Abstract
Maintaining functional protein homeostasis (proteostasis) is a constant challenge in the face of limited protein-folding capacity, environmental threats, and aging. Cells have developed several quality-control mechanisms that assist nascent polypeptides to fold properly, clear misfolded molecules, respond to the accumulation of protein aggregates, and deposit potentially toxic conformers in designated sites. Proteostasis collapse can lead to the development of diseases known as proteinopathies. Here we delineate the current knowledge on the different layers of protein quality-control mechanisms at the organelle and cellular levels with an emphasis on the prion protein (PrP). We also describe how protein quality control is integrated at the organismal level and discuss future perspectives on utilizing proteostasis maintenance as a strategy to develop novel therapies for the treatment of proteinopathies.
Cells have developed quality-control mechanisms to maintain protein homeostasis. Failure of any of these mechanisms can lead to proteinopathies—including prion diseases.
THE MATURATION OF NASCENT POLYPEPTIDES IN THE SECRETORY PATHWAY
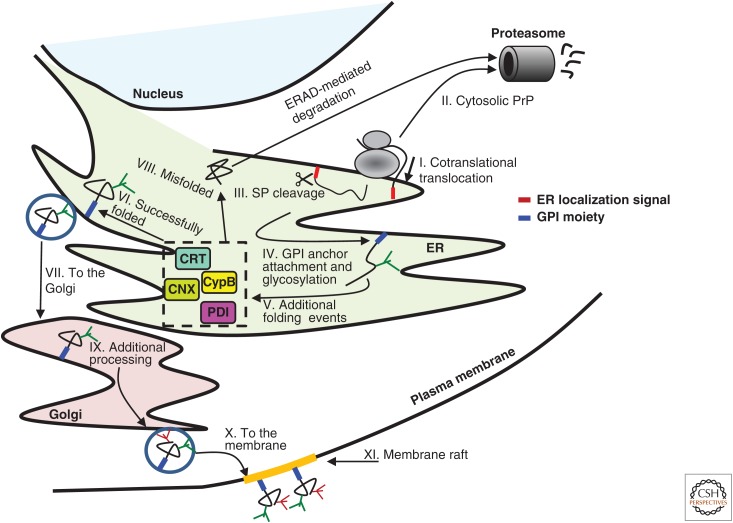
Nascent polypeptides undergo a complex, multistep process of maturation to attain their correct spatial structure, obtain proper posttranslational modifications, and become functional proteins. Arrays of specialized chaperones assist newly synthesized cytosolic (Hartl and Hayer-Hartl 2002) and secreted molecules (Hebert and Molinari 2007) to complete this process successfully and form proper intra- and intermolecular interactions. Because the prion protein (PrP) is a secreted protein, here we focus on the mechanisms that support protein folding, maturation, and quality control within the secretory pathway (Fig. 1). Like other secreted proteins, PrP bears a short endoplasmic reticulum (ER) localization signal in its N-terminus, which is first translated by the ribosome. The appearance of the signal peptide, and its recognition by the signal recognition particle (SRP) (Lauffer et al. 1985), mediates an interaction between the ribosome and the ER channel protein sec61p (Sanders et al. 1992). The bound ribosome cotranslationally translocates the nascent PrP molecule into the ER lumen (Fig. 1I). Molecules that fail to enter the ER are degraded by the proteasome (II) (Drisaldi et al. 2003). Upon entry into the ER, the signal peptide is cleaved by a signal peptidase (III), oligosaccharides are attached to asparagine residues of the molecule, and a glycosylphosphatidylinositol (GPI) anchor is added to the protein’s C-terminus (IV) (Stahl et al. 1990). A specialized set of ER folding chaperones then catalyzes the folding of the new PrP molecule (V). The ER chaperones calnexin (Wang et al. 2010) and calreticulin (Shiraishi et al. 2011) interact with PrP and assist its folding. Likewise, the protein disulfide-isomerase (PDI) catalyzes the formation of the single cysteine bond in the sequence of PrP (between residues Cys178–Cys213 of the murine protein), and the ER-resident cis–trans isomerase cyclophilin B utilizes specific proline residues to convert the protein from the cis to trans position (Cohen and Taraboulos 2003). Finally, the new PrP molecule (VI) is shuttled to the Golgi apparatus (VII), whereas molecules that failed to fold properly are retro-translocated to the cytosol and degraded by proteasomes (VIII). In the Golgi apparatus, PrP molecules undergo additional maturation steps (IX) and are then transported to the cell surface (X), where they are anchored to unique lipid assemblies known as lipid rafts (XI) (Naslavsky et al. 1997).
Figure 1.
Folding and quality control of nascent prion protein (PrP) molecules. As a secreted protein, PrP bears an endoplasmic reticulum (ER)-localization signal (red) that mediates its cotranslational translocation into the ER (I). A fraction of the nascent PrP molecules stays cytosolic and is designated for degradation by the ubiquitin-proteasome system (II). In the ER, the localization signal is cleaved (III), and a glycosylphosphatidylinositol (GPI) anchor and glycans are attached to the protein (IV). Next, the molecule undergoes a series of chaperone-assisted folding events that involve the formation of a single cysteine–cysteine bridge, cis–trans isomerization by cyclophilin B, and calnexin/calreticulin-assisted folding (V). Successfully folded molecules (VI) are transported to the Golgi apparatus (VII) for further processing, whereas molecules that failed to fold properly are directed for degradation by the ER-associated degradation mechanism (VIII). After additional maturation steps at the Golgi apparatus (IX), mature PrP molecules are shuttled to the cell surface (X), where they are presented on membrane rafts (XI). SP, Signal peptide.
Despite the assistance and supervision of the specialized network of chaperones, subsets of nascent polypeptides fail to fold properly and often expose hydrophobic domains that are otherwise buried within the core of the correctly processed protein. ER-resident chaperones recognize these misfolded molecules, preclude their shuttle to the Golgi (Ellgaard and Helenius 2003), and designate them for degradation by the ER-associated degradation (ERAD) mechanism. This culling process is constantly performed by a highly conserved set of ERAD components that mediate the retro-translocation of misfolded polypeptides to the cytosol, mostly promote their ubiquitination by specialized E3 ubiquitin ligases, and confer their digestion by the proteasome (for review, see Ruggiano et al. 2014). Like many other aggregation-prone proteins, subpopulations of PrP species misfold during the maturation process and are designated by the ERAD for proteasomal degradation (Ma and Lindquist 2001; Yedidia et al. 2001).
Although the main topological form of the prion protein is anchored to the membrane via a C-terminal GPI, other proteins require the insertion of transmembrane domains into the lipid bilayer in the desired and functional orientation. Recent data display that the upregulation of a ubiquitin-dependent ER resident intramembrane protease RHBDL4 upon ER stress leads to cleavage of unstable membrane proteins, which are subsequently degraded by canonical ERAD (Fleig et al. 2012). This mechanism promotes the clearance of membrane-integrated misfolded proteins.
Under normal and unstressed conditions, folding, quality control, and degradation mechanisms maintain protein homeostasis (proteostasis) (Balch et al. 2008). However, when stress is applied, either in late stages of life or when a load of mutated, aggregation-prone proteins challenge the proteostasis network, misfolded molecules evade the cellular surveillance mechanisms and form toxic aggregates. Uncontrolled accumulation of aggregated proteins underlies the development of diseases that are collectively known as proteinopathies (Paulson 1999). Neurodegenerative maladies such as Alzheimer’s disease (AD), Parkinson’s disease (PD) (Selkoe 2003), Huntington’s disease (HD) (Bates 2003), amyotrophic lateral sclerosis (ALS) (Ticozzi et al. 2011), and other neurodegenerative diseases caused by PrP aggregation constitute a subgroup of late onset (Amaducci and Tesco 1994) proteinopathies.
Aggregation of PrP triggers the development of at least four clinically distinct human neurological disorders: Creutzfeldt–Jakob disease (CJD), which onsets either sporadically, as a mutation-linked familial disease or as an infectious malady; Gerstmann–Sträussler–Scheinker syndrome (GSS) and fatal familial insomnia (FFI), which exclusively manifest as mutation-linked illnesses; and kuru, which was transmitted among individuals who participated in cannibalistic rituals (for review, see Aguzzi and Calella 2009).
Because the accumulation of toxic protein aggregates (proteotoxicity) presents major risks to cellular and organismal functionality and viability, cells have developed several defense mechanisms to respond to the accumulation of hazardous protein conformers, attempting to detoxify them and restore proteostasis.
CELLULAR UNFOLDED PROTEIN RESPONSES
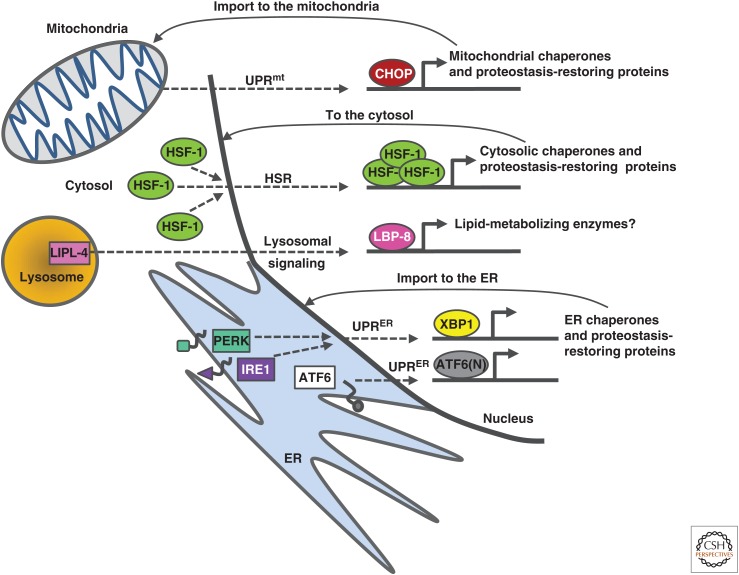
Different cellular organelles respond to the accumulation of precarious, misfolded proteins by activating stress programs that signal to the nucleus and modulate gene expression (Fig. 2). These changes reduce the expression of various genes to alleviate aggregation load and induce the production of chaperones, which act in concert to restore proteostasis. The heat-shock response (HSR) is the first mechanism that was identified as a cellular response to the accumulation of misfolded proteins in the cytosol (Lindquist 1986; Morimoto 1998). Upon exposure to heat, the heat-shock factor 1 (HSF-1), which under unstressed conditions is retained in the cytosol, trimerizes, enters the nucleus, and induces the expression of different genes, including those that encode for the subset of heat-shock proteins (HSPs) (Sarge et al. 1993). HSPs assist the refolding of damaged proteins and help the cell regain functionality.
Figure 2.
Cellular unfolded protein response mechanisms. The accumulation of misfolded proteins activates organelle-specific complex mechanisms that modulate gene expression in an attempt to restore proteostasis. Upon accumulation of misfolded proteins within the cytosol, heat-shock factor 1 (HSF-1) trimerizes, enters the nucleus, and activates the expression of various genes, including the subset of chaperones of the heat-shock protein group. Protein misfolding in the mitochondria activates the mitochondrial unfolded protein response (UPRmt), which activates the expression of genes that encode for mitochondrial chaperones. This expression is promoted by transcription factors such as CHOP. Similarly, at least three signaling cascades can respond to the accrual of unfolded proteins within the ER. The ER unfolded protein response (UPRER) mechanisms are based on the sensing of folding stress membrane proteins (ATF, IRE1, and PERK), the migration of transcription factors into the nucleus (such as ATF6[N] and XBP1), and the induction of chaperone-encoding genes. Lysosomes also signal to activate gene expression in the nucleus of Caenorhabditis elegans. The lysosomal acid lipase LIPL-4 signals to confer the nuclear localization of the lipid chaperone LBP-8, which induces gene expression. This pathway was shown to promote longevity; however, its possible roles in the maintenance of protein quality control are yet to be explored.
The accumulation of misfolded proteins within the ER activates at least four stress-response pathways, collectively known as the ER unfolded protein response (UPRER). Three highly conserved UPRER mechanisms share common principles. Upon sensing an accumulation of misfolded proteins within the ER lumen, ATF6, IRE1, and PERK initiate the migration of transcription factors (ATF6[N], XBP1, and ATF4, respectively) to the nucleus, where they modulate gene expression programs. Although all transcription factors elevate the expression of genes encoding for proteins that increase the folding capacity of the ER, PERK and IRE1 also reduce the production of proteins that require the assistance of ER chaperones to mature properly (for review, see Walter and Ron 2011). The observation that the PERK pathway is activated in mice that express aggregation-prone PrP species directly links PrP toxicity and the UPRER (Herrmann et al. 2015). A fourth UPRER pathway that is activated when the canonical mechanisms are blocked was identified in the nematode Caenorhabditis elegans (Urano et al. 2002).
A signaling pathway that responds to proteostasis perturbations also exists in the mitochondria. Like the UPRER, the mitochondrial unfolded protein response (UPRmt) senses protein-folding imbalance and induces the expression of genes that encode for mitochondrial chaperones, which act to restore proteostasis (Martinus et al. 1996; Haynes and Ron 2010).
Recently, a lysosome-to-nucleus mechanism was discovered in C. elegans (Folick et al. 2015). This signaling pathway shares key features with the HSR and UPR pathways. It is activated by a lysosomal lipid chaperone and modulates gene expression in the nucleus. However, it is yet to be determined whether it responds to the accumulation of aggregated proteins.
Various experimental findings indicate that the aforementioned stress response mechanisms have limited capacity, and under a heavy load of misfolded proteins, they cannot prevent the accumulation of hazardous species. Thus, to avert disastrous damage, cells have developed mechanisms that remove toxic aggregates from the cellular environment and deposit them in designated sites.
THE DEPOSITION OF AGGREGATES IS A HALLMARK OF NEURODEGENERATIVE PROTEINOPATHIES
The deposition of hazardous protein aggregates in cellular sites emerges as an additional arm of the cellular protein quality-control network, which is activated when the proteostasis network is overwhelmed. Cells actively accumulate disease-linked aggregates in different types of specialized deposition sites. Accordingly, the presence of deposition sites that contain aggregated proteins is a neuropathological hallmark of various neurodegenerative disorders (Soto 2003).
According to the amyloid hypothesis (Hardy and Higgins 1992), AD ensues from the proteolytic digestion of the amyloid precursor protein (APP) by the β and γ secretases, which release the family of aggregation-prone amyloid-β peptides (Aβ). Small Aβ oligomers are thought to be the most toxic species (Shankar et al. 2008; Cohen et al. 2009). Thus, their assembly into large aggregates of lower toxicity that are deposited in designated sites is probably protective. This notion is strongly supported by the observation that the inhibition of the insulin growth factor 1 (IGF-1)-signaling pathway protects worms (Cohen et al. 2006) and mice (Cohen et al. 2009) from Aβ toxicity while inducing the hyperaggregation of this toxic peptide.
It is important to note that the amyloid hypothesis has been seriously challenged by studies showing that, in some cases, AD develops as a result of loss of γ-secretase activity (Ben-Gedalya et al. 2015; Xia et al. 2015). Moreover, in many cases, brains of individuals who suffered from AD contain no excess of Aβ (Szaruga et al. 2015), and transmissibility experiments strongly suggest that different, prion-like Aβ strains exist (Meyer-Luehmann et al. 2006). Together, these studies propose that different mechanisms underlie AD, and protein aggregation is not necessarily deleterious.
PD, the most prevalent movement disorder, is pathologically characterized by the presence of cytosolic inclusions known as Lewy bodies (LBs) (Holdorff 2002). LBs contain aggregated α-synuclein (Spillantini et al. 1997), attract proteasomes (McNaught et al. 2002), and react with ubiquitin antibodies (Love et al. 1988). These observations suggest that LBs serve as quality-control compartments that enable efficient digestion of toxic α-synuclein assemblies. Recent studies show that α-synuclein prion-like assemblies cause multiple system atrophy, which shares many features observed in PD (Watts et al. 2013; Prusiner et al. 2015).
The deposition of aggregated PrP species in rod-like amyloidogenic structures is a common feature of CJD (Prusiner 1998) and other prion disorders (Salmona et al. 2003). Similarly to Aβ, small PrP oligomeric structures are the most infectious prion species (Silveira et al. 2005). In addition, the stabilization of PrP fibrils was found to reduce infectivity (Margalith et al. 2012). Together, these observations suggest that the assemblage of PrP oligomers to create large prion rods reduces toxicity.
The idea that hyperaggregation and sequestration of small oligomers reduce toxicity is further supported by the finding that chaperones that exhibit protective properties of disaggregation when the concentrations of aggregative proteins are low induce aggregation when the challenge of aggregation is increased (Shorter and Lindquist 2004).
Because of the limitations in the research of human brain tissues, it was necessary to develop laboratory models to investigate the biological and metabolic features of protein aggregate sequestration and deposition sites. These cellular models have enabled us to address the question of whether deposition sites actually serve as protective entities or perhaps as sources of toxicity.
CELLULAR DEPOSITION SITES
The formation of aggregate deposition sites in cultured cells was achieved by the overexpression of aggregation-prone proteins, the inhibition of protein degradation mechanisms, or the combination of both. An early study unveiled that the inhibition of proteasomes results in the accumulation of dense proteinaceous material, which cross-reacted with ubiquitin antibodies, in a juxtanuclear localization. The formation of these “proteolysis centers” was prevented by the inhibition of protein synthesis, and they were dispersed by the disruption of microtubules (Wojcik et al. 1996). These discoveries indicate that when cultured cells fail to clear damaged proteins, which under normal conditions are digested by proteasomes, these molecules are convoyed to a designated cellular location in a microtubule-dependent manner. Later, it was reported that concurrent overexpression of disease-linked, mutated, aggregation-prone proteins, and proteasome inhibition result in a similar phenomenon of protein deposition in a cytosolic, nucleus-adjacent location. These sites, which are termed aggresomes (Johnston et al. 1998), contain ubiqutinated proteins, are located at the microtubule-organizing center (MTOC), and are confined by collapsed fibers that are labeled by antibodies against the intermediate filament protein vimentin. The association of aggresomes with neurodegenerative maladies was demonstrated by accumulation of the familial AD (fAD)-linked, mutated presenilin 1 (PS1) carrying the A246E mutation (PS1 is an active component of the γ-secretase complex) in these structures (Johnston et al. 1998). Interestingly, PS1 molecules that harbor other fAD-causing mutations accumulate within the ER upon proteasome inhibition (Ben-Gedalya et al. 2015), showing that distinct conformers of the same protein can be sorted to distinct cellular deposition sites. Toxic PrP species (Kristiansen et al. 2005), disease-causing PrP mutants (Cohen and Taraboulos 2003; Mishra et al. 2003), and PD-associated, aggregated α-synuclein (Tanaka et al. 2004; Wong et al. 2008) were also shown to be deposited in aggresomes of mammalian cells, further linking these sites with human illnesses. However, the question of whether the accumulation of aggregated proteins in an aggresome is protective or deleterious to the cell remains largely unanswered.
If aggresomes serve as protein quality-control centers, it is expected that molecular chaperones and components of cellular degradation machineries will be attracted to these structures. Several research groups examined these assumptions and found that α-synuclein-containing aggresomes (McNaught et al. 2002) and aggresomes that harbor aggregates of GSS-linked, mutated PrP (Ben-Gedalya et al. 2011) attract folding chaperones and proteasomes. We further scrutinized this question by testing whether the deposition of aggregated PrP in aggresomes enabled its digestion. We fused PrP and yellow fluorescent protein (PrP-YFP), induced the formation of PrP-containing aggresomes, and followed the dynamics of the chimeric, fluorescently tagged protein aggregates within these deposits. Using live-imaging techniques, we found that PrP-YFP molecules are highly mobile within the aggresomes, and these structures exhibit a high rate of molecular exchange with the cytosol (Ben-Gedalya et al. 2011). Yet, although these indirect observations support the notion that the aggregate deposition sites are protective entities, it was required to directly test the relationships between these sites and cell survival.
A direct indication for the protective roles of deposition sites was provided by a live imaging technique. Visualization of neurons that either contain deposition sites that harbor aggregated, HD-linked polyQ stretches or lack such structures, unveiled that cells that contain deposition sites exhibit higher survival rates compared with their counterparts that do not contain such foci (Arrasate et al. 2004). This finding coincides with the finding that soluble polyQ oligomers, rather than large fibrils, activate the UPRER (Leitman et al. 2013). Nevertheless, a recent article that describes long-term surveillance of deposition sites containing aggregated α-synuclein indicated that over time these deposits are associated with cell death (Osterberg et al. 2015). This apparent contradiction can be explained by at least two models. One suggests that different types of deposition sites exhibit different properties (some are protective, whereas others are poisonous). An alternative explanation proposes that deposition sites are initially protective entities; however, over time, they become sources of toxicity that eventually lead to cell death (Ben-Gedalya and Cohen 2012).
AGGREGATE DEPOSITION SITES—BEYOND AGGRESOMES
To better characterize the nature of protein deposition sites, several groups used fluorescently tagged, aggregation-prone proteins and advanced microscopic techniques.
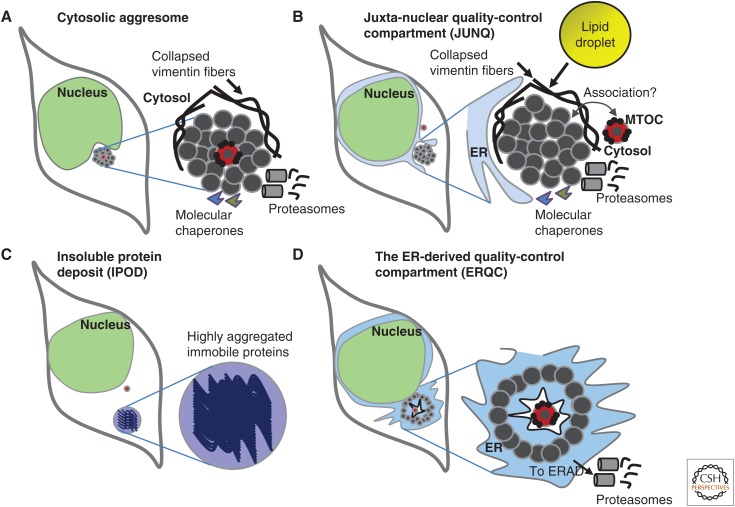
Using yeast and cultured mammalian cells that overexpress aggregation-prone proteins, it was discovered that different aggregative proteins are triaged to two distinct types of deposition sites that concurrently exist within a single cell (Kaganovich et al. 2008). One site that was found to be located in a juxtanuclear localization and to function as a dynamic quality-control deposition structure was termed the juxtanuclear quality control compartment (JUNQ). The JUNQ and aggresome share basic features (Fig. 3). They both are located proximal to the nucleus, contain highly mobile protein aggregates, exhibit a high rate of molecular exchange with the cytosol, and recruit molecular chaperones and proteasomes (Garcia-Mata et al. 1999; Ben-Gedalya et al. 2011). Interestingly, vimentin fibers are also important in the regulation of JUNQ and were found to control an asymmetric inheritance of JUNQ in mammalian cells (Ogrodnik et al. 2014). Unlike aggresomes, the JUNQ does not necessarily co-localize with the yeast’s spindle pole body (equivalent to the MTOC of mammalian cells) or with the MTOC (Kaganovich et al. 2008).
Figure 3.
Aggregated proteins are deposited in cellular sites. (A) The overwhelming of protein quality-control mechanisms by massively overexpressing certain aggregation-prone proteins, proteasome inhibition, or the impairment of chaperone activity leads to the deposition of aggregated proteins in juxtanuclear inclusion bodies known as aggresomes. Aggresomes are confined by collapsed vimentin fibers, co-localize with the microtubule-organizing center (MTOC), attract chaperones and proteasomes, and serve as protein quality-control centers. (B) The juxtanuclear quality control compartment (JUNQ) shares key features with the aggresome. It localizes next to the nucleus, possibly associated with the MTOC, and serves as a dynamic inclusion body. Lipid droplets (LDs) are found in close proximity with the JUNQ of yeast that overexpress aggregation-prone proteins. LDs secrete sterols that assist in clearing protein aggregates. (C) Terminally aggregated proteins accumulate in an insoluble protein deposit (IPOD). Proteins within the IPOD exhibit a low rate of molecular exchange with the cytosol and are highly immobile. (D) Under certain circumstances, proteins that aggregate within the ER are deposited in the ER-derived quality-control compartment (ERQC), which can serve as a platform for proteasome-mediated protein degradation.
A recent study proposes that the JUNQ resides within the nucleus and serves as a deposition center for nuclear as well as for cytosolic aggregated proteins (Miller et al. 2015). Accordingly, this site was termed the intranuclear quality-control compartment (INQ). Another study, which was based on a yeast screen, unveiled that lipid droplets are associated with the JUNQ and play key functional roles in the clearance of its content. This mechanism is proposed to be based on the secretion of hydroxyl sterols (Moldavski et al. 2015). Together, these studies indicate that different subtypes of dynamic quality-control compartments exist within cells and raise interesting questions. For instance, why is a certain aggregative protein deposited in a specific type of site, whereas other proteins are targeted to sites displaying different features? In addition, are there cell-type-specific preferences for the formation of a certain type of deposition site?
Another type of cellular deposition site, which is called an insoluble protein deposit (IPOD), contains immobile proteins, shows a low rate of exchange with the cytosol, is located away from the nucleus, and does not recruit proteasomes. The IPOD sequesters terminally aggregated, nondegradable proteins (Kaganovich et al. 2008). The recruitment of the fluorescently tagged, autophagy-related protein ATG8 to the IPOD increased the prospects that this site was a pre-autophagosome that later mediated the digestion of its content by the lysosome. This argument may be strengthened by the finding that autophagy is crucial for the clearance of some neurodegeneration-linked protein aggregates (Wong and Cuervo 2010); however, the possibility that IPODs are linked to autophagy requires further elucidation.
The export of proteins that misfold in the ER to the cytosol is probably a prerequisite for their deposition in aggresomes, the JUNQ, or the IPOD. However, in some cases, misfolded proteins are retained in the ER and accumulate in a suborganelle that contains misfolded ER-resident proteins and that is known as the ER-derived quality-control compartment (ERQC) (Kamhi-Nesher et al. 2001). Recently, the ERQC was shown to serve as an intermediate stage for ERAD substrates, indicating that this suborganelle plays a cytoprotective role (Leitman et al. 2014).
AD-linked, mutated PS1 that bears either the P264L or P267S substitution has been recently found to accumulate in the ERQC (Ben-Gedalya et al. 2015). This observation shows a direct link between this deposition site and certain cases of familial AD.
The aforementioned stress response and aggregate deposition mechanisms mediate interorganelle communication and transport within the individual, stressed cell. Therefore, it is commonly thought that each cell responds to stress in an autonomous manner. However, accumulating evidence shows that intertissue signaling cascades play a key role in the orchestration of protein quality control in the organism.
PROTEIN QUALITY CONTROL AT THE ORGANISMAL LEVEL
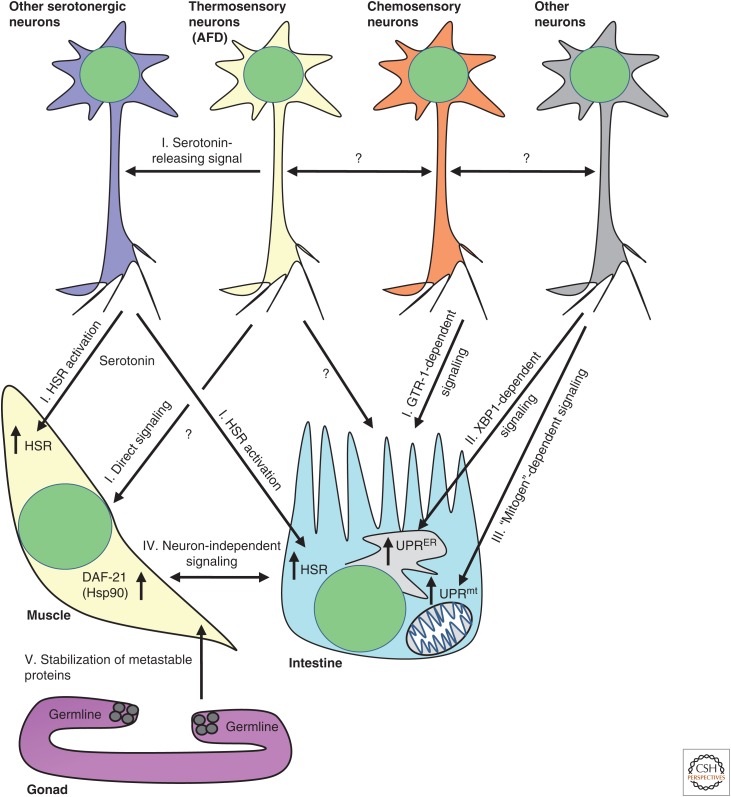
The transparency of C. elegans and the availability of genetic tools applicable in this organism enabled concurrent gene knockdown in a tissue-specific manner while visualizing the expression of fluorescent proteins in other tissues. This approach allowed the characterization of an intricate nexus of intertissue communication mechanisms that regulate stress responses and protein quality control at the organismal level (Fig. 4). One example of an intertissue link is the connection between the AFD neurons, their neighboring AIY interneurons, and distal tissues. These neurons are known to be crucial for heat sensing (Mori and Ohshima 1995). This function is dependent on the activity of the guanylyl cyclase gcy-8 in AFD neurons (Inada et al. 2006) and the LIM homeobox gene ttx-3 in AIY cells (Hobert et al. 1997). The activity of this neural circuit was shown not to be restricted to thermosensation but needed for HSR activation, because the knockdown of either gcy-8 or ttx-3 averts the expression of HSPs in remote tissues of heat-stressed animals (Prahlad et al. 2008). These findings indicate that the HSR is regulated at the organismal level by neuron-to-soma signaling. Serotonin is involved in this communication mechanism (Tatum et al. 2015), which not only activates the HSR but also modulates proteotoxicity in distal tissues (Prahlad and Morimoto 2011; Teixeira-Castro et al. 2015). The HSR activating network involves additional types of neurons. We found that gtr-1, which encodes a G-protein-coupled receptor that is expressed in chemosensory neurons, is also critically needed for HSR activation in non-neuronal tissues (Maman et al. 2013), and signals that originate from these neurons differentially control the activation of HSR-promoting transcription factors (Volovik et al. 2014).
Figure 4.
The regulation of proteostasis at the organismal level. Studies in nematodes unveiled that the proteostasis of somatic tissues is regulated by neuron-dependent and -independent manners. Thermosensory neurons (AFD) activate the heat-shock response (HSR) in the intestine upon exposure to heat (I). Serotonin and chemosensory neurons are involved in this activation. The HSR-coordinating, interneuronal communication mechanisms are largely unexplored. Neurons were also found to control the activity of the endoplasmic reticulum unfolded protein response (UPRER) (II) and mitochondrial unfolded protein response (UPRmt) (III) in distal tissues. Intestine and muscle cells exchange signals to activate the chaperone Hsp90 (DAF-21 in the nematode) when metastable proteins fail to fold properly (IV). Signals from germ cells regulate proteostasis in muscle cells (V).
The regulation of protein quality-control mechanisms by neurons is not limited to the HSR. The expression of a constitutively active isoform of the transcription factor XBP (XBPs) in neurons activates the UPRER in distal tissues (Taylor and Dillin 2013). Further evidence supporting the roles of neurons in the regulation of UPRER was recently provided by the report that the overactivation of IRE1 in ASI neurons promotes apoptosis of the worm’s germ cells (Levi-Ferber et al. 2014), cells whose ablation confers proteostasis in muscle cells (Shemesh et al. 2013). Similarly, UPRmt is activated in the nematode intestine by the inhibition of the electron transport chain in neurons (Durieux et al. 2011).
Proteostasis is also maintained in the nematode by a neuron-independent signaling mechanism. The expression of misfolding-prone proteins in muscle cells was shown to activate a communication mechanism that leads to the elevation of Hsp90 expression in muscle, intestinal, and pharyngeal cells (van Oosten-Hawle et al. 2013). This study shows that cell nonautonomous activation of hsp-90 is regulated by direct communication between somatic tissues.
Taken together, these findings show that intertissue communication coordinates the activation of proteostasis-maintaining mechanisms at the organismal level. Moreover, these findings present new research opportunities to develop treatments for proteinopathies based on the orchestration of proteostasis (Carvalhal Marques et al. 2015).
THE THERAPEUTIC POTENTIAL OF PROTEOSTASIS STABILIZATION
Because proteostasis failure underlies the development of various proteinopathies, the stabilization of proteostasis has a great potential to efficiently restore proteome integrity, postpone the manifestation of these maladies, and slow their progression once they have emerged. Several mechanisms can be targeted to rebalance the proteostasis network (for review, see Powers et al. 2009). First, small molecules that serve as chemical chaperones can directly assist nascent metastable proteins to fold properly and be located appropriately. This approach is suitable foremost for diseases that emanate from the loss of function of a single mutated protein. One prominent example of such a compound is Ivacaftor, which was approved for the treatment of certain cases of cystic fibrosis (CF). These cases stem from the mutation G551D in the sequence of the CF transmembrane conductance regulator (CFTR), which does not affect the protein’s localization but impedes its activity. Ivacaftor binds the mutated CFTR molecules, modulates their spatial structure, enhances their activity, and alleviates CFTR symptoms (Van Goor et al. 2009). Recently, Ivacaftor was approved for the treatment of CF patients who carry additional mutations that impair CF folding (Carter et al. 2015).
Reducing the levels of toxic oligomers by stabilizing large amyloidogenic fibrils has emerged as an additional therapeutic approach. Accordingly, compounds that specifically bind amyloids were shown to alleviate neurodegeneration-linked toxicity in nematode-based models (Alavez et al. 2011).
Next, compounds that activate stress-response mechanisms have the potential to restore proteostasis and serve as treatments for proteinopathies. Proteotoxicity models of C. elegans and mammalian cultured cells serve as efficient platforms for the screening of compound libraries and the investigation of specific drugs. Several promising HSR-activating compounds have been identified in such a screen (Calamini et al. 2012); however, this approach requires careful examination, because the activation of HSF-1 supports the progression of various types of cancer (Dai et al. 2007). UPRER activation has also been proposed as a proteostasis-restoring intervention that may have therapeutic potential (Halliday and Mallucci 2014). Similarly, it is conceivable that UPRmt activation may be useful for the treatment of certain mitochondrial syndromes.
Finally, it is apparent that compounds that target pathways that regulate aging have the potential to simultaneously activate several mechanisms that maintain protein quality control and rebalance proteostasis in the entire organism (Alavez and Lithgow 2012). As part of this approach, we discovered that NT219, a potent inhibitor of the IGF-1 signaling cascade that controls aging, protects model nematodes from phenotypes that stem from neurodegeneration-linked proteotoxicity (El-Ami et al. 2014). A careful analysis unveiled that although it enhances the expression of certain folding chaperones, NT219 reduces the activity of protein-degradation cellular machineries (Moll et al. 2016). These surprising results show the complexity of the proteostasis-maintaining mechanisms.
CONCLUDING REMARKS
The understanding that the maintenance of protein quality control is crucial for health and our deepening knowledge on the complexity of the proteostasis network provide new opportunities for the development of novel therapies for presently incurable proteinopathies. Accordingly, we will probably witness significant efforts to develop proteostasis-restoring drugs that function at all levels: assisting the folding of a single mutated protein aiming to restore its functionality, activating stress-response pathways, and modulating the mechanisms that control aging. The variable nature of neurodegenerative disorders and their complex effects on different brain regions suggest that, in the future, combinations of compounds will be personally tailored to restore proteostasis according to the needs of specific patients.
It is also important to intensify basic research to further explore the proteostasis network. With this in mind, it will be imperative to comprehensively examine whether specific types of deposition sites serve as protective entities or sources of toxicity, how lipid droplets contribute to proteostasis, and what roles cytoskeletal components play in protein deposition (Ogrodnik et al. 2014) and quality control (Baird et al. 2014).
Footnotes
Editor: Stanley B. Prusiner
Additional Perspectives on Prion Biology available at www.cshperspectives.org
REFERENCES
- Aguzzi A, Calella AM. 2009. Prions: Protein aggregation and infectious diseases. Physiol Rev 89: 1105–1152. [DOI] [PubMed] [Google Scholar]
- Alavez S, Lithgow GJ. 2012. Pharmacological maintenance of protein homeostasis could postpone age-related disease. Aging Cell 11: 187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alavez S, Vantipalli MC, Zucker DJ, Klang IM, Lithgow GJ. 2011. Amyloid-binding compounds maintain protein homeostasis during ageing and extend lifespan. Nature 472: 226–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaducci L, Tesco G. 1994. Aging as a major risk for degenerative diseases of the central nervous system. Curr Opin Neurol 7: 283–286. [DOI] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. 2004. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431: 805–810. [DOI] [PubMed] [Google Scholar]
- Baird NA, Douglas PM, Simic MS, Grant AR, Moresco JJ, Wolff SC, Yates JR III, Manning G, Dillin A. 2014. HSF-1-mediated cytoskeletal integrity determines thermotolerance and life span. Science 346: 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. 2008. Adapting proteostasis for disease intervention. Science 319: 916–919. [DOI] [PubMed] [Google Scholar]
- Bates G. 2003. Huntingtin aggregation and toxicity in Huntington’s disease. Lancet 361: 1642–1644. [DOI] [PubMed] [Google Scholar]
- Ben-Gedalya T, Cohen E. 2012. Quality control compartments coming of age. Traffic 13: 635–642. [DOI] [PubMed] [Google Scholar]
- Ben-Gedalya T, Lyakhovetsky R, Yedidia Y, Bejerano-Sagie M, Kogan NM, Karpuj MV, Kaganovich D, Cohen E. 2011. Cyclosporin-A-induced prion protein aggresomes are dynamic quality-control cellular compartments. J Cell Sci 124: 1891–1902. [DOI] [PubMed] [Google Scholar]
- Ben-Gedalya T, Moll L, Bejerano-Sagie M, Frere S, Cabral WA, Friedmann-Morvinski D, Slutsky I, Burstyn-Cohen T, Marini JC, Cohen E. 2015. Alzheimer’s disease-causing proline substitutions lead to presenilin 1 aggregation and malfunction. EMBO J 34: 2820–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calamini B, Silva MC, Madoux F, Hutt DM, Khanna S, Chalfant MA, Saldanha SA, Hodder P, Tait BD, Garza D, et al. 2012. Small-molecule proteostasis regulators for protein conformational diseases. Nat Chem Biol 8: 185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter S, Kelly S, Caples E, Grogan B, Doyle J, Gallagher CG, McKone EF. 2015. Ivacaftor as salvage therapy in a patient with cystic fibrosis genotype F508del/R117H/IVS8-5T. J Cyst Fibros 14: e4–e5. [DOI] [PubMed] [Google Scholar]
- Carvalhal Marques F, Volovik Y, Cohen E. 2015. The roles of cellular and organismal aging in the development of late-onset maladies. Annu Rev Pathol 10: 1–23. [DOI] [PubMed] [Google Scholar]
- Cohen E, Taraboulos A. 2003. Scrapie-like prion protein accumulates in aggresomes of cyclosporin A-treated cells. EMBO J 22: 404–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. 2006. Opposing activities protect against age-onset proteotoxicity. Science 313: 1604–1610. [DOI] [PubMed] [Google Scholar]
- Cohen E, Paulsson JF, Blinder P, Burstyn-Cohen T, Du D, Estepa G, Adame A, Pham HM, Holzenberger M, Kelly JW, et al. 2009. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell 139: 1157–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Whitesell L, Rogers AB, Lindquist S. 2007. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell 130: 1005–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drisaldi B, Stewart RS, Adles C, Stewart LR, Quaglio E, Biasini E, Fioriti L, Chiesa R, Harris DA. 2003. Mutant PrP is delayed in its exit from the endoplasmic reticulum, but neither wild-type nor mutant PrP undergoes retrotranslocation prior to proteasomal degradation. J Biol Chem 278: 21732–21743. [DOI] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. 2011. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 144: 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Ami T, Moll L, Carvalhal Marques F, Volovik Y, Reuveni H, Cohen E. 2014. A novel inhibitor of the insulin/IGF signaling pathway protects from age-onset, neurodegeneration-linked proteotoxicity. Aging Cell 13: 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L, Helenius A. 2003. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol 4: 181–191. [DOI] [PubMed] [Google Scholar]
- Fleig L, Bergbold N, Sahasrabudhe P, Geiger B, Kaltak L, Lemberg MK. 2012. Ubiquitin-dependent intramembrane rhomboid protease promotes ERAD of membrane proteins. Mol Cell 47: 558–569. [DOI] [PubMed] [Google Scholar]
- Folick A, Oakley HD, Yu Y, Armstrong EH, Kumari M, Sanor L, Moore DD, Ortlund EA, Zechner R, Wang MC. 2015. Aging. Lysosomal signaling molecules regulate longevity in Caenorhabditis elegans. Science 347: 83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata R, Bebok Z, Sorscher EJ, Sztul ES. 1999. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J Cell Biol 146: 1239–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday M, Mallucci GR. 2014. Targeting the unfolded protein response in neurodegeneration: A new approach to therapy. Neuropharmacology 76: 169–174. [DOI] [PubMed] [Google Scholar]
- Hardy JA, Higgins GA. 1992. Alzheimer’s disease: The amyloid cascade hypothesis. Science 256: 184–185. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. 2002. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science 295: 1852–1858. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Ron D. 2010. The mitochondrial UPR—Protecting organelle protein homeostasis. J Cell Sci 123: 3849–3855. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Molinari M. 2007. In and out of the ER: Protein folding, quality control, degradation, and related human diseases. Physiol Rev 87: 1377–1408. [DOI] [PubMed] [Google Scholar]
- Herrmann US, Sonati T, Falsig J, Reimann RR, Dametto P, O’Connor T, Li B, Lau A, Hornemann S, Sorce S, et al. 2015. Prion infections and anti-PrP antibodies trigger converging neurotoxic pathways. PLoS Pathog 11: e1004662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O, Mori I, Yamashita Y, Honda H, Ohshima Y, Liu Y, Ruvkun G. 1997. Regulation of interneuron function in the C. elegans thermoregulatory pathway by the ttx-3 LIM homeobox gene. Neuron 19: 345–357. [DOI] [PubMed] [Google Scholar]
- Holdorff B. 2002. Friedrich Heinrich Lewy (1885–1950) and his work. J Hist Neurosci 11: 19–28. [DOI] [PubMed] [Google Scholar]
- Inada H, Ito H, Satterlee J, Sengupta P, Matsumoto K, Mori I. 2006. Identification of guanylyl cyclases that function in thermosensory neurons of Caenorhabditis elegans. Genetics 172: 2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR. 1998. Aggresomes: A cellular response to misfolded proteins. J Cell Biol 143: 1883–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J. 2008. Misfolded proteins partition between two distinct quality control compartments. Nature 454: 1088–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamhi-Nesher S, Shenkman M, Tolchinsky S, Fromm SV, Ehrlich R, Lederkremer GZ. 2001. A novel quality control compartment derived from the endoplasmic reticulum. Mol Biol Cell 12: 1711–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen M, Messenger MJ, Klohn PC, Brandner S, Wadsworth JD, Collinge J, Tabrizi SJ. 2005. Disease-related prion protein forms aggresomes in neuronal cells leading to caspase activation and apoptosis. J Biol Chem 280: 38851–38861. [DOI] [PubMed] [Google Scholar]
- Lauffer L, Garcia PD, Harkins RN, Coussens L, Ullrich A, Walter P. 1985. Topology of signal recognition particle receptor in endoplasmic reticulum membrane. Nature 318: 334–338. [DOI] [PubMed] [Google Scholar]
- Leitman J, Ulrich Hartl F, Lederkremer GZ. 2013. Soluble forms of polyQ-expanded huntingtin rather than large aggregates cause endoplasmic reticulum stress. Nat Commun 4: 2753. [DOI] [PubMed] [Google Scholar]
- Leitman J, Shenkman M, Gofman Y, Shtern NO, Ben-Tal N, Hendershot LM, Lederkremer GZ. 2014. Herp coordinates compartmentalization and recruitment of HRD1 and misfolded proteins for ERAD. Mol Biol Cell 25: 1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi-Ferber M, Salzberg Y, Safra M, Haviv-Chesner A, Bülow HE, Henis-Korenblit S. 2014. It’s all in your mind: Determining germ cell fate by neuronal IRE-1 in C. elegans. PLoS Genet 10: e1004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S. 1986. The heat-shock response. Annu Rev Biochem 55: 1151–1191. [DOI] [PubMed] [Google Scholar]
- Love S, Saitoh T, Quijada S, Cole GM, Terry RD. 1988. Alz-50, ubiquitin and tau immunoreactivity of neurofibrillary tangles, Pick bodies and Lewy bodies. J Neuropathol Exp Neurol 47: 393–405. [DOI] [PubMed] [Google Scholar]
- Ma J, Lindquist S. 2001. Wild-type PrP and a mutant associated with prion disease are subject to retrograde transport and proteasome degradation. Proc Natl Acad Sci 98: 14955–14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maman M, Carvalhal Marques F, Volovik Y, Dubnikov T, Bejerano-Sagie M, Cohen E. 2013. A neuronal GPCR is critical for the induction of the heat shock response in the nematode C. elegans. J Neurosci 33: 6102–6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalith I, Suter C, Ballmer B, Schwarz P, Tiberi C, Sonati T, Falsig J, Nyström S, Hammarström P, Aslund A, et al. 2012. Polythiophenes inhibit prion propagation by stabilizing prion protein (PrP) aggregates. J Biol Chem 287: 18872–18887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinus RD, Garth GP, Webster TL, Cartwright P, Naylor DJ, Høj PB, Hoogenraad NJ. 1996. Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. Eur J Biochem 240: 98–103. [DOI] [PubMed] [Google Scholar]
- McNaught KS, Shashidharan P, Perl DP, Jenner P, Olanow CW. 2002. Aggresome-related biogenesis of Lewy bodies. Eur J Neurosci 16: 2136–2148. [DOI] [PubMed] [Google Scholar]
- Meyer-Luehmann M, Coomaraswamy J, Bolmont T, Kaeser S, Schaefer C, Kilger E, Neuenschwander A, Abramowski D, Frey P, Jaton AL, et al. 2006. Exogenous induction of cerebral β-amyloidogenesis is governed by agent and host. Science 313: 1781–1784. [DOI] [PubMed] [Google Scholar]
- Miller SB, Ho CT, Winkler J, Khokhrina M, Neuner A, Mohamed MY, Guilbride DL, Richter K, Lisby M, Schiebel E, et al. 2015. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J 34: 778–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra RS, Bose S, Gu Y, Li R, Singh N. 2003. Aggresome formation by mutant prion proteins: The unfolding role of proteasomes in familial prion disorders. J Alzheimers Dis 5: 15–23. [DOI] [PubMed] [Google Scholar]
- Moldavski O, Amen T, Levin-Zaidman S, Eisenstein M, Rogachev I, Brandis A, Kaganovich D, Schuldiner M. 2015. Lipid droplets are essential for efficient clearance of cytosolic inclusion bodies. Dev Cell 33: 603–610. [DOI] [PubMed] [Google Scholar]
- Moll L, Ben-Gedalya T, Reuveni H, Cohen E. 2016. The inhibition of IGF-1 signaling promotes proteostasis by enhancing protein aggregation and deposition. FASEB J 30: 1656–1669. [DOI] [PubMed] [Google Scholar]
- Mori I, Ohshima Y. 1995. Neural regulation of thermotaxis in Caenorhabditis elegans. Nature 376: 344–348. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. 1998. Regulation of the heat shock transcriptional response: Cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev 12: 3788–3796. [DOI] [PubMed] [Google Scholar]
- Naslavsky N, Stein R, Yanai A, Friedlander G, Taraboulos A. 1997. Characterization of detergent-insoluble complexes containing the cellular prion protein and its scrapie isoform. J Biol Chem 272: 6324–6331. [DOI] [PubMed] [Google Scholar]
- Ogrodnik M, Salmonowicz H, Brown R, Turkowska J, Sredniawa W, Pattabiraman S, Amen T, Abraham AC, Eichler N, Lyakhovetsky R, et al. 2014. Dynamic JUNQ inclusion bodies are asymmetrically inherited in mammalian cell lines through the asymmetric partitioning of vimentin. Proc Natl Acad Sci 111: 8049–8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterberg VR, Spinelli KJ, Weston LJ, Luk KC, Woltjer RL, Unni VK. 2015. Progressive aggregation of α-synuclein and selective degeneration of lewy inclusion-bearing neurons in a mouse model of parkinsonism. Cell Rep 10: 1252–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson HL. 1999. Protein fate in neurodegenerative proteinopathies: Polyglutamine diseases join the (mis)fold. Am J Hum Genet 64: 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. 2009. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem 78: 959–991. [DOI] [PubMed] [Google Scholar]
- Prahlad V, Morimoto RI. 2011. Neuronal circuitry regulates the response of Caenorhabditis elegans to misfolded proteins. Proc Natl Acad Sci 108: 14204–14209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahlad V, Cornelius T, Morimoto RI. 2008. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science 320: 811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. 1998. Prions. Proc Natl Acad Sci 95: 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB, Woerman AL, Mordes DA, Watts JC, Rampersaud R, Berry DB, Patel S, Oehler A, Lowe JK, Kravitz SN, et al. 2015. Evidence for α-synuclein prions causing multiple system atrophy in humans with parkinsonism. Proc Natl Acad Sci 112: E5308–E5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiano A, Foresti O, Carvalho P. 2014. Quality control: ER-associated degradation: Protein quality control and beyond. J Cell Biol 204: 869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmona M, Morbin M, Massignan T, Colombo L, Mazzoleni G, Capobianco R, Diomede L, Thaler F, Mollica L, Musco G, et al. 2003. Structural properties of Gerstmann–Sträussler–Scheinker disease amyloid protein. J Biol Chem 278: 48146–48153. [DOI] [PubMed] [Google Scholar]
- Sanders SL, Whitfield KM, Vogel JP, Rose MD, Schekman RW. 1992. Sec61p and BiP directly facilitate polypeptide translocation into the ER. Cell 69: 353–365. [DOI] [PubMed] [Google Scholar]
- Sarge KD, Murphy SP, Morimoto RI. 1993. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol 13: 1392–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ. 2003. Folding proteins in fatal ways. Nature 426: 900–904. [DOI] [PubMed] [Google Scholar]
- Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA, Rowan MJ, Lemere CA, et al. 2008. Amyloid-β protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat Med 14: 837–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh N, Shai N, Ben-Zvi A. 2013. Germline stem cell arrest inhibits the collapse of somatic proteostasis early in Caenorhabditis elegans adulthood. Aging Cell 12: 814–822. [DOI] [PubMed] [Google Scholar]
- Shiraishi N, Inai Y, Hirano Y, Ihara Y. 2011. Calreticulin inhibits prion protein PrP-(23-98) aggregation in vitro. Biosci Biotechnol Biochem 75: 1625–1627. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. 2004. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science 304: 1793–1797. [DOI] [PubMed] [Google Scholar]
- Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, Caughey B. 2005. The most infectious prion protein particles. Nature 437: 257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C. 2003. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat Rev Neurosci 4: 49–60. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. 1997. α-Synuclein in Lewy bodies. Nature 388: 839–840. [DOI] [PubMed] [Google Scholar]
- Stahl N, Baldwin MA, Burlingame AL, Prusiner SB. 1990. Identification of glycoinositol phospholipid linked and truncated forms of the scrapie prion protein. Biochemistry 29: 8879–8884. [DOI] [PubMed] [Google Scholar]
- Szaruga M, Veugelen S, Benurwar M, Lismont S, Sepulveda-Falla D, Lleo A, Ryan NS, Lashley T, Fox NC, Murayama S, et al. 2015. Qualitative changes in human γ-secretase underlie familial Alzheimer’s disease. J Exp Med 212: 2003–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Kim YM, Lee G, Junn E, Iwatsubo T, Mouradian MM. 2004. Aggresomes formed by α-synuclein and synphilin-1 are cytoprotective. J Biol Chem 279: 4625–4631. [DOI] [PubMed] [Google Scholar]
- Tatum MC, Ooi FK, Chikka MR, Chauve L, Martinez-Velazquez LA, Steinbusch HW, Morimoto RI, Prahlad V. 2015. Neuronal serotonin release triggers the heat shock response in C. elegans in the absence of temperature increase. Curr Biol 25: 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Dillin A. 2013. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell 153: 1435–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira-Castro A, Jalles A, Esteves S, Kang S, da Silva Santos L, Silva-Fernandes A, Neto MF, Brielmann RM, Bessa C, Duarte-Silva S, et al. 2015. Serotonergic signalling suppresses ataxin 3 aggregation and neurotoxicity in animal models of Machado–Joseph disease. Brain 138: 3221–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticozzi N, Tiloca C, Morelli C, Colombrita C, Poletti B, Doretti A, Maderna L, Messina S, Ratti A, Silani V. 2011. Genetics of familial amyotrophic lateral sclerosis. Arch Ital Biol 149: 65–82. [DOI] [PubMed] [Google Scholar]
- Urano F, Calfon M, Yoneda T, Yun C, Kiraly M, Clark SG, Ron D. 2002. A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. J Cell Biol 158: 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goor F, Hadida S, Grootenhuis PD, Burton B, Cao D, Neuberger T, Turnbull A, Singh A, Joubran J, Hazlewood A, et al. 2009. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc Natl Acad Sci 106: 18825–18830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosten-Hawle P, Porter RS, Morimoto RI. 2013. Regulation of organismal proteostasis by transcellular chaperone signaling. Cell 153: 1366–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volovik Y, Moll L, Marques FC, Maman M, Bejerano-Sagie M, Cohen E. 2014. Differential regulation of the heat shock factor 1 and DAF-16 by neuronal nhl-1 in the nematode C. elegans. Cell Rep 9: 2192–2205. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. 2011. The unfolded protein response: From stress pathway to homeostatic regulation. Science 334: 1081–1086. [DOI] [PubMed] [Google Scholar]
- Wang W, Chen R, Luo K, Wu D, Huang L, Huang T, Xiao G. 2010. Calnexin inhibits thermal aggregation and neurotoxicity of prion protein. J Cell Biochem 111: 343–349. [DOI] [PubMed] [Google Scholar]
- Watts JC, Giles K, Oehler A, Middleton L, Dexter DT, Gentleman SM, DeArmond SJ, Prusiner SB. 2013. Transmission of multiple system atrophy prions to transgenic mice. Proc Natl Acad Sci 110: 19555–19560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik C, Schroeter D, Wilk S, Lamprecht J, Paweletz N. 1996. Ubiquitin-mediated proteolysis centers in HeLa cells: Indication from studies of an inhibitor of the chymotrypsin-like activity of the proteasome. Eur J Cell Biol 71: 311–318. [PubMed] [Google Scholar]
- Wong E, Cuervo AM. 2010. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci 13: 805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ES, Tan JM, Soong WE, Hussein K, Nukina N, Dawson VL, Dawson TM, Cuervo AM, Lim KL. 2008. Autophagy-mediated clearance of aggresomes is not a universal phenomenon. Hum Mol Genet 17: 2570–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D, Watanabe H, Wu B, Lee SH, Li Y, Tsvetkov E, Bolshakov VY, Shen J, Kelleher RJ III. 2015. Presenilin-1 knockin mice reveal loss-of-function mechanism for familial Alzheimer’s disease. Neuron 85: 967–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedidia Y, Horonchik L, Tzaban S, Yanai A, Taraboulos A. 2001. Proteasomes and ubiquitin are involved in the turnover of the wild-type prion protein. EMBO J 20: 5383–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]