles, together with bound cargo, to move through this block by deforming an elastic pore. In such a model, IFT cargo binding would constitute the key selectivity step in determining which proteins can enter the cilium.
Several lines of evidence suggest that the ciliary pore and the nuclear pore share components and machinery (Dishinger et al. 2010; Kee et al. 2012). Importin and the small GTPase Ran—two proteins that contribute to importing proteins into the nucleus through nuclear pores—also contribute to transporting cargo with ciliary targeting sequences into cilia (Dishinger et al. 2010). However, the mechanistic similarity of the nuclear and ciliary pores is not yet firmly established (Breslow et al. 2013).
5. CONTROLLING CILIARY LENGTH BY REGULATING TRANSPORT AND DYNAMICS
Cells are thought to regulate the lengths of cilia and flagella actively, presumably to generate lengths appropriate for their functions. These lengths are characteristic of each different cell type and narrowly distributed, typically showing a distribution more sharply peaked than the best-fit Gaussian (Kannegaard et al. 2014). If cilia are severed, they grow back to a length similar to their pre-severing length (Rosenbaum et al. 1969). The pace of regeneration slows as the length increases, suggesting that growth rates are modulated as a function of length. Finally, and perhaps most strikingly, if one flagellum is severed from biflagellated Chlamydomonas cells, it grows back, while the other flagellum shortens, until the two flagella are equal in length (Coyne and Rosenbaum 1970; Ludington et al. 2012). These observations suggest that some unknown pathway controls the length of cilia.
Organelle size control is a matter of general interest for investigators concerned with any organelle (Rafelski and Marshall 2008; Chan and Marshall 2012), and cilia provide a particularly simple model organelle because their size varies in a single dimension—the length (Randall 1969; Wemmer and Marshall 2007). Furthermore, possession of cilia and flagella of abnormal length has been implicated in a number of human diseases characterized by ciliary dysfunction, including Meckel syndrome (Tammachote et al. 2009), BBS (Mokrzan et al. 2007), and retinal degeneration (Omori et al. 2010; Özgül et al. 2011).
The length of a cilium could, in principle, be regulated during initial assembly, so that cilia are built to a particular length and then remain in a static, nondynamic state. However, cilia are dynamic structures, undergoing constant turnover of tubulin (Stephens 1995, 1997, 2000; Marshall and Rosenbaum 2001; Song and Dentler 2001), so that, even when their lengths appear stable, the ciliary microtubules constantly disassemble tubulin from their tips and replace it with newly assembled tubulin (Marshall and Rosenbaum 2001). The fact that the cilium is a dynamic structure, undergoing constant turnover, means that its size must be regulated by the balance of assembly and disassembly. When the cilium is too short, assembly must predominate over disassembly to elongate the cilium to the correct length; if the cilium is too long, disassembly must predominate to cause the cilium to shorten. A key question is whether the cell regulates assembly, disassembly, or perhaps both, as a function of ciliary length.
Here, we first consider whether the assembly rate might be a function of length. IFT appears to limit the assembly rate, based on the experimental observation that a partial reduction in IFT rate leads to shorter flagella (Marshall and Rosenbaum 2001). Biochemical studies of IFT protein content within flagella (Marshall et al. 2005) and quantitative imaging of IFT proteins tagged with green fluorescent protein (GFP) (Engel et al. 2009) showed that the total quantity of IFT particles within a flagellum is independent of length. In an initial model (Marshall and Rosenbaum 2001), this constant quantity was interpreted as indicating a fixed set of IFT particles that circulated back and forth from the base to the tip, carrying assembly precursors out to the tip, and then returning for another load. In such a scenario, each particle requires a time proportional to the length L to arrive at the tip, and a time proportional to L to return to the base to obtain more cargo. As the round-trip transit time for each particle is proportional to L, such a particle will arrive at the tip with a frequency proportional to 1/L (given that speed is approximately independent of length [Engel et al. 2009]). Thus, the rate of cargo transport drops with the length of the flagella.
Combining these assumptions leads to a simple equation for flagellar length dynamics:
| (1) |
In this balance-point model, L is length, A and D describe the rates of IFT and disassembly, respectively, P is the total flagellar precursor protein pool, and the cell is assumed to have two flagella. This equation has a unique stable steady-state length for which the assembly and disassembly rates balance. This model accounts for the kinetics of flagellar regeneration (Marshall and Rosenbaum 2001), the dependence of flagellar length on the number of flagella per cell (Marshall et al. 2005), and the ability of biflagellate cells to equalize the lengths of their flagella after one is severed (Marshall and Rosenbaum 2001; Ludington et al. 2012). The original version of this model proposed that a fixed number of IFT particles are loaded into the cilium or flagellum when it forms and then recirculates back and forth without entering or exiting the ciliary compartment. Such an “initial bolus” mechanism would ensure that the number of IFT proteins is constant, thus causing the frequency of IFT particle arrival at the flagellar tip to scale as 1/L. However, photobleaching experiments suggest that IFT particles are not trapped inside a particular flagellum (Ludington et al. 2015), so some mechanism must actively regulate the quantity of IFT proteins inside the flagellum to maintain a constant total number of IFT particles, independent of changes in length. A regulatory mechanism is needed because if IFT particles are simply injected into the flagellum at a constant rate, either with a fixed periodicity or randomly with a constant average rate of injection per unit time, then the number of IFT particles inside the flagellum would not be independent of length. For example, if a single particle is injected, it must move out to the tip and then return back to the base, spending a “dwell time” inside the flagellum that is proportional to the length. If the flagellum were twice as long, then the dwell time that each particle spends inside the flagellum would double. If particles enter the flagellum at a constant rate independent of length, but each particle spends a time inside the flagellum that is proportional to the length, then the total quantity of particles inside the flagellum must increase in proportion to the length. However, this is absolutely not the case (Marshall and Rosenbaum 2001; Marshall et al. 2005; Engel et al. 2009). The total number of IFT particles inside the flagellum, at steady state, is given by the product of the rate at which particles are injected into the flagellum, which we will call the injection rate r, and the dwell time of each particle within the flagellum, which is proportional to the length L. So the total number of particles at steady state is proportional to rL. The only way this quantity can be independent of length is if r ∼ 1/L. Remarkably, quantitative total internal reflection fluorescence (TIRF) microscopy showed that the rate of injection of IFT particles into the flagellum is indeed proportional to 1/L (Ludington et al. 2013). It thus appears that some mechanism must regulate IFT injection as a function of length. Quantitative measurement of IFT protein accumulation at the basal body suggests that this accumulation, referred to as “IFT recruitment,” is length-dependent. It has been proposed, therefore, that the injection of IFT into the cilium or flagellum is regulated by controlling recruitment to the basal bodies as a function of length. But how is this regulation achieved?
The molecular pathways that sense length and regulate IFT injection are not known, but might be found by using genetics to find mutants that alter the steady-state length. If the balance-point model (Equation 1) is correct, alteration of IFT regulation should lead to changes in steady-state flagellar length. A combination of genetic and small-molecule inhibitor studies has identified six kinases that have been proposed to play a role in flagellar length modulation in Chlamydomonas, possibly acting as part of the system that regulates IFT: long flagella proteins LF2 (Tam et al. 2007), LF4 (Berman et al. 2003), and LF5 (Tam et al. 2013); Aurora-like protein CALK (Pan et al. 2004); NIMA-related kinase 2 (CNK2) (Bradley and Quarmby 2005); and glycogen synthase kinase 3 (GSK3) (Wilson and Lefebvre 2004). Might any of these kinases regulate IFT injection? Three of these kinases, LF2, CALK, and CNK2, are known to regulate flagellar disassembly rather than assembly (Marshall and Rosenbaum 2001; Pan et al. 2004; Cao et al. 2013; Hilton et al. 2013), and so they are not candidates to regulate IFT. GSK3 might be the target of lithium, a length-increasing compound, but this is still hypothetical (Wilson and Lefebvre 2004). The function of LF5 is not known. Another candidate kinase is protein kinase A (PKA) as pharmacological activation of PKA in mouse cells increases IFT speed (Besschetnova et al. 2010). But PKA also increases ciliary length and, as IFT speed is a function of length (Engel et al. 2009), the speed increase following PKA activation might be a side effect of the ciliary elongation.
Currently, the LF4 kinase, a flagella-localized member of the mitogen-activated protein (MAP) kinase family, is the primary candidate to regulate IFT injection because a null mutation in the LF4 gene (which causes flagella to become twice wild-type length) increases both IFT recruitment at basal bodies and injection into flagella (Ludington et al. 2013). This phenotype suggests that part of the normal function of the LF4 kinase is to down-regulate IFT injection. However, in the lf4 null mutant, IFT injection is still proportional to 1/L (Ludington et al. 2013). Thus, LF4 exerts some control over injection, but it is not required for either the measuring of length or the transmission of length information to the injection machinery. Thus, LF4 acts as a “control knob” for the feedback system—much like the temperature control knob on a household thermostat—rather than being a crucial component of the feedback control system itself.
Genetic and inhibitor studies have indicated a role for actin and myosin in regulating IFT recruitment as a function of length. When either actin or myosin was perturbed, IFT recruitment did not increase in short flagella compared with wild-type unperturbed cells. In the most extreme case, treatment of cells with the myosin inhibitor blebbistatin completely eliminated the length dependence of IFT recruitment (Avasthi et al. 2014). Thus, the actin cytoskeleton contributes in some way to the molecular pathways that sense length and regulate IFT recruitment.
Current models of IFT generally have assumed that cargo binding is a simple process determined by the affinity of cargo for different IFT proteins but otherwise not regulated. This assumption is implicit in Equation 1 above. However, the work of Wren and coworkers shows that at least one IFT cargo, the motility-related protein DRC4, is loaded onto IFT particles at a level that correlates with flagellar length (Wren et al. 2013). The investigators presented a modification of the balance-point model (Equation 1), with cargo loading explicitly regulated by length, and showed that this modified model can account for flagellar length regulation. They also presented an argument that length regulation of cargo binding is necessary to account for the kinetics of flagellar regeneration and that, without such regulation, flagellar assembly would be too slow. This conclusion appears to rest on the assumption that DRC4 incorporation and tubulin incorporation occur at equal rates. But, as DRC4 is involved in motility and is not a structural component that determines flagellar length, this assumption must currently be viewed as purely hypothetical. Thus, it is not possible to conclude that length-regulated cargo binding is required to explain flagellar assembly kinetics.
More recently, quantitative analysis of tubulin transport in Chlamydomonas flagella has shown that the occupancy of IFT for tubulin (that is, the fraction of IFT particles carrying at least one tubulin dimer) is a decreasing function of flagellar length (Craft et al. 2015). Such a dependence could, in principle, be explained by the increase in tubulin synthesis during flagellar regeneration; however, Craft and colleagues showed that the length dependence was flagellum-autonomous, such that, in a cell with two flagella of different lengths, the longer flagellum showed a lower occupancy of IFT particles by tubulin than the short flagellum. It thus seems clear that loading of tubulin as a cargo onto the IFT complex is a second way in which IFT function is regulated by length, in addition to the length dependence of IFT injection referred to above.
As discussed above, steady-state length depends on the balance of assembly and disassembly so that regulation of length can be achieved by regulating either assembly or disassembly, or both. Given that the rates of IFT and, therefore, assembly clearly depend on length, the next issue is whether disassembly might also be regulated as a function of length. The continuous disassembly does not appear to be mediated simply by spontaneous depolymerization of microtubules in the axoneme because isolated axonemes are extremely stable and can be kept in the cold (conditions that normally cause rapid depolymerization) indefinitely. It is thus thought that some active machinery must catalyze the removal of subunits from the tip of the axoneme. Indeed, members of the kinesin-13 family of depolymerizing kinesins have been found to be localized at the tips of cilia, and the effect of their depletion by RNA interference (RNAi) indicates that they are involved in active shortening of cilia (Blaineau et al. 2007; Dawson et al. 2007; Piao et al. 2009). The rate of an enzymatic reaction is typically governed by the intrinsic activity of the enzyme and the concentration of its substrate. For a depolymerizing kinesin, its substrate is the assembled microtubule. For depolymerases localized at the tip, the local concentration of the microtubules is independent of length. We would therefore expect that the depolymerization rate would be constant, and not a function of length. Indeed, when IFT is inactivated using mutants in the fla10 kinesin, flagella shorten at a constant rate, suggesting that depolymerization of flagellar microtubules is length independent (Marshall and Rosenbaum 2001).
Because enzymes mediate disassembly, their regulation could control disassembly. For such regulation of disassembly to produce a stable-length control system, disassembly rates should increase with the length of the cilium, as shown by genetic studies in Chlamydomonas (Hilton et al. 2013). Two candidates have emerged to explain this behavior.
Depolymerizing kinesins with processive movement are candidates for disassembly factors with length-dependent activity. It has been shown in in vitro experiments that the activity of a depolymerizing kinesin-8 increases with the length of the microtubule. This effect appears to arise because kinesin-8 can initially bind anywhere along the side of microtubule, after which it then moves rapidly and processively to the microtubule tip, where it catalyzes disassembly. Longer microtubules can bind more kinesin-8 molecules per unit time, which then move to the tip and catalyze more rapid disassembly (Varga et al. 2009). Whether such a mechanism is active inside cilia or flagella has not been determined.
A second candidate component that could mediate length-dependent disassembly is the Ser/Thr protein kinase CNK2. Genetic analysis has shown that CNK2 is part of the feedback pathway by which cilium length regulates disassembly. The length-dependent stimulation of disassembly is lost when CNK2 is mutated (Hilton et al. 2013). This suggests that the length feedback is not directly mediated by motility of the depolymerizing kinesins themselves but, rather, by a length-sensing pathway of some sort, which measures the length of the cilium and adjusts the activity of the depolymerization system. However, the nature of this pathway is obscure, because we do not know the substrates of CNK2 or the molecular signals that regulate its activity. An even more fundamental question is whether this pathway regulates length under normal conditions as the experiments demonstrating length-dependent disassembly were performed in mutants with abnormally long cilia. Whether small decreases in length would suppress disassembly is not currently known.
6. CONCLUSION
Cilia and flagella are microtubule-based structures of immense complexity whose assembly and maintenance requires the constant action of motor proteins moving IFT particles. These organelles can be expected to continue to serve as model systems to study the coordination of assembly, turnover, and transport, as well as the self-assembly and size regulation of complex cytoskeletal structures. A major challenge for future progress is understanding how the many different pathways that impinge on ciliary length—including IFT, length-dependent disassembly, and expression of genes encoding ciliary proteins—are all coordinated to produce a cilium of the appropriate size for a given cell type. Another future challenge is to understand how the length of cilia affects their function, both as generators of fluid flow and as sensors of the environment. These challenges are of more than academic interest. Given that human ciliopathies are often accompanied by alterations in ciliary length, understanding the pathways that control ciliogenesis and length might provide novel therapeutic strategies in which, for example, a defect in the IFT machinery could be at least partially corrected by altering the rate of ciliary disassembly. Cilia and IFT thus provide an example of how a systems-level understanding of a cytoskeletal organelle might hold out the promise for new ways to treat disease.
Acknowledgments
Work on flagellar assembly and length control in the authors’ laboratory has been supported by National Institutes of Health grant R01 GM097017 and by the W.M. Keck Foundation. We thank members of the Marshall laboratory, past and present, for many enlightening discussions, as well as Joel Rosenbaum and members of his laboratory for setting us on our course.
Footnotes
Editors: Thomas D. Pollard and Robert D. Goldman
Additional Perspectives on The Cytoskeleton available at www.cshperspectives.org
REFERENCES
*Reference is in this collection.
- Ahmed NT, Gao C, Lucker BF, Cole DG, Mitchell DR. 2008. ODA16 aids axonemal outer row dynein assembly through an interaction with the intraflagellar transport machinery. J Cell Biol 183: 313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avasthi P, Onishi M, Karpiak J, Yamamoto R, Mackinder L, Jonikas MC, Sale WS, Shoichet B, Pringle JR, Marshall WF. 2014. Actin is required for IFT regulation in Chlamydomonas reinhardtii. Curr Biol 24: 2025–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano JL, Mitsuma N, Beales PL, Katsanis N. 2006. The ciliopathies: An emerging class of human genetic disorders. Annu Rev Genomics Hum Genet 7: 125–148. [DOI] [PubMed] [Google Scholar]
- Berbari NF, Johnson AD, Lewis JS, Askwith CC, Mykytyn K. 2008a. Identification of ciliary localization sequences within the third intracellular loop of G protein-coupled receptors. Mol Biol Cell 19: 1540–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari NF, Lewis JS, Bishop GA, Askwith CC, Mykytyn K. 2008b. Bardet–Biedl syndrome proteins are required for the localization of G protein–coupled receptors to primary cilia. Proc Natl Acad Sci 105: 4242–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari NF, O’Connor AK, Haycraft CJ, Yoder BK. 2009. The primary cilium as a complex signaling center. Curr Biol 19: R526–R535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman SA, Wilson NF, Haas NA, Lefebvre PA. 2003. A novel MAP kinase regulates flagellar length in Chlamydomonas. Curr Biol 13: 1145–1149. [DOI] [PubMed] [Google Scholar]
- Besschetnova TY, Kolpakova-Hart E, Guan Y, Zhou J, Olsen BR, Shah JV. 2010. Identification of signaling pathways regulating primary cilium length and flow-mediated adaptation. Curr Biol 20: 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S, Cajanek L, Fort C, Blisnick T, Weber K, Taschner M, Mizuno N, Lamla S, Bastin P, Nigg EA, et al. 2013. Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science 341: 1009–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GA, Berbari NF, Lewis J, Mykytyn K. 2007. Type III adenylyl cyclase localizes to primary cilia throughout the adult mouse brain. J Comp Neurol 505: 562–571. [DOI] [PubMed] [Google Scholar]
- Blaineau C, Tessier M, Dubessay P, Tasse L, Crobu L, Pagès M, Bastien P. 2007. A novel microtubule-depolymerizing kinesin involved in length control of a eukaryotic flagellum. Curr Biol 17: 778–782. [DOI] [PubMed] [Google Scholar]
- Bloodgood RA. 1977. Motility occurring in association with the surface of the Chlamydomonas flagellum. J Cell Biol 75: 983–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloodgood RA. 1988. Gliding motility and the dynamics of flagellar membrane glycoproteins in Chlamydomonas reinhardtii. J Protozool 35: 552–558. [DOI] [PubMed] [Google Scholar]
- Bradley BA, Quarmby LM. 2005. A NIMA-related kinase, Cnk2p, regulates both flagellar length and cell size in Chlamydomonas. J Cell Sci 118: 3317–3326. [DOI] [PubMed] [Google Scholar]
- Brazelton WJ, Amundsen CD, Silflow CD, Lefebvre PA. 2001. The bld1 mutation identifies the Chlamydomonas osm-6 homolog as a gene required for flagellar assembly. Curr Biol 11: 1591–1594. [DOI] [PubMed] [Google Scholar]
- Breslow DK, Koslover EF, Seydel F, Spakowitz AJ, Nachury MV. 2013. An in vitro assay for entry into cilia reveals unique properties of the soluble diffusion barrier. J Cell Biol 203: 129–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Meng D, Wang L, Bei S, Snell WJ, Pan J. 2013. Activation loop phosphorylation of a protein kinase is a molecular marker of organelle size that dynamically reports flagellar length. Proc Natl Acad Sci 110: 12337–12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. 2002. The phagotrophic origin of eukaryotes and phylogenetic classification of protozoa. Int J Syst Evol Microbiol 52: 297–354. [DOI] [PubMed] [Google Scholar]
- Chan YH, Marshall WF. 2012. How cells know the size of their organelles. Science 337: 1186–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B, Liu P, Chinn Y, Chalouni C, Komuves LG, Hass PE, Sandoval W, Peterson AS. 2012. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat Cell Biol 14: 61–72. [DOI] [PubMed] [Google Scholar]
- Cole DG, Cande WZ, Baskin RJ, Skoufias DA, Hogan CJ, Scholey JM. 1992. Isolation of a sea urchin egg kinesin-related protein using peptide antibodies. J Cell Sci 101: 291–301. [DOI] [PubMed] [Google Scholar]
- Cole DG, Chinn SW, Wedaman KP, Hall K, Vuong T, Scholey JM. 1993. Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature 366: 268–270. [DOI] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. 1998. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol 141: 993–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne B, Rosenbaum JL. 1970. Flagellar elongation and shortening in Chlamydomonas. II. Re-utilization of flagellar proteins. J Cell Biol 47: 777–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft JM, Harris JA, Hyman S, Kner P, Lechtreck KF. 2015. Tubulin transport by IFT is upregulated during ciliary growth by a cilium-autonomous mechanism. J Cell Biol 208: 223–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craige B, Tsao C-C, Diener DR, Hou Y, Lechtreck K-F, Rosenbaum JL, Witman GB. 2010. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J Cell Biol 190: 927–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson SC, Sagolla MS, Mancuso JJ, Woessner DJ, House SA, Fritz-Laylin L, Cande WZ. 2007. Kinesin-13 regulates flagellar, interphase, and mitotic microtubule dynamics in Giardia intestinalis. Eukaryot Cell 6: 2354–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentler W. 2005. Intraflagellar transport (IFT) during assembly and disassembly of Chlamydomonas flagella. J Cell Biol 170: 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dishinger JF, Kee HL, Jenkins PM, Fan S, Hurd TW, Hammond JW, Truong YN-T, Margolis B, Martens JR, Verhey KJ. 2010. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-β2 and RanGTP. Nat Cell Biol 12: 703–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle WE, Robinson JF, Kneist A, Sirerol-Piquer MS, Frints SGM, Corbit KC, Zaghloul NA, Zaghloul NA, van Lijnschoten G, Mulders L, et al. 2011. Disruption of a ciliary B9 protein complex causes Meckel syndrome. Am J Hum Genet 89: 94–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond IA. 2012. Cilia functions in development. Curr Opin Cell Biol 24: 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK. 1995. Flagellar assembly in two hundred and fifty easy-to-follow steps. Trends Genet 11: 398–404. [DOI] [PubMed] [Google Scholar]
- Efimenko E, Blacque OE, Ou G, Haycraft CJ, Yoder BK, Scholey JM, Leroux MR, Swoboda P. 2006. Caenorhabditis elegans DYF-2, an orthologue of human WDR19, is a component of the intraflagellar transport machinery in sensory cilia. Mol Biol Cell 17: 4801–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel BD, Ludington WB, Marshall WF. 2009. Intraflagellar transport particle size scales inversely with flagellar length: Revisiting the balance-point length control model. J Cell Biol 187: 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel BD, Ishikawa H, Wemmer KA, Geimer S, Wakabayashi K-I, Hirono M, Craige B, Pazour GJ, Witman GB, Kamiya R, et al. 2012. The role of retrograde intraflagellar transport in flagellar assembly, maintenance, and function. J Cell Biol 199: 151–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, Tuft RA, Fogarty KE, Pazour GJ. 2006. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell 17: 3781–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, Li L, Vucica Y, Pazour GJ. 2010. The cytoplasmic tail of fibrocystin contains a ciliary targeting sequence. J Cell Biol 188: 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis SS, Sfakianos J, Lo B, Mellman I. 2011. A hierarchy of signals regulates entry of membrane proteins into the ciliary membrane domain in epithelial cells. J Cell Biol 193: 219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, Kuo A, Parede A, Chapman J, Pham J, et al. 2010. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140: 631–642. [DOI] [PubMed] [Google Scholar]
- Gaertig J, Wloga D. 2008. Ciliary tubulin and its post-translational modifications. Curr Top Dev Biol 85: 83–113. [DOI] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Corbit KC, Sirerol-Piquer MS, Ramaswami G, Otto EA, Noriega TR, Seol AD, Robinson JF, Bennett CL, Josifova DJ, et al. 2011. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet 43: 776–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng L, Okuhara D, Yu Z, Tian X, Cai Y, Shibazaki S, Somlo S. 2006. Polycystin-2 traffics to cilia independently of polycystin-1 by using an N-terminal RVxP motif. J Cell Sci 119: 1383–1395. [DOI] [PubMed] [Google Scholar]
- Gibbons BH, Asai DJ, Tang WJ, Hays TS, Gibbons IR. 1994. Phylogeny and expression of axonemal and cytoplasmic dynein genes in sea urchins. Mol Biol Cell 5: 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilula NB, Satir P. 1972. The ciliary necklace. A ciliary membrane specialization. J Cell Biol 53: 494–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz SC, Anderson KV. 2010. The primary cilium: A signalling centre during vertebrate development. Nat Rev Genet 11: 331–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissom PM, Vaisberg EA, McIntosh JR. 2002. Identification of a novel light intermediate chain (D2LIC) for mammalian cytoplasmic dynein 2. Mol Biol Cell 13: 817–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Händel M, Schulz S, Stanarius A, Schreff M, Erdtmann-Vourliotis M, Schmidt H, Wolf G, Höllt V. 1999. Selective targeting of somatostatin receptor 3 to neuronal cilia. Neuroscience 89: 909–926. [DOI] [PubMed] [Google Scholar]
- Hao L, Thein M, Brust-Mascher I, Civelekoglu-Scholey G, Lu Y, Acar S, Prevo B, Shaham S, Scholey JM. 2011. Intraflagellar transport delivers tubulin isotypes to sensory cilium middle and distal segments. Nat Cell Biol 13: 790–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycraft CJ, Schafer JC, Zhang Q, Taulman PD, Yoder BK. 2003. Identification of CHE-13, a novel intraflagellar transport protein required for cilia formation. Exp Cell Res 284: 251–263. [DOI] [PubMed] [Google Scholar]
- He M, Subramanian R, Bangs F, Omelchenko T, Liem KF, Kapoor TM, Anderson KV. 2014. The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat Cell Biol 16: 663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton LK, Gunawardene K, Kim JW, Schwarz MC, Quarmby LM. 2013. The kinases LF4 and CNK2 control ciliary length by feedback regulation of assembly and disassembly rates. Curr Biol 23: 2208–2214. [DOI] [PubMed] [Google Scholar]
- Hou Y, Pazour GJ, Witman GB. 2004. A dynein light intermediate chain, D1bLIC, is required for retrograde intraflagellar transport. Mol Biol Cell 15: 4382–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Milenkovic L, Jin H, Scott MP, Nachury MV, Spiliotis ET, Nelson WJ. 2010. A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science 329: 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert MC, Weihbrecht K, Searby CC, Li Y, Pope RM, Sheffield VC, Seo S. 2012. ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc Natl Acad Sci 109: 19691–19696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Setou M. 2010. Unique post-translational modifications in specialized microtubule architecture. Cell Struct Funct 35: 15–22. [DOI] [PubMed] [Google Scholar]
- Insinna C, Humby M, Sedmak T, Wolfrum U, Besharse JC. 2009. Different roles for KIF17 and kinesin II in photoreceptor development and maintenance. Dev Dyn 238: 2211–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini C, Babaev-Khaimov V, Sassaroli M, Piperno G. 2001. Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J Cell Biol 153: 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini C, Li L, Esparza JM, Dutcher SK. 2009. Retrograde intraflagellar transport mutants identify complex A proteins with multiple genetic interactions in Chlamydomonas reinhardtii. Genetics 183: 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF. 2011. Ciliogenesis: Building the cell’s antenna. Nat Rev Mol Cell Biol 12: 222–234. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Ide T, Yagi T, Jiang X, Hirono M, Sasaki H, Yanagisawa H, Wemmer KA, Stainier DY, Qin H, et al. 2014. TTC26/DYF13 is an intraflagellar transport protein required for transport of motility-related proteins into flagella. Elife 3: e01566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, White SR, Shida T, Schulz S, Aguiar M, Gygi SP, Bazan JF, Nachury MV. 2010. The conserved Bardet–Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 141: 1208–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA, Rosenbaum JL. 1992. Polarity of flagellar assembly in Chlamydomonas. J Cell Biol 119: 1605–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA, Rosenbaum JL. 1993. Flagellar regeneration in Chlamydomonas: A model system for studying organelle assembly. Trends Cell Biol 3: 156–161. [DOI] [PubMed] [Google Scholar]
- Kannegaard E, Rego EH, Schuck S, Feldman JL, Marshall WF. 2014. Quantitative analysis and modeling of katanin function in flagellar length control. Mol Biol Cell 25: 3686–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keady BT, Samtani R, Tobita K, Tsuchya M, San Agustin JT, Follit JA, Jonassen JA, Subramanian R, Lo CW, Pazour GJ. 2012. IFT25 links the signal-dependent movement of Hedgehog components to intraflagellar transport. Dev Cell 22: 940–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee HL, Dishinger JF, Blasius TL, Liu C-J, Margolis B, Verhey KJ. 2012. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat Cell Biol 14: 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Shindo A, Park TJ, Oh EC, Ghosh S, Gray RS, Lewis RA, Johnson CA, Attie-Bittach T, Katsanis N, et al. 2010. Planar cell polarity acts through septins to control collective cell movement and ciliogenesis. Science 329: 1337–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. 1993. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci 90: 5519–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Beech PL, Rosenbaum JL. 1995. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J Cell Biol 131: 1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laib JA, Marin JA, Bloodgood RA, Guilford WH. 2009. The reciprocal coordination and mechanics of molecular motors in living cells. Proc Natl Acad Sci 106: 3190–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-C, Niewiadomski P, Lin B, Nakamura H, Phua SC, Jiao J, Levchenko A, Inoue T, Rohatgi R, Inoue T. 2013. Chemically inducible diffusion trap at cilia reveals molecular sieve-like barrier. Nat Chem Biol 9: 437–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loktev AV, Jackson PK. 2013. Neuropeptide Y family receptors traffic via the Bardet–Biedl syndrome pathway to signal in neuronal primary cilia. Cell Rep 5: 1316–1329. [DOI] [PubMed] [Google Scholar]
- Ludington WB, Shi LZ, Zhu Q, Berns MW, Marshall WF. 2012. Organelle size equalization by a constitutive process. Curr Biol 22: 2173–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludington WB, Wemmer KA, Lechtreck KF, Witman GB, Marshall WF. 2013. Avalanche-like behavior in ciliary import. Proc Natl Acad Sci 110: 3925–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludington WB, Ishikawa H, Serebrenik YV, Ritter A, Hernandez-Lopez RA, Gunzenhauser J, Kannegaard E, Marshall WF. 2015. A systematic comparison of mathematical models for inherent measurement of ciliary length: How a cell can measure length and volume. Biophys J 108: 1361–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malicki J, Avidor-Reiss T. 2014. From the cytoplasm into the cilium: Bon voyage. Organogenesis 10: 138–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Rosenbaum JL. 2001. Intraflagellar transport balances continuous turnover of outer doublet microtubules: Implications for flagellar length control. J Cell Biol 155: 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Qin H, Rodrigo Brenni M, Rosenbaum JL. 2005. Flagellar length control system: Testing a simple model based on intraflagellar transport and turnover. Mol Biol Cell 16: 270–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DR. 2007. The evolution of eukaryotic cilia and flagella as motile and sensory organelles. Adv Exp Med Biol 607: 130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokrzan EM, Lewis JS, Mykytyn K. 2007. Differences in renal tubule primary cilia length in a mouse model of Bardet–Biedl syndrome. Nephron Exp Nephrol 106: e88–e96. [DOI] [PubMed] [Google Scholar]
- Molla-Herman A, Ghossoub R, Blisnick T, Meunier A, Serres C, Silbermann F, Emmerson C, Romeo K, Bourdoncle P, Schmitt A, et al. 2010. The ciliary pocket: An endocytic membrane domain at the base of primary and motile cilia. J Cell Sci 123: 1785–1795. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peränen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, et al. 2007. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129: 1201–1213. [DOI] [PubMed] [Google Scholar]
- Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. 1998. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95: 829–837. [DOI] [PubMed] [Google Scholar]
- Omori Y, Zhao C, Saras A, Mukhopadhyay S, Kim W, Furukawa T, Sengupta P, Veraksa A, Malicki J. 2008. Elipsa is an early determinant of ciliogenesis that links the IFT particle to membrane-associated small GTPase Rab8. Nat Cell Biol 10: 437–444. [DOI] [PubMed] [Google Scholar]
- Omori Y, Chaya T, Katoh K, Kajimura N, Sato S, Muraoka K, Ueno S, Koyasu T, Kondo M, Furukawa T. 2010. Negative regulation of ciliary length by ciliary male germ cell associated kinase (MAK) is required for retinal photoreceptor survival. Proc Natl Acad Sci 107: 22671–22676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski LE, Dutcher SK, Lo CW. 2011. Cilia and models for studying structure and function. Proc Am Thorac Soc 8: 423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ounjai P, Kim KD, Liu H, Dong M, Tauscher AN, Witkowska HE, Downing KH. 2013. Architectural insights into a ciliary partition. Curr Biol 23: 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özgül RK, Siemiatkowska AM, Yücel D, Myers CA, Collin RW, Zonneveld MN, Beryozkin A, Banin E, Hoyng CB, van den Born LI, et al. 2011. Exome sequencing and cis-regulatory mapping identifies mutations in MAK, a gene encoding a regulator of ciliary length, as a cause of retinitis pigmentosa. Am J Hum Genet 89: 253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Wang Q, Snell WJ. 2004. An aurora kinase is essential for flagellar disassembly in Chlamydomonas. Dev Cell 6: 445–451. [DOI] [PubMed] [Google Scholar]
- Patel-King RS, Gilberti RM, Hom EFY, King SM. 2013. WD60/FAP163 is a dynein intermediate chain required for retrograde intraflagellar transport in cilia. Mol Biol Cell 24: 2668–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Wilkerson CG, Witman GB. 1998. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT). J Cell Biol 141: 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Witman GB. 1999. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J Cell Biol 144: 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, Witman GB, Cole DG. 2000. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene Tg737, are required for assembly of cilia and flagella. J Cell Biol 151: 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone CA, Tritschler D, Taulman P, Bower R, Yoder BK, Porter ME. 2003. A novel dynein light intermediate chain colocalizes with the retrograde motor for intraflagellar transport at sites of axoneme assembly in Chlamydomonas and mammalian cells. Mol Biol Cell 14: 2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao T, Luo M, Wang L, Guo Y, Li D, Li P, Snell WJ, Pan J. 2009. A microtubule depolymerizing kinesin functions during both flagellar disassembly and flagellar assembly in Chlamydomonas. Proc Natl Acad Sci 106: 4713–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigino G, Geimer S, Lanzavecchia S, Paccagnini E, Cantele F, Diener DR, Rosenbaum JL, Lupetti P. 2009. Electron-tomographic analysis of intraflagellar transport particle trains in situ. J Cell Biol 187: 135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Mead K. 1997. Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc Natl Acad Sci 94: 4457–4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Bower R, Knott JA, Byrd P, Dentler W. 1999. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol Biol Cell 10: 693–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Diener DR, Geimer S, Cole DG, Rosenbaum JL. 2004. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J Cell Biol 164: 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafelski SM, Marshall WF. 2008. Building the cell: Design principles of cellular architecture. Nat. Rev Mol Cell Biol 9: 593–602. [DOI] [PubMed] [Google Scholar]
- Randall J. 1969. The flagellar apparatus as a model organelle for the study of growth and morphopoiesis. Proc R Soc Lond B Biol Sci. 173: 31–62. [DOI] [PubMed] [Google Scholar]
- Rompolas P, Pedersen LB, Patel-King RS, King SM. 2007. Chlamydomonas FAP133 is a dynein intermediate chain associated with the retrograde intraflagellar transport motor. J Cell Sci 120: 3653–3665. [DOI] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. 2002. Intraflagellar transport. Nat Rev Mol Cell Biol 3: 813–825. [DOI] [PubMed] [Google Scholar]
- Rosenbaum JL, Moulder JE, Ringo DL. 1969. Flagellar elongation and shortening in Chlamydomonas. The use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J Cell Biol 41: 600–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer JC, Haycraft CJ, Thomas JH, Yoder BK, Swoboda P. 2003. XBX-1 encodes a dynein light intermediate chain required for retrograde intraflagellar transport and cilia assembly in Caenorhabditis elegans. Mol Biol Cell 14: 2057–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey JM. 2003. Intraflagellar transport. Annu Rev Cell Dev Biol 19: 423–443. [DOI] [PubMed] [Google Scholar]
- Shih SM, Engel BD, Kocobas F, Bilyard T, Gennerich A, Marshall WF, Yildiz A. 2013. Intraflagellar transport drives flagellar surface motility. Elife 2: e00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla V, Reiter JF. 2006. The primary cilium as the cell’s antenna: Signaling at a sensory organelle. Science 313: 629–633. [DOI] [PubMed] [Google Scholar]
- Snow JJ, Ou G, Gunnarson AL, Walker MRS, Zhou HM, Brust-Mascher I, Scholey JM. 2004. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat Cell Biol 6: 1109–1113. [DOI] [PubMed] [Google Scholar]
- Song L, Dentler WL. 2001. Flagellar protein dynamics in Chlamydomonas. J Biol Chem 276: 29754–29763. [DOI] [PubMed] [Google Scholar]
- Stephens RE. 1995. Selective incorporation of architectural proteins into terminally differentiated molluscan gill cilia. J Exp Zool 274: 300–309. [DOI] [PubMed] [Google Scholar]
- Stephens RE. 1997. Synthesis and turnover of embryonic sea urchin ciliary proteins during selective inhibition of tubulin synthesis and assembly. Mol. Biol. Cell 8: 2187–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RE. 2000. Preferential incorporation of tubulin into the junctional region of ciliary outer doublet microtubules: A model for treadmilling by lattice dislocation. Cell Motil Cytoskel 47: 130–140. [DOI] [PubMed] [Google Scholar]
- Sun Z, Amsterdam A, Pazour GJ, Cole DG, Miller MS, Hopkins N. 2004. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development 131: 4085–4093. [DOI] [PubMed] [Google Scholar]
- Tam LW, Wilson NF, Lefebvre PA. 2007. A CDK-related kinase regulates the length and assembly of flagella in Chlamydomonas. J Cell Biol 176: 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam LW, Ranum PT, Lefebvre PA. 2013. CDKL5 regulates flagellar length and localizes to the base of the flagella in Chlamydomonas. Mol Biol Cell 24: 588–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammachote R, Hommerding CJ, Sinders RM, Miller CA, Czarnecki PG, Leightner AC, Salisbury JL, Ward CJ, Torres VE, Gattone VH, et al. 2009. Ciliary and centrosomal defects associated with mutation and depletion of the Meckel syndrome genes MKS1 and MKS3. Hum Mol Genet 18: 3311–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi K, Yamazaki Y, Nishida T, Watanabe S, Kunimoto K, Ishikawa H, Tsukita S. 2013. Two appendages homologous between basal bodies and centrioles are formed using distinct Odf2 domains. J Cell Biol 203: 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao C-C, Gorovsky MA. 2008. Tetrahymena IFT122A is not essential for cilia assembly but plays a role in returning IFT proteins from the ciliary tip to the cell body. J Cell Sci 121: 428–436. [DOI] [PubMed] [Google Scholar]
- Varga V, Leduc C, Bormuth V, Diez S, Howard J. 2009. Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell 138: 1174–1183. [DOI] [PubMed] [Google Scholar]
- Veland IR, Awan A, Pedersen LB, Yoder BK, Christensen ST. 2009. Primary cilia and signaling pathways in mammalian development, health and disease. Nephron Physiol 111: 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Vertii A, Hehnly H, Doxsey S. 2016. The centrosome, a multitalented renaissance organelle. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a025049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira OV, Gaus K, Verkade P, Fullekrug J, Vaz WLC, Simons K. 2006. FAPP2, cilium formation, and compartmentalization of the apical membrane in polarized Madin–Darby canine kidney (MDCK) cells. Proc Natl Acad Sci 103: 18556–18561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincensini L, Blisnick T, Bastin P. 2011. 1001 model organisms to study cilia and flagella. Biol Cell Auspices Eur Cell Biol Organ 103: 109–130. [DOI] [PubMed] [Google Scholar]
- *.Viswanadha R, Sale WS, Porter ME. 2016. Ciliary motility: Regulation of axonemal dynein motors. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmer KA, Marshall WF. 2007. Flagellar length control in Chlamydomonas—A paradigm for organelle size regulation. Int Rev Cytol 260: 175–212. [DOI] [PubMed] [Google Scholar]
- Williams CL, Li C, Kida K, Inglis PN, Mohan S, Semenec L, Bialas NJ, Stupay RM, Chen N, Blacque OE, et al. 2011. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J Cell Biol 192: 1023–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NF, Lefebvre PA. 2004. Regulation of flagellar assembly by glycogen synthase kinase 3 in Chlamydomonas reinhardtii. Eukaryot Cell 3: 1307–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CR, Rosenbaum JL. 2014. Proteins of the ciliary axoneme are found on cytoplasmic membrane vesicles during growth of cilia. Curr Biol 24: 1114–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren KN, Craft JM, Tritschler D, Schauer A, Patel DK, Smith EF, Porter ME, Kner P, Lechtreck KF. 2013. A differential cargo-loading model of ciliary length regulation by IFT. Curr Biol 23: 2463–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S-I, Egerer J, Fuchs E, Haas AK, Barr FA. 2007. Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol 178: 363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann KW. 1898. Beiträge zur Kenntniss einiger Drüsen und Epithelien. Arch Für Mikrosk Anat 52: 552–706. [Google Scholar]