Abstract
Dysfunctionality of the p53 tumor suppressor is a major cause of therapeutic drug resistance in cancer. Recently we reported that mutant, but otherwise functional, p53V172F was inactivated in cisplatin-resistant 2780CP/Cl-16 and 2780CP/Cl-24 human ovarian tumor cells by increased recruitment of the inhibitor MDM4. The current study demonstrates that, unlike cisplatin, platinum analogs oxaliplatin and DACH-diacetato-dichloro-Pt(IV) (DAP), strongly stabilize and activate p53V172F in resistant cells, as indicated by prolonged p53 half-life and transactivation of targets p21 (CDKN1A) and MDM2. This increase in MDM2 reduced MDM4 levels in cell lysates as well as the p53 immunocomplex and prevented reversion of p53 to the inactive p53-MDM2-MDM4 bound state. Phosphorylation of p53 at Ser15 was demonstrated by all three drugs in sensitive A2780 and corresponding resistant 2780CP/Cl-16 and 2780CP/Cl-24 cell lines. However, cisplatin induced Ser20 phosphorylation in A2780 cells only, but not in resistant cells; in contrast, both DAP and oxaliplatin induced this phosphorylation in all three cell lines. The inference that Ser20 phosphorylation is more important for p53 activation was confirmed by ectopic expression of a phosphomimetic (S20D) mutant p53 that displayed reduced binding, relative to wild-type p53, to both MDM2 and MDM4 in p53-knockout A2780 cells. In consonance, temporal studies demonstrated drug-induced Ser15 phosphorylation coincided with p53 stabilization, whereas Ser20 phosphorylation coincided with p53 transactivation.
Implications
Cisplatin fails to activate the pathway involved in phosphorylating mutant p53V172F at Ser20 in resistant cells, but this phosphorylation is restored by oxaliplatin and DAP that reactivates p53 function and circumvents cisplatin resistance.
Keywords: cisplatin resistance, p53, V172F missense mutation, Serine-20 phosphorylation, MDM2, MDM4
Introduction
Among DNA damaging antitumor agents, cisplatin is perhaps the most effective. It is active against a number of cancers, including ovarian cancer, which is the most lethal gynecological cancer in United States (1–3). However, responses are short-lived as tumors acquire resistance to cisplatin and the resultant tumor progression leads to the high mortality rate (1, 2). A major cause of cisplatin resistance is loss of p53 function (4). Tumor cells with dysfunctional p53 fail to activate the apoptotic cell death program and this failure induces cellular tolerance to DNA damage (5). Dysfunctionality of p53 can occur through several mechanisms, including overexpression of p53 inhibitors MDM2 and MDM4 (6–8). Under basal conditions, the activity of p53 protein is inhibited by its interaction with MDM2 and the closely-related MDM4 (4). Both MDM2 and MDM4 bind to each other to form heterodimers through the C-terminal RING domain and bind independently to p53 through their N-termini to inhibit p53 transactivity. This interaction between MDM2 and MDM4 activates the E3 ligase activity of MDM2 and induces proteosomal degradation of p53 to suppress its endogenous cellular levels below the threshold required for transactivation of target genes, such as the cyclin-dependent kinase (CDK) inhibitor p21 (9–11). The inhibitory inter-relationship between the two homologs is regulated by their relative levels, with increased expression of MDM4 inhibiting both degradation and transactivity of p53 (10, 12). The complex interplay is further demonstrated by the observation that MDM2 induced in a p53-dependent manner by DNA-damaging agents then promotes MDM4 degradation as part of the facile cytotoxic response (9, 10, 13).
Stabilization and activation of p53 by DNA-damaging drugs, including cisplatin, are mediated through post-translational modification, with Ser15 and Ser20 phosphorylation reported to be tightly linked to the process (6, 14, 15). Phosphorylation at these two sites enables p53 to dissociate from MDM2 and/or MDM4 as a prerequisite for p53 transactivity, which is facilitated by concomitant binding of the freed phospho-p53 to transcription co-activators, such as p300 that enables post-translational acetylation of p53 at specific lysine residues (4, 14, 15). Moreover, phosphorylation of p53 is important to the cytotoxic process as failure to modify specific sites, such as Ser20, by mutations or deletions of upstream kinases is associated with loss of tumor sensitivity to a wide variety of therapeutic drugs (4).
Loss of p53 function can also occur through mutations in the DNA binding domain that can abolish normal p53 transactivity. In this regard, we have recently reported that cisplatin resistant human ovarian 2780CP/Cl-16 and 2780CP/Cl-24 tumor cells harbor mutant p53V172F, which may be important as it appears to select for cisplatin resistance (16). However, the missense V172F mutation is not directly inactivating, as the mutant p53 protein in its free state retains full transactivity, but its function is inhibited by mutation-driven elevation in MDM4 binding (16). Not surprisingly, disruption of the tri-protein p53-MDM2-MDM4 super-complex re-sensitizes resistant cells to cisplatin (16). Based on this observation, the present study was undertaken to examine whether specific drugs through their independent DNA damage response are capable of disrupting the super-complex and, thereby, restoring p53 function and circumventing cisplatin resistance as an important mode of their action. The experimental and FDA-approved anticancer agents DACH-diacetato-dichloro-Pt(IV) (DAP) (17–19) and oxaliplatin (19), respectively, were selected to test the concept as these platinum-based analogs are considered to be distinct from parental cisplatin but the underlying basis has not been defined. Our results indicate that DAP and oxaliplatin circumvent cisplatin resistance in 2780CP/Cl-16 and 2780CP/Cl-24 tumor cells by activating p53V172F, but this requires phosphorylation of p53 at the critical Ser20 site.
Material and Methods
Cell culture and reagents
Tissue culture conditions for A2780, 2780CP/Cl-16, 2780CP/Cl-24, HCT-116 p53+/+ and HCT-116 p53−/− cells have been described in our previous publications (16, 20). 2780CP/Cl-24 cells, which harbor temperature-sensitive mutant p53V172F, were also cultured at the permissive temperature of 32.5°C for at least 7 days to permit restoration of wild-type p53 function, as described previously (16). The control and p53−/− A2780 clones were selected from cells transfected by appropriate plasmids using the CRISPR system and made available to us by Dr. Michelle Martinez-Rivera (MD Anderson Cancer Center). Expression of p53 in clones was confirmed by Western blot analysis. Protein A beads (sc-2001) and antibodies for p53 (DO-7: sc-47698; FL-393: sc-6243), p21 (sc-6246), MDM2 (sc-13161), MDM4 (sc-374147) and β-actin (sc-47778) were purchased from Santa Cruz. Phospho-specific antibodies for p53-Ser15 (9284) and p53-Ser20 (9287) were purchased from Cell Signaling, MTT (19265) from Affymetrix, Lipofectamine® 2000 (11668019) from Life Technologies, Halt™ Protease & Phosphatase inhibitor (1861280) from Thermo Scientific and cisplatin (C2210000), MG132 (C2211), oxaliplatin (O9512) and cycloheximide (C1988) from Sigma. DAP was synthesized in-house, as previously described (21). The five p53 plasmids (WT, S20A, S20D, S15A and S15D) were kind gifts from Dr. Mickey C.-T. Hu (Stanford University School of Medicine). For maximal stability, stock solutions of cisplatin were prepared in saline, with oxaliplatin and DAP prepared in water.
Western blot and immunoprecipitation
Cells were harvested in a lysis buffer, as reported previously (16). For immunoblotting, 20–50 µg of protein from each sample was resolved by SDS-PAGE and transferred to nitrocellulose membrane. Membranes were then incubated overnight at 4°C with primary antibodies, washed and appropriate secondary antibodies conjugated to horseradish peroxidase (HRP) were then added to visualize the protein band by ECL fluorescence detection (Bio-Rad). DO-7 was used as the primary p53 antibody in immunoblots throughout. For densitometric quantitation, the X-ray films were scanned and the signals analyzed by the Image J software. Normalized protein expressions were plotted using GraphPad Prism computer program.
For immunoprecipitation, cell lysates were incubated overnight at 4°C with primary antibodies, and Protein A beads then added. Beads were centrifuged 1 hour later and washed three times with the immunocomplex wash buffer, as described before (16). Samples were then mixed with SDS loading buffer (2×) and boiled for 10 minutes. The supernatants were subjected to Western blot analysis, as described above.
Transfection studies
For studies with phosphomimetic p53, A2780 p53−/− cells were transected for 48 hours with empty vector (2 µg), WT-p53 (1 µg), S15A (2 µg), S20A (2 µg), S15D (1 µg) and S20D (1 µg) plasmids using Lipofectamine 2000. Transfections were adjusted to provide similar p53 expression levels. Cells were then treated with 2 µM MG132 for an additional 6 hours. Interactions between p53 and MDM4 and MDM2 were examined by immunoprecipitation, as described above. siRNA transfections were performed as previously described (16). For mutant p53V172F study, A2780 p53−/− cells were similarly transfected with 0.5 µg of WT-p53 or V172F-p53 plasmid. After 24 hr, cells were treated with cisplatin, DAP or oxaliplatin and incubated for a further 24 hours, after which time the cells were prepared for analysis by Western blot.
MTT cell growth inhibition assay
MTT assay was conducted as described before (16). In brief, cells in 100 µL media were plated into 96-well plates and exposed to the drugs 24 hours later. For this, stock drug solutions were serially diluted in ice-cold media and an equal volume (100 µL) added immediately to the wells. Growth inhibition was measured 5 days later. Specifically, MTT (3 mg/ml) was added to each well and incubated for 4 hours at 37°C. The medium was then replaced with 100 µL DMSO to dissolve the reduced formazan product. The absorbance of each well was determined at 570 nm in a microplate reader. The IC50 value was then determined by fitting the concentration versus growth inhibition data to a 4-parameter sigmoidal curve using the GraphPad Prism computer program.
Half-life assay
The half-life of p53 was determined as reported previously (16). Briefly, cells treated with each platinum drug were exposed to 4 µM cycloheximide (CHX). Cells were then sampled at timed intervals for up to 8 hours and subjected to immunoblot analysis using anti-p53 antibody. The intensity of the bands was determined using the Image J software. The half-life was determined by fitting the data to a monoexponential decay equation with a weighting factor of 1/y using the GraphPad Prism computer program. The half-life was calculated from the equation 0.693/α, where α is the decay rate constant.
Results
DAP and oxaliplatin overcome cisplatin resistance in a p53 dependent manner
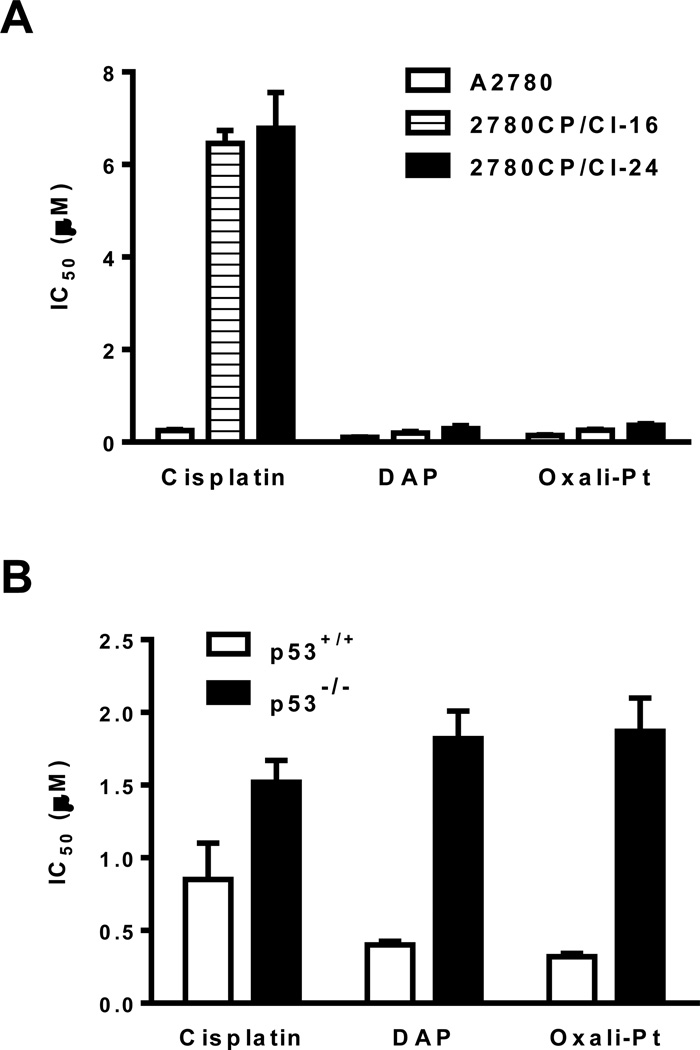
The sensitive A2780 and related cisplatin-resistant 2780CP/Cl-16 and 2780CP/Cl-24 human ovarian tumor cells were used in the study, and we have recently demonstrated that these three tumor models have functional p53, despite the presence of a V172F missense mutation in resistant cells (16). Since selected cisplatin-resistant tumor cell lines are reported to lack cross resistance to DAP and oxaliplatin (17–19), it was possible that this characteristic may be due to the ability of these agents to stabilize and activate p53 where cisplatin cannot. To examine this possibility at relevant drug concentrations, it was first necessary to establish the relative sensitivities of the cell lines to these platinum analogs by determining their IC50 (drug concentration inhibiting cell growth by 50%). The results in Figure 1A demonstrate that the IC50s of cisplatin in 2780CP/Cl-16 (6.5 ± 0.28 µM) and 2780CP/Cl-24 (6.8 ± 0.77 µM) cells were about 25-fold greater than in A2780 cells (0.25 ± 0.024 µM). In comparison, the IC50s of DAP and oxaliplatin in 2780CP/Cl-16 (DAP, 0.19 ± 0.038 µM; oxaliplatin, 0.26 ± 0.014 µM) and 2780CP/Cl-24 cells (DAP, 0.29 ± 0.062 µM; oxaliplatin, 0.36 ± 0.035 µM) were similar to those in A2780 cells (DAP, 0.10 ± 0.003 µM; oxaliplatin, 0.15 ± 0.010 µM) that indicated a low 2- to 3-fold cross-resistance of resistant cells to these analogs. To conclusively establish the mode of action of DAP and oxaliplatin and, specifically, the role of p53 in the circumvention of cisplatin resistance by these analogs, we also determined the IC50 values in HCT-116 cells proficient (p53+/+) or deficient (p53−/−) for p53. As in A2780 cells, DAP (0.40 ± 0.027 µM) and oxaliplatin (0.32 ± 0.022 µM) demonstrated greater potency than cisplatin (0.85 ± 0.25 µM) in HCT-116 p53+/+ cells, but a 2- to 6-fold loss in potency was demonstrated in HCT-116 p53−/− to each of these drugs (Figure 1B). Our data clearly show that all three drugs were dependent on p53 for their cytotoxicity.
Figure 1.
Cytotoxicity of cisplatin and analogs is p53-dependent. IC50 values of cisplatin, DAP and oxaliplatin were determined in sensitive A2780 and resistant 2780CP/Cl-16 and 2780CP/Cl-24 cells (A) or HCT-116 p53−/− and HCT-116 p53+/+ cells (B) using the MTT assay. The results are shown as mean ± SD of three independent experiments.
DAP and oxaliplatin induce p53 by increasing p53 half life
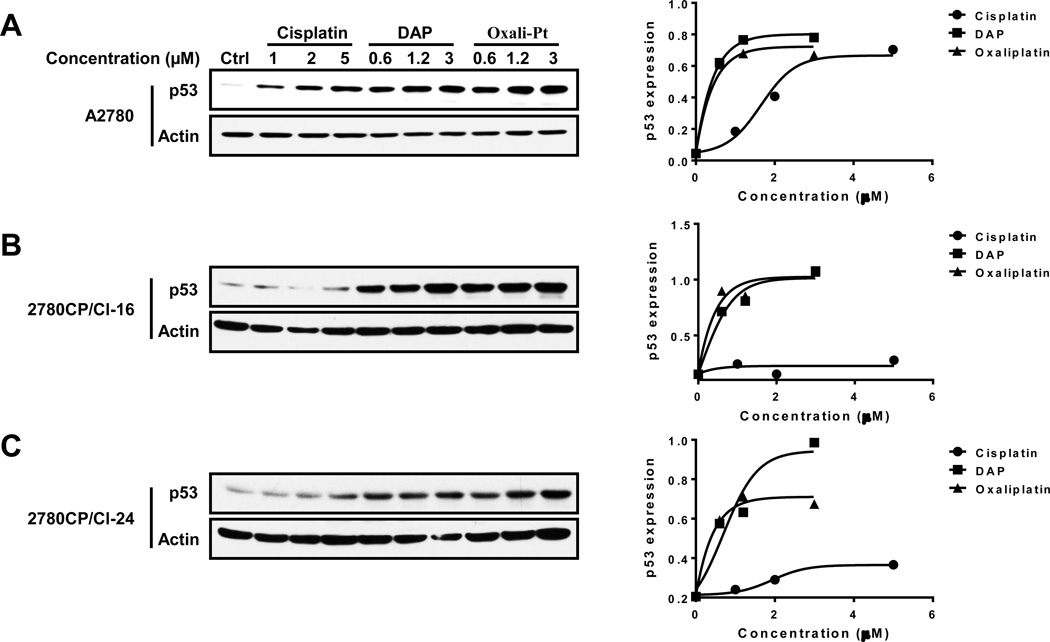
Based on the demonstration that activity of cisplatin, DAP and oxaliplatin is p53-dependent, it was appropriate to consider whether the differential cytotoxic response of these compounds in cisplatin-resistant cells can be explained at the level of p53 stabilization. We, therefore, prepared immunoblots of lysates from cells exposed to the platinum drugs for 24 hr using concentrations that were equivalent to about 5, 10 and 25 × IC50 levels in A2780 cells. The results show that cisplatin elevated p53 levels in sensitive A2780 cells, but failed to do so in resistant 2780CP/Cl-16 and 2780CP/Cl-24 cells (Figure 2A–C). DAP and oxaliplatin, on the other hand, robustly increased p53 levels in both sensitive and resistant tumor cells. When the bands in immunoblots were quantified by densitometry and plotted against drug concentration, it was clear that the increase in p53 by DAP and oxaliplatin was similar in all three cell lines (Figure 2A–C). Moreover, it was also apparent from results in A2780 cells that these two agents were more effective than cisplatin at inducing p53, and this was particularly evident at the low equitoxic drug concentration equivalent to about 5 × IC50 (cisplatin, 1.0 µM; analogs, 0.6 µM).
Figure 2.
DAP and oxaliplatin stabilize p53 in a dose-dependent manner in resistant 2780CP/Cl-16 and 2780CP/Cl-24 cells. A2780 (A), 2780CP/Cl-16 (B) and 2780CP/Cl-24 (C) cells were exposed to increasing concentrations of cisplatin, DAP or oxaliplatin for 24 hours. Cells were then collected and p53 expression levels determined by Western blot analysis. The p53 and β-actin signals in the immunoblots were quantified by densitometry, the ratios of p53/β-actin were then plotted against drug concentration.
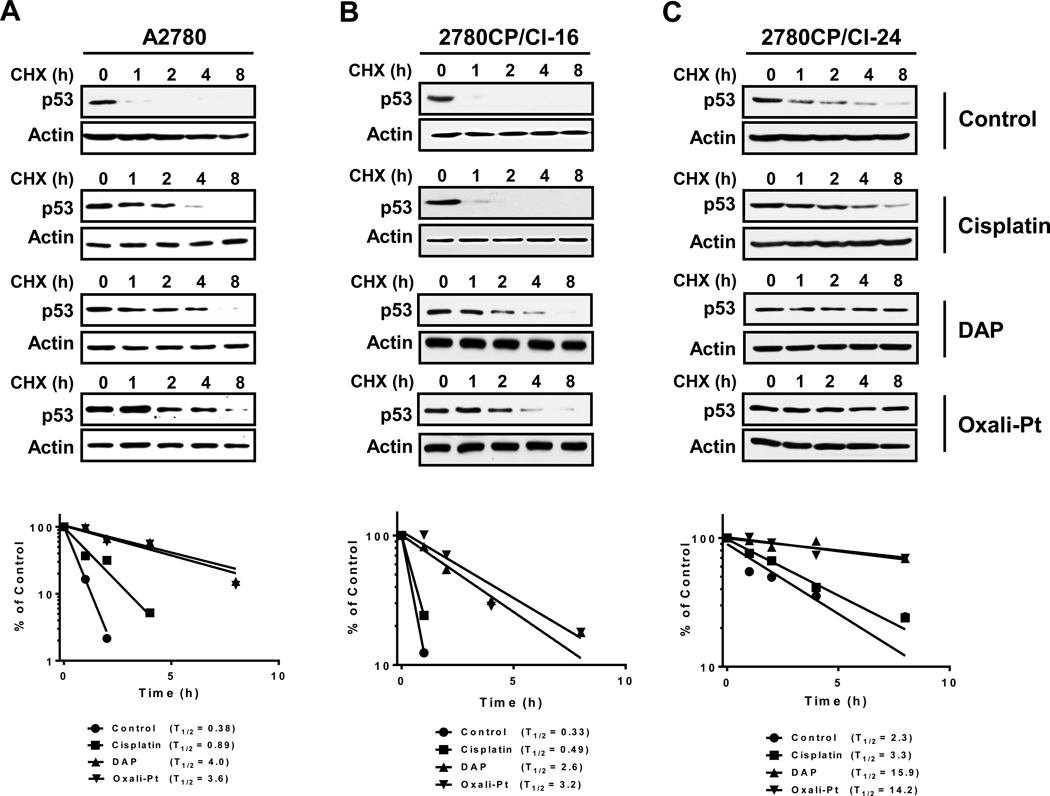
Levels of p53 are tightly regulated by the proteasome pathway, and inhibition of degradation by DNA-damaging agents leads to its stabilization (6, 10, 22). Therefore, to understand the differential effects between cisplatin and DAP or oxaliplatin on p53 levels in both sensitive and resistant cells, p53 half-life was determined using cycloheximide (CHX) to inhibit translation and allow accurate assessment of p53 decay. Cells treated with cisplatin (A2780, 1 µM; 2780CP/Cl-16 or 2780CP/Cl-24, 2 µM), DAP (A2780, 0.6 µM; 2780CP/Cl-16 or 2780CP/Cl-24, 1.2 µM) or oxaliplatin (A2780, 0.6 µM; 2780CP/Cl-16 or 2780CP/Cl-24, 1.2 µM) for 24 hours were then exposed to CHX for up to 8 hours. Cells sampled at timed intervals were immunoblotted for p53, and the bands quantified by densitometry and plotted against time to assess the p53 half-life (T1/2). The results in Figure 3 demonstrate that the relative half-life of p53 in control cells was 2780CP/Cl-24 > A2780 > 2780CP/Cl-16, and these differences confirmed our previous observation (16). With cisplatin, p53 was stabilized in sensitive A2780 cells, with a 2.3-fold increase in T1/2 of p53 vs. control (Figure 3A), but the increase in resistant cells was minimal (<1.5-fold) (Figures 3B and 3C). In contrast, substantial stabilization was observed with the analogs across the three cell lines, and this was reflected in a corresponding 6- to 10-fold increase in T1/2 (Figures 3A–C). These results demonstrate that DAP and oxaliplatin, but not cisplatin, can effectively increase the half-life of p53 as an index of its stabilization in resistant cells.
Figure 3.
Determination of p53 half-life in tumor cell lines exposed to platinum drugs. A2780 (A), 2780CP/Cl-16 (B) and 2780CP/Cl-24 (C) cells treated with cisplatin, DAP or oxaliplatin for 24 hours were additionally exposed to 4 µM cycloheximide (CHX) and then harvested at the indicated times. Cell lysates were prepared and resolved by SDS-PAGE, and p53 levels examined by immunoreaction. The p53 and β-actin signals in the immunoblots were quantified by densitometry, the ratios of p53/β-actin were normalized to time zero and plotted against time.
DAP and oxaliplatin induce MDM4 degradation in a MDM2-dependent manner
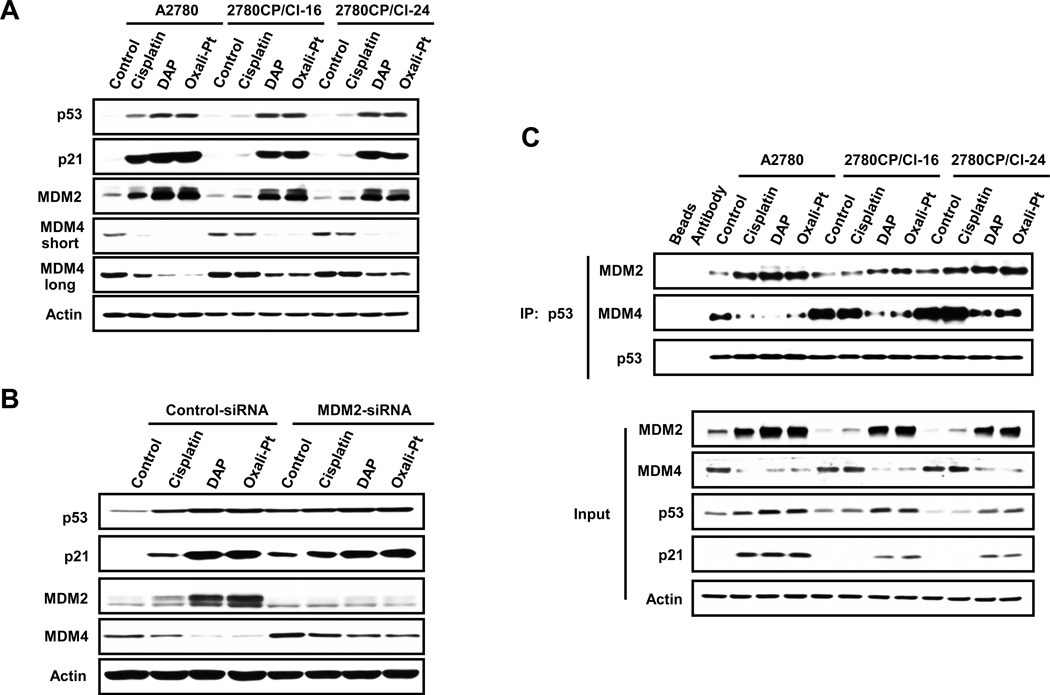
Failure of cisplatin to stabilize and transactivate p53 in 2780CP/Cl-16 and 2780CP/Cl-24 cells was recently ascribed by us to elevated MDM4 binding to p53 (16), and this provided the rationale to examine whether DAP and oxaliplatin were stabilizing and activating p53 in 2780CP/Cl-16 and 2780CP/Cl-24 cells by modulating levels of this endogenous inhibitor. The immunoblot in Figure 4A indicates that, unlike cisplatin, both DAP and oxaliplatin functionally activated p53 in these cisplatin-resistant cell lines, as demonstrated by increases in the two downstream targets of p53, namely p21 and MDM2. Conversely, DAP and oxaliplatin reduced MDM4 levels, with cisplatin failing to impact this protein in 2780CP/Cl-16 and 2780CP/Cl-24 cells. All three drugs, however, increased p53, p21 and MDM2 in sensitive A2780 cells, with concomitant decreases in MDM4. This apparent inverse relationship between MDM2 and MDM4 may be due to MDM2-dependent proteosomal degradation of MDM4, as has been proposed (23). To examine this possibility in our studies, A2780 cells exposed to MDM2 siRNA were treated the next day with each of the three platinum drugs and analyzed 24 hours later for specific proteins by immunoblot analysis. As shown in Figure 4B, knockdown of MDM2 moderately increased MDM4 in control drug-naive cells. As in Figure 4A, MDM2 increases in control-siRNA cells exposed to platinum drugs were associated with reduction in MDM4 levels, but these MDM4 reductions were attenuated when MDM2 was knocked down. Thus, MDM4 levels are directly regulated by MDM2 expression. It is notable that basal levels of p53 and p21 were increased by MDM2 siRNA, but this is to be expected since MDM2 knock-down is known to stabilize and activate p53 (16).
Figure 4.
Differential effects of cisplatin, DAP and Oxaliplatin on p53, MDM2 and MDM4 in resistant cells. (A) A2780, 2780CP/Cl-16 and 2780CP/Cl-24 cells were treated with cisplatin, DAP and oxaliplatin for 24 hours, harvested and then subjected to Western blot analysis using the indicated antibodies. (B) A2780 cells transfected with control-siRNA or MDM2-siRNA were treated with cisplatin, DAP or oxaliplatin for 24 hours, harvested and lysates prepared for Western blot analysis using the indicated antibodies. (C) A2780, 2780CP/Cl-16 and 2780CP/Cl-24 cells treated with cisplatin, DAP and oxaliplatin for 18 hours and then exposed to MG132 for another 6 hours. Cells were then harvested and lysates prepared and immunoprecipitated with the p53 antibody (FL-393). The proteins were resolved by SDS-PAGE and immunoblotted with the indicated antibodies (DO-7 for p53). Whole cell lysates were also similarly resolved and are represented as “input.”
Activity of p53 is also tightly regulated by its binding to MDM2 and MDM4. Since enhanced MDM4 binding to p53 to generate a super-complex was reported by us to inhibit p53 transactivity as a major mechanism of cisplatin resistance in 2780CP/Cl-16 and 2780CP/Cl-24 cells (16), it was necessary to investigate the composition of the molecular complex between p53, MDM2 and MDM4 and explain differences in effects between cisplatin and DAP or oxaliplatin in resistant cells. In Figure 4C, the tri-protein complex was monitored by immunoprecipitation of p53. The “input” immunoblot of lysates is consistent with results shown in Figure 4A, and this demonstrates reproducible molecular response to the platinum drugs in the three cell lines. Increased binding of MDM4 to p53 in control immunocomplex from resistant cells also confirms our earlier report. In general, however, relative levels of MDM2 and MDM4 in the p53 immunocomplex reflected those observed in the lysate. Thus, cisplatin increased MDM2 in the immunocomplex from A2780 cells and concomitantly diminished MDM4, with no such changes being observed in 2780CP/Cl-16 and 2780CP/Cl-24 cells. On the other hand, both DAP and oxaliplatin generally increased MDM2 and decreased MDM4 in lysates of sensitive and resistant cells. In consonance, the two analogs increased MDM2, but substantially reduced MDM4, binding to p53 compared to controls in all three cell lines that coincided with transactivation of p21. The slightly lower transactivation of this protein in resistant cells is consistent with the small residual cross-resistance to DAP and oxaliplatin seen in Figure 1A.
DAP and oxaliplatin, but not cisplatin, phosphorylate p53 at Ser20 in resistant cells
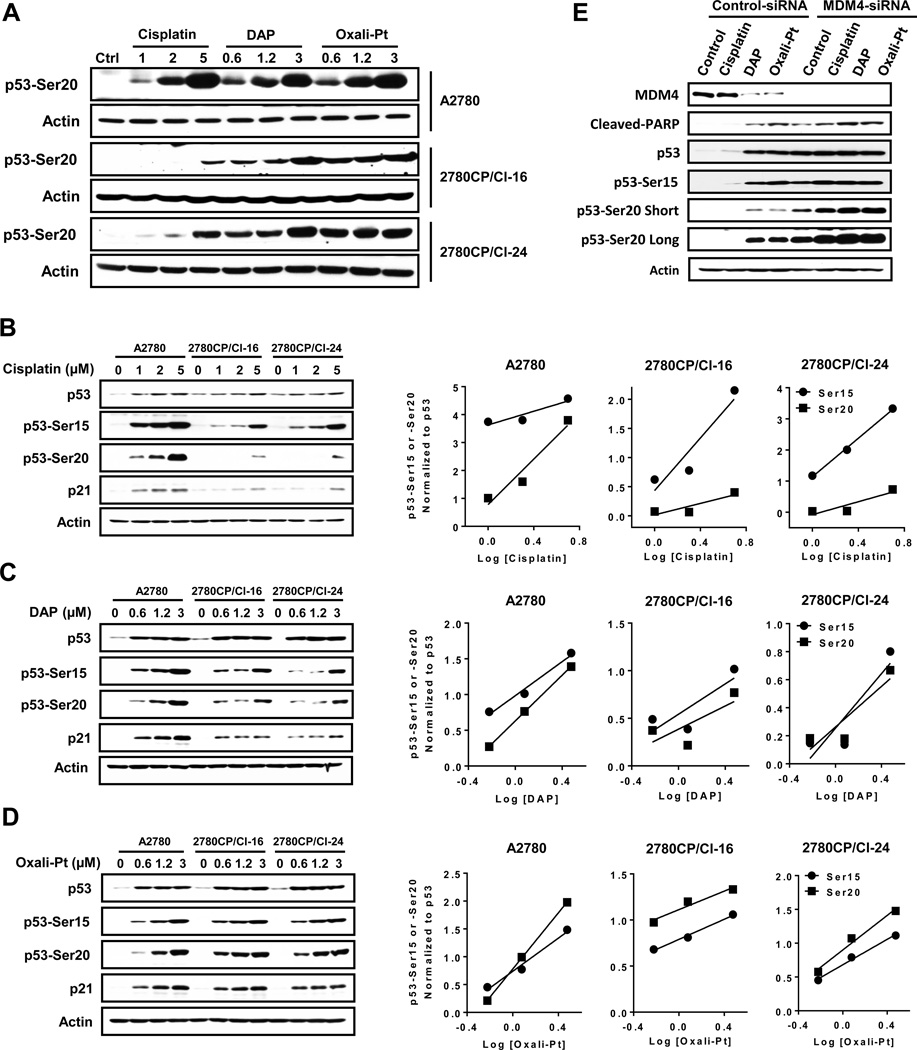
To better understand differential stabilization and activation of p53 through DNA damage-dependent regulation of the p53-MDM2-MDM4 complex by the specific platinum drugs requires an examination of the role of post-translational modification. In this regard, phosphorylation of Ser15 and Ser20 are reported to be principal sites associated with p53 stabilization and/or activation (14, 22, 24, 25). Both sensitive and resistant cells were exposed to cisplatin, DAP or oxaliplatin for 24 hours, and p53 phosphorylation monitored by immunoblot using phospho-specific antibodies. As shown in Figure 5A, dose-dependent increases in p53 phosphorylation at Ser20 in sensitive A2780 cells were similar for the three drugs, and this validated that drug concentrations equivalent to about 5, 10 or 25 × IC50 levels produced comparable levels of Ser20 phosphorylation with each drug. However, in resistant 2780CP/Cl-16 and 2780CP/Cl-24 cells, Ser20 phosphorylation by cisplatin was severely attenuated as compared to DAP or oxaliplatin. To better appreciate this difference and whether it extended to Ser15 phosphorylation, phospho-p53 immunoblots were prepared to permit direct comparisons between the three cell lines for each drug. These are shown in Figures 5B–D. With cisplatin, severe attenuation of Ser20 phosphorylation was confirmed in resistant cell lines compared to A2780 sensitive cells, and this is readily apparent at the highest drug concentration (Figure 5B). In contrast, dose-dependent phosphorylation at Ser15 was attenuated to a lesser extent in resistant cells. The difference between Ser15 and Ser20 phosphorylation in resistant cells is best observed in corresponding linear regression plots of drug concentrations against phospho-p53 levels normalized to total p53: plots for Ser15 and Ser20 converge with increasing cisplatin concentrations in A2780 cells, whereas they diverge in resistant 2780CP/Cl-16 and 2780CP/Cl-24 cells. This is a clear indication that the capacity of resistant cells to support induction of Ser20 phosphorylation with cisplatin is substantially reduced relative to Ser15, and correlates directly with attenuated p21 induction with cisplatin (Figure 5B). DAP and oxaliplatin, on the other hand, induced phosphorylation of p53 at both Ser15 and Ser20 in each cell line to a similar extent in a concentration-dependent manner (Figures 5C and 5D). The associated dose-response plots for Ser15 and Ser20 phosphorylation in general had similar slopes, which indicate that the phosphorylation machinery for both sites in sensitive and resistant cell lines remain equally responsive to DAP and oxaliplatin, and this correlated directly with p21 expression. Indeed, considering the data collectively with all three drugs, it is possible to generalize that phosphorylation of p53 on Ser20 closely parallels expression of p21.
Figure 5.
Post-translational phosphorylation of p53 by platinum drugs in ovarian tumor cells. A2780, 2780CP/Cl-16 and 2780CP/Cl-24 cells were treated with increasing concentrations of cisplatin, DAP or oxaliplatin for 24 hours, and lysates then prepared for Western blot analysis using the Ser20 phospho-specific antibody for direct comparison of effects of the three drugs in each cell line (A). The lysates from cells similarly treated with cisplatin (B), DAP (C) or oxaliplatin (D) were analyzed using the indicated antibodies to compare the effects of each drug in the three cell lines. The Ser15 and Ser20 signals in the immunoblots were quantified by densitometry and the ratios of Ser15:total p53 and Ser20:total p53 determined and plotted against concentration using linear regression analysis. In an independent study, 2780CP/Cl-24 cells transfected with control- or MDM4-siRNA were then treated with cisplatin, DAP or oxaliplatin for 24 hours. Cells were then harvested and subjected to Western blot using the indicated antibodies (E). In some cases, “short” and “long” exposures were used to visualize the proteins.
We have previously demonstrated that increased MDM4 binding to mutant p53V172F may induce cisplatin resistance, but post-translational modification of p53 was not investigated (16). Since increased MDM4 binding may restrict cisplatin-induced Ser20 phosphorylation and prevent activation of p53 function, this possibility was tested by MDM4 knockdown with siRNA. In 2780CP/Cl-24 cells, MDM4 knockdown alone resulted in p53 stabilization and increased levels of cleaved PARP as a biomarker of apoptosis (Fig. 5E). Interestingly, apoptosis correlated with induction of p53 phosphorylation at both Ser15 and Ser20 sites, which suggests that these sites are protected by MDM4. As expected, in 2780CP/Cl-24 cells exposed to control siRNA followed by cisplatin 24 hr later, total p53 and phospho-Ser15 were induced, but phospho-Ser20 was not. In addition, there was no overt reduction of MDM4 or increase in cleaved PARP in these control cells exposed to cisplatin. However, after MDM4 knockdown, cisplatin exposure further increased phosphorylation of the two sites and cleavage of PARP (Fig. 5E), which suggests drug-inducible phosphorylation at these sites is restricted in the presence of MDM4. In contrast, and consistent with data in Figures 5C and 5D, both DAP and oxaliplatin are capable of inducing Ser15 and Ser20 phosphorylation and inducing PARP cleavage in MDM4-proficient as well as MDM4-deficient cells. These results with the platinum analogs indicate that increased MDM4 binding does not entirely restrict mutant p53V172F in 2780CP/Cl-24 cells from phosphorylation at Ser20 of p53 and suggests that the machinery involved in phosphorylation of this site with cisplatin fails to become fully engaged and, thereby, reduces cytotoxic sensitivity to cisplatin.
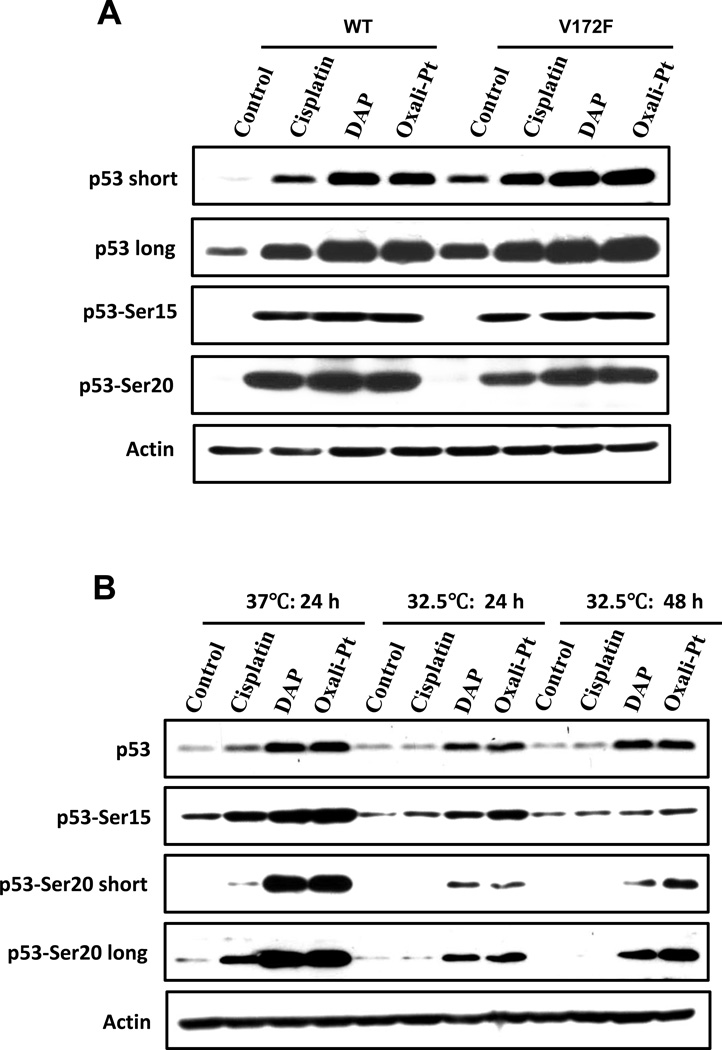
To support the conclusion that reduced phosphorylation of Ser20 with cisplatin demonstrated above is likely not due solely to mutant p53V172F, we obtained two pieces of evidence in the present study. First, our study documents that there were only small differences in phosphorylation at Ser20 or Ser15 with cisplatin, DAP and oxaliplatin in p53-knockout A2780 cells ectopically expressing wild-type p53 or p53V172F (Fig. 6A). Second, studies with 2780CP/Cl-24 cells grown at 32.5°C to permit conversion of temperature-sensitive mutant p53V172F to wild-type conformation did not result in any increase in Ser20 phosphorylation with cisplatin, even at the longer culture time of 48 hr to compensate for reduced response at the lower temperature (Fig. 6B). These data confirm failure of cisplatin to activate the pathway involved in Ser20 phosphorylation of mutant p53 in resistant cells, and suggest that drug-induced phosphorylation at Ser20 of p53 by the platinum analogs overcomes the negative effect of increased binding of MDM4 and, thereby, permits activation of mutant p53.
Figure 6.
Ser20 phosphorylation of p53V172F by platinum drugs. (A) p53-knockout A2780 cells transfected with wild type (WT) or mutant (V172F) p53 were treated the next day with cisplatin, DAP or oxaliplatin for 24 hours, then lysed and immunoblotted with the indicated antibodies. (B) 2780CP/Cl-24 cells cultured at 37°C or 32.5°C were treated with cisplatin, DAP or oxaliplatin for 24 hours (37°C and 32.5°C) or 48 hours (32.5°C), harvested and analyzed by Western blot. For p53-Ser20, “short” and “long” exposures were used to visualize the protein.
Ser20 phosphorylation inhibits MDM2 and MDM4 binding to p53
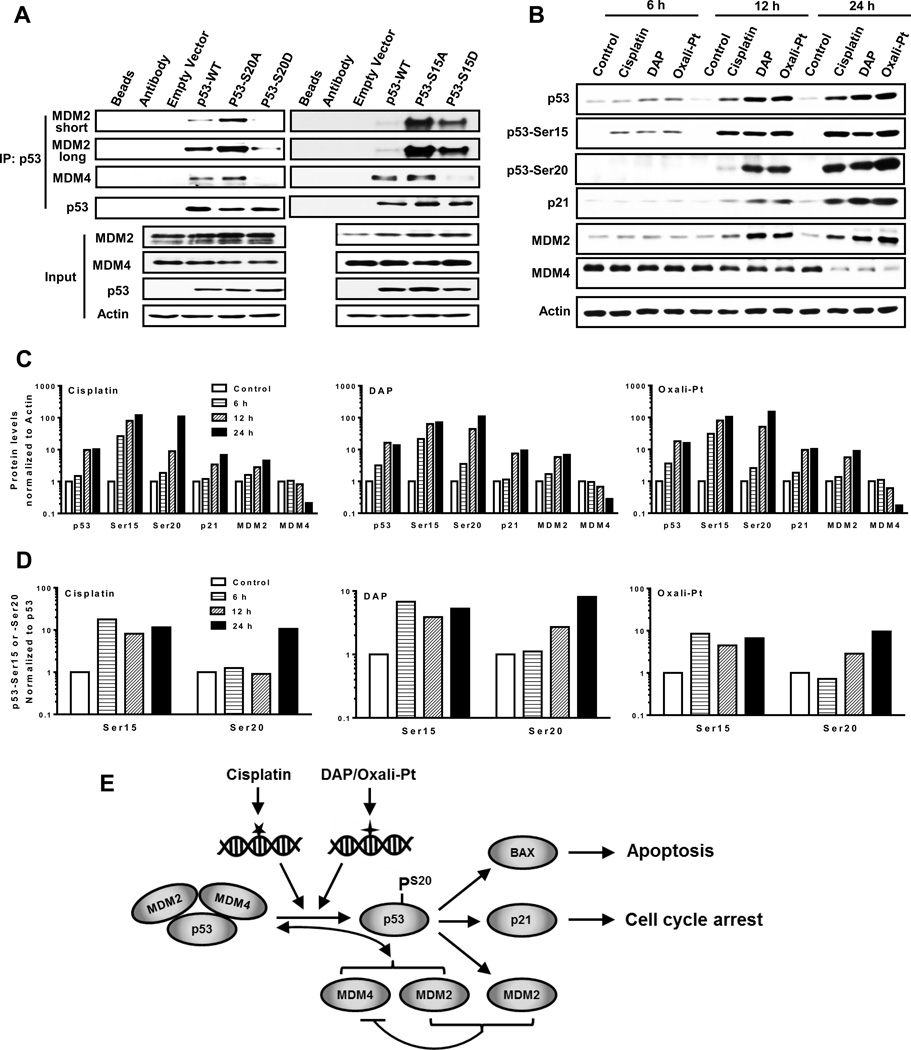
Our data indicates that phosphorylation at Ser20 activates p53, likely by inhibiting its binding to either MDM2, MDM4, or both. To validate this understanding and compare with Ser15 phosphorylation, specific p53 constructs were examined: wild-type p53, mutant p53-S15A and p53-S20A that cannot be phosphorylated, and mutant phosphomimetic p53-S15D and p53-S20D. A2780 cells engineered for p53 knockout were transfected with the five p53 constructs for 48 hours, harvested and processed for p53 immunoprecipitation. As shown in Figure 7A, expression of the p53 constructs in the lysate (input) were similar and did not substantially impact MDM2 and MDM4 levels. Immunoprecipitates indicated that ectopic wild-type p53 bound both MDM2 and MDM4, but this binding to p53-S20A mutant was relatively greater. In contrast, binding of MDM2 and MDM4 to p53-S20D mutant was markedly lower than to wild-type p53. Unlike p53-S20D, however, binding between p53-S15D and MDM2 was considerably greater relative to wild-type p53, although it was lower than with p53-S15A (Fig. 7A). On the other hand, binding of p53-S15D to MDM4 was much lower than wild-type p53 or mutant p53-S15A. These results indicate that phosphorylation of p53 at Ser20 may have a greater role than Ser15 in p53 activation since it inhibits p53 binding to both MDM2 and MDM4.
Figure 7.
The effect of Ser15 and Ser20 phosphorylation on MDM2 and MDM4 binding to p53. (A) A2780 p53−/− cells were transfected with empty vector, wild type p53 (p53-WT) and p53 mutant S20A, S20D, S15A or S15D for 48 hours. Cell lysates were then prepared and immunoprecipitated with the p53 antibody (FL-393), the proteins resolved by SDS-PAGE and immunoblotted with the indicated antibodies (DO-7 for p53). Whole cell lysates were used as input. (B) A2780 cells treated with cisplatin, DAP or oxaliplatin were harvested after 6, 12 or 24 hr treatment. Cell lysates were then prepared and subjected to Western blot analysis using the indicated antibodies. The signals from immunoblots in (B) were quantified by densitometry, and the ratios of indicated proteins to β-Actin were normalized to control and plotted for each platinum drug (C). The ratios of Ser15:total p53 and Ser20:total p53 were also determined and plotted similarly (D). (E) Schematic representation and summary of the distinct DNA damage induced by cisplatin and DAP or oxaliplatin that result in activation of independent pathways for phosphorylating p53 at Ser20, induction of MDM4 degradation and disruption of the p53-MDM2-MDM4 tri-protein complex in sensitive tumor cells. In resistant 2780CP/Cl-16 and 2780CP/Cl-24 cells, however, Ser20 phosphorylation by cisplatin is absent, and this disrupts normal p53 function and induces resistance. DAP and oxaliplatin, by virtue of their ability to use an independent pathway to restore Ser20 phosphorylation, activate p53 function and circumvent cisplatin resistance. Transactivation of MDM2 is important for inducing degradation of MDM4 in order to prevent re-formation of the inhibitory p53-MDM2-MDM4 complex and this, therefore, may prolong p53 activity. The stoichiometry of this complex is unknown and is presented here as a 1:1:1 complex for demonstration purposes only.
To consolidate the finding that activation of p53 is likely due to phosphorylation of p53 at Ser20, we examined temporal relationship between p53 phosphorylation and MDM2, MDM4 and p21 expressions in A2780 cells. All three drugs demonstrated an early DNA damage response, as indicated by Ser15 phosphorylation at 6 hr that increased with time (Figures 7B and 7C). Ser20 phosphorylation, on the other hand, became detectable at 12 hr with cisplatin, but was prominently upregulated with DAP and oxaliplatin at this time. By 24 hr, Ser20 phosphorylation was similar with all three drugs. Changes over time in Ser15 and Ser20 phosphorylation relative to total p53 were quantified by densitometry and the results are presented in Figure 7D. These results indicate that normalized phospho-Ser15 levels increase at 6 hr and remain constant thereafter, but phospho-Ser20 appears later and continues to increase up to at least 24 hr. More importantly, the temporal increase in Ser20 phosphorylation coincides directly with time-dependent increases in expression of MDM2 and p21 with each drug (Figures 7B and 7C), with decreases in MDM4 becoming prominent at 24 hr following MDM2 increases. These results indicate that Ser15 phosphorylation correlates with p53 stabilization, whereas Ser20 phosphorylation is associated with activation of p53, with the resultant increase in MDM2 then leading to reduction in MDM4 levels.
Discussion
We have previously reported that 2780CP/Cl-16 and 2780CP/Cl-24 tumor cells that harbor mutant p53V172F are resistant to cisplatin due to increased number of inhibitory MDM4 molecules bound to the tumor suppressor, and not mutation-based inactivation (16). In the present study, we have confirmed that this mutant p53 functions as wild-type, but we have also demonstrated that cisplatin cannot activate the mutant p53 in resistant cells due to its inability to induce phosphorylation of p53 at Ser20, although it could still induce Ser15 phosphorylation. In contrast, the platinum analogs DAP and oxaliplatin phosphorylate at Ser20 and Ser15 sites to stabilize and activate the mutant p53, and this is associated with circumvention of cisplatin resistance. The difference between cisplatin and DAP or oxaliplatin in their ability to induce Ser20 phosphorylation is not due to reduced accumulation of cisplatin in resistant cells, which has been documented as a mechanism of cisplatin resistance (5), since cross-reduction in uptake of DAP and oxaliplatin is similarly observed in resistant cells (19, 26).
Dysfunctionality of the p53 tumor suppressor has been documented to induce resistance to chemotherapy in cancer cells, and this is a major reason for treatment failure (4). Apart from mutations of p53 in the DNA binding domain, overexpression of MDM2 and/or MDM4 is also reported to result in p53 loss-of-function phenotype by increasing its proteosomal degradation and/or inhibiting its transcriptional activity (7–10, 27–29). This negative impact on p53 may likely be due to overexpressed MDM2 or MDM4 increasing the stoichiometry of their binding to the p53 molecule. Knockdown of MDM2 or inhibiting its binding to p53 using small molecule antagonists, such as Nutlin-3a and RITA, in MDM2-overexpressing tumor cells are reported to activate p53 and induce cell death, both in vitro and in vivo (30, 31). Accumulating evidence also demonstrates that overexpression of MDM4 can be a mechanism of resistance to chemotherapeutic agents (32–34), and several MDM4 inhibitors, therefore, have been explored preclinically for their potential to induce cell death (13, 28, 29, 33). However, in 2780CP/Cl-16 and 2780CP/Cl-24 cells, the enhanced MDM4 binding to mutant p53V172F was not due to elevated cellular expression levels of this protein (16). Moreover, in the present study, we demonstrate that the two structurally-distinct platinum-based analogs, DAP and oxaliplatin, activated p53 whether the binding of this inhibitor to p53 was high, as in resistant cells, or low, as in parental A2780 cells. This ability to activate p53 in all three cell lines is consistent with the analogs demonstrating similar potencies in these tumor models. Cisplatin, on the other hand, was only effective in activating p53 in sensitive A2780 cells, and its inability to disrupt the tri-protein p53-MDM2-MDM4 complex and activate p53 in 2780CP/Cl-16 and 2780CP/Cl-24 cells is the likely basis for these cells becoming resistant to this drug.
Post-translational modifications, such as phosphorylation, acetylation and ubiquitination, play important roles in regulating p53 function (6, 14, 15). Specifically, the stability and activity of p53 induced by antitumor agents are highly associated with phosphorylation at Ser15 and Ser20 (14, 22, 24, 25). However, which site is important in regulating p53 stability vs. activity is still not clearly defined. Based on our present investigations, it is possible to shed some light on the different roles of these two sites in regulating p53. Our temporal studies with cisplatin, DAP and oxaliplatin in A2780 cells demonstrate that the concordant increase in total p53 and phospho-Ser15 form at 6–24 hr is suggestive of Ser15 phosphorylation playing an important role in stabilizing p53. Phosphorylation of Ser20, on the other hand, appeared at 12 hr post-treatment together with concomitant increases in p21 and MDM2, and this indicates that Ser20 phosphorylation is consistent with p53 activation. These results are in keeping with reports that Ser20 phosphorylation can inhibit binding between MDM2 and p53 that then allows recruitment of the transcription coactivator p300 to p53 (22, 24, 25, 35, 36). Although evidence indicates that MDM4 can also bind to p53 at the transactivation domain and block p53 from interacting with p300 (10, 12, 15, 29, 31), our data with phosphomimetic p53 demonstrate that phosphorylation of Ser15 substantially inhibits binding of p53 to MDM4 only, whereas Ser20 phosphorylation inhibits binding to both MDM2 and MDM4 and, thereby, appears to be the stronger activator of the tumor suppressor. However, the possibility exists that inhibition of MDM4 binding to p53 requires cooperation between drug-induced Ser15 and Ser20 phosphorylation. On the other hand, the observation that Ser15 phosphorylation with cisplatin, DAP and oxaliplatin is similar in resistant cells supports the conclusion that downregulation of phosphorylation at Ser20 site with cisplatin is a critical event that prevents p53 activation and induces cisplatin resistance.
It is important to note that resistant 2780CP/Cl-16 and 2780CP/Cl-24 tumor cells not only lack phosphorylation at Ser20 of p53 in response to DNA damage by cisplatin, but also harbor mutant p53V172F. We have previously demonstrated that this mutation increases binding of MDM4 to p53 (16), and it is possible that the mutation works in concert with loss of Ser20 phosphorylation to induce cisplatin resistance. Importantly, phosphorylation at Ser20 by DAP or oxaliplatin is a seminal event activating both wild-type p53 and mutant p53V172F. However, cisplatin also induced phosphorylation of p53V172F that was ectopically expressed in A2780 cells, and this confirms a failure of cisplatin to upregulate Ser20 phosphorylation of endogenous mutant p53V172F in resistant cells only. At this point, it is unknown if a specific kinase fails to be activated by cisplatin in resistant cells to prevent Ser20 phosphorylation and, conversely, if a distinctly different kinase is activated by the two platinum analogs to restore this phosphorylation. The implication that independent kinases may likely be involved is based on our previous report that cisplatin is dependent on ATR, Chk1 and Chk2 for activating DNA damage-dependent signaling pathways, whereas DAP is independent of these kinases (37). Thus, additional studies may be needed to identify the key kinase associated with induction of Ser20 phosphorylation with each platinum drug. Nevertheless, some evidence already exists to support our conjecture that kinase dysfunction can be associated with loss of p53 function and drug resistance. For instance, dysfunctionality of ATM in B-cell chronic lymphocytic leukemia, which predominantly harbors wild-type p53, correlates with resistance to fludarabine and poor prognosis in patients (38–40). Similarly, 83% of tumor specimens from patients with NSCLC, a highly refractory cancer with a poor survival rate, and 23% of clinical ovarian cancers are reported to be Chk2-defective (41–44). Although not investigated in these reports, it is tempting to speculate, based on our studies, that loss of Ser20 phosphorylation may have induced refractoriness in these cancers.
It is noteworthy that MDM4 down-regulation by the three platinum drugs in the present study was also observed only when p53 became activated, as indicated by p21 and MDM2 up-regulations. MDM4 decreases by antitumor agents have been reported previously and are likely due to MDM2-dependent ubiquitination and proteosomal degradation (36, 45). However, it is uncertain why MDM4 must be degraded as part of an overall DNA damage response; it is possible that loss of MDM4 may extend the period of p53 transcriptional function even when MDM2 levels continue to increase as a result.
Structural considerations indicate that DNA adducts of cisplatin are different from those of DAP or oxaliplatin (46) and, consequently, these adducts transduce DNA damage signaling along independent pathways, which converge to activate p53 via Ser20 phosphorylation and, thereby, release bound MDM2 and MDM4, as summarized schematically in Figure 7E. The activated p53 then transactivates specific genes, such as the cell cycle CDK inhibitor p21 and the pro-apoptotic Bax. In addition, p53 transactivates MDM2, which promotes degradation of MDM4, and this may prolong p53 activity by preventing conversion to the inactive tri-protein p53-MDM2-MDM4 state. Based on this understanding, we further conclude that increased binding to p53V172F of MDM4 in conjunction with loss of Ser20 phosphorylation raise the threshold for cisplatin to activate p53, and this leads to cisplatin resistance. However, the tri-protein complex can be disrupted by DAP- or oxaliplatin-mediated restoration of this post-translational modification, which activates p53 and promotes high activity of these analogs in both sensitive and cisplatin-resistant cell lines. An additional significant finding from our study is that mutation of p53 does not necessarily lead to a direct loss-of-function phenotype, and that in some cases, as exemplified here with the p53V172F mutant, the wild-type p53 function can be harnessed to induce tumor cell killing by matching a specific drug to the right actionable phosphorylation pathway and/or kinase targeting the critical Ser20 site of p53.
Acknowledgments
The research support from the U.S. Public Health Service grants CA160687 to ZHS and Support Grant CA16672 to MD Anderson Cancer Center awarded by the National Cancer Institute, and in part from the Megan McBride Franz Endowed Research Fund, is gratefully acknowledged. We thank Dr. Mickey C.-T. Hu (Stanford University School of Medicine) for the kind gift of p53 plasmids. We also thank Michelle Martinez-Rivera for providing the A2780 p53-knockout clone.
Footnotes
Conflict of Interest
The authors have no conflicts of interest.
Author Contributions
XX, GH and ZHS designed and/or conducted the experiments, and analyzed and interpreted the data. XX and ZHS wrote the initial draft of the manuscript, and GH contributed to revision of the final version.
References
- 1.Agarwal R, Kaye SB. Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat Rev Cancer. 2003;3:502–516. doi: 10.1038/nrc1123. [DOI] [PubMed] [Google Scholar]
- 2.Yap TA, Carden CP, Kaye SB. Beyond chemotherapy: targeted therapies in ovarian cancer. Nat Rev Cancer. 2009;9:167–181. doi: 10.1038/nrc2583. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong DK, Bundy B, Wenzel L, et al. Intraperitoneal cisplatin and paclitaxel in ovarian cancer. New England Journal of Medicine. 2006;354:34–43. doi: 10.1056/NEJMoa052985. [DOI] [PubMed] [Google Scholar]
- 4.Martinez-Rivera M, Siddik ZH. Resistance and gain-of-resistance phenotypes in cancers harboring wild-type p53. Biochemical Pharmacology. 2012;83:1049–1062. doi: 10.1016/j.bcp.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 6.Kruse JP, Gu W. Modes of p53 Regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller PAJ, Vousden KH. Mutant p53 in Cancer: New Functions and Therapeutic Opportunities. Cancer Cell. 2014;25:304–317. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soussi T, Lozano G. p53 mutation heterogeneity in cancer. Biochemical and Biophysical Research Communications. 2005;331:834–842. doi: 10.1016/j.bbrc.2005.03.190. [DOI] [PubMed] [Google Scholar]
- 9.Perry ME. The Regulation of the p53-mediated Stress Response by MDM2 and MDM4. Cold Spring Harbor Perspectives in Biology. 2010:2. doi: 10.1101/cshperspect.a000968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang XJ, Jiang XJ. Mdm2 and MdmX partner to regulate p53. Febs Letters. 2012;586:1390–1396. doi: 10.1016/j.febslet.2012.02.049. [DOI] [PubMed] [Google Scholar]
- 11.Pei DS, Zhang YP, Zheng JN. Regulation of p53: a collaboration between Mdm2 and MdmX. Oncotarget. 2012;3:228–235. doi: 10.18632/oncotarget.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabbatini P, McCormick F. MDMX inhibits the p300/CBP-mediated acetylation of p53. Dna and Cell Biology. 2002;21:519–525. doi: 10.1089/104454902320219077. [DOI] [PubMed] [Google Scholar]
- 13.Marine JCW, Dyer MA, Jochemsen AG. MDMX: from bench to bedside. Journal of Cell Science. 2007;120:371–378. doi: 10.1242/jcs.03362. [DOI] [PubMed] [Google Scholar]
- 14.Maclaine NJ, Hupp TR. The regulation of p53 by phosphorylation: A model for how distinct signals Integrate into the p53 pathway. Aging-Us. 2009;1:490–502. doi: 10.18632/aging.100047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nature Reviews Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 16.Xie X, Lozano G, Siddik ZH. Heterozygous p53 mutation in cisplatin-resistant human tumor cells promotes MDM4 recruitment and decreases stability and transactivity of p53. Oncogene. 2016;35:4798–4806. doi: 10.1038/onc.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagopian GS, Mills GB, Khokhar AR, Bast RC, Siddik ZH. Expression of p53 in cisplatin-resistant ovarian cancer cell lines: Modulation with the novel platinum analogue (1R, 2R–diaminocyclohexane)(trans-diacetato)-(dichloro)platinum(IV) Clinical Cancer Research. 1999;5:655–663. [PubMed] [Google Scholar]
- 18.Siddik ZH, Hagopian GS, Thai G, Tomisaki S, Toyomasu T, Khokhar AR. Role of p53 in the ability of 1,2-diaminocyclohexane-diacetato-dichloro-Pt(IV) to circumvent cisplatin resistance. Journal of Inorganic Biochemistry. 1999;77:65–70. doi: 10.1016/s0162-0134(99)00144-0. [DOI] [PubMed] [Google Scholar]
- 19.Arambula JF, Sessler JL, Siddik ZH. A texaphyrin-oxaliplatin conjugate that overcomes both pharmacologic and molecular mechanisms of cisplatin resistance in cancer cells. Medchemcomm. 2012;3:1275–1281. doi: 10.1039/C2MD20206A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He G, Siddik ZH, Huang Z, et al. Induction of p21 by p53 following DNA damage inhibits both Cdk4 and Cdk2 activities. Oncogene. 2005;24:2929–2943. doi: 10.1038/sj.onc.1208474. [DOI] [PubMed] [Google Scholar]
- 21.Khokhar AR, al Baker S, Siddik ZH. Synthesis and antitumor activity of 1,2-diaminocyclohexane platinum(IV) complexes. J Inorg Biochem. 1994;54:39–47. doi: 10.1016/0162-0134(94)85122-0. [DOI] [PubMed] [Google Scholar]
- 22.Bode AM, Dong ZG. Post-translational modification of p53 in tumorigenesis. Nature Reviews Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 23.Kawai H, Wiederschain D, Kitao H, Stuart J, Tsai KKC, Yuan ZM. DNA damage-induced MDMX degradation is mediated by MDM2. Journal of Biological Chemistry. 2003;278:45946–45953. doi: 10.1074/jbc.M308295200. [DOI] [PubMed] [Google Scholar]
- 24.Appella E, Anderson CW. Post-translational modifications and activation of p53 by genotoxic stresses. European Journal of Biochemistry. 2001;268:2764–2772. doi: 10.1046/j.1432-1327.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- 25.Lavin MF, Gueven N. The complexity of p53 stabilization and activation. Cell Death and Differentiation. 2006;13:941–950. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- 26.Kido Y, Khokhar AR, al Baker S, Siddik ZH. Modulation of cytotoxicity and cellular pharmacology of 1,2-diaminocyclohexane platinum (IV) complexes mediated by axial and equatorial ligands. Cancer Res. 1993;53:4567–4572. [PubMed] [Google Scholar]
- 27.Cheok CF, Verma CS, Baselga J, Lane DP. Translating p53 into the clinic. Nature Reviews Clinical Oncology. 2011;8:25–37. doi: 10.1038/nrclinonc.2010.174. [DOI] [PubMed] [Google Scholar]
- 28.Li Q, Lozano G. Molecular Pathways: Targeting Mdm2 and Mdm4 in Cancer Therapy. Clinical Cancer Research. 2013;19:34–41. doi: 10.1158/1078-0432.CCR-12-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toledo F, Wahl GM. MDM2 and MDM4: p53 regulators as targets in anticancer therapy. International Journal of Biochemistry & Cell Biology. 2007;39:1476–1482. doi: 10.1016/j.biocel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chene P. Inhibiting the p53-MDM2 interaction: An important target for cancer therapy. Nature Reviews Cancer. 2003;3:102–109. doi: 10.1038/nrc991. [DOI] [PubMed] [Google Scholar]
- 31.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nature Reviews Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam S, Lodder K, Teunisse AF, Rabelink MJ, Schutte M, Jochemsen AG. Role of Mdm4 in drug sensitivity of breast cancer cells. Oncogene. 2010;29:2415–2426. doi: 10.1038/onc.2009.522. [DOI] [PubMed] [Google Scholar]
- 33.Gembarska A, Luciani F, Fedele C, et al. MDM4 is a key therapeutic target in cutaneous melanoma. Nature Medicine. 2012;18:1239. doi: 10.1038/nm.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin G, Cook S, Cui B, et al. HDMX regulates p53 activity and confers chemoresistance to 3-bis(2-chloroethyl)-1-nitrosourea. Neuro Oncol. 2010;12:956–966. doi: 10.1093/neuonc/noq045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bode AM, Dong ZG. Post-translational modification of p53 in tumorigenesis. Nature Reviews Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 36.Chen LH, Gilkes DM, Pan Y, Lane WS, Chen JD. ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. Embo Journal. 2005;24:3411–3422. doi: 10.1038/sj.emboj.7600812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He G, Kuang J, Khokhar AR, Siddik ZH. The impact of S- and G2-checkpoint response on the fidelity of G1-arrest by cisplatin and its comparison to a non-cross-resistant platinum(IV) analog. Gynecol Oncol. 2011;122:402–409. doi: 10.1016/j.ygyno.2011.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kojima K, Konopleva M, McQueen T, O’Brien S, Plunkett W, Andreeff M. Mdm2 inhibitor Nutlin-3a induces p53-mediated apoptosis by transcription-dependent and transcription-independent mechanisms and may overcome Atm-mediated resistance to fludarabine in chronic lymphocytic leukemia. Blood. 2006;108:993–1000. doi: 10.1182/blood-2005-12-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lavin MF, Kozlov S. DNA damage-induced signalling in ataxia-telangiectasia and related syndromes. Radiotherapy and Oncology. 2007;83:231–237. doi: 10.1016/j.radonc.2007.04.032. [DOI] [PubMed] [Google Scholar]
- 40.Ripolles L, Ortega M, Ortuno F, et al. Genetic abnormalities and clinical outcome in chronic lymphocytic leukemia. Cancer Genetics and Cytogenetics. 2006;171:57–64. doi: 10.1016/j.cancergencyto.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 41.MacLaren A, Slavin D, McGowan CH. Chk2 Protects against Radiation-Induced Genomic Instability. Radiation Research. 2009;172:463–472. doi: 10.1667/RR1603.1. [DOI] [PubMed] [Google Scholar]
- 42.Takai H, Naka K, Okada Y, et al. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. Embo Journal. 2002;21:5195–5205. doi: 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang PL, Wang J, Gao WY, Yuan BZ, Rogers J, Reed E. CHK2 kinase expression is down-regulated due to promoter methylation in non-small cell lung cancer. Molecular Cancer. 2004:3. doi: 10.1186/1476-4598-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams LH, Choong D, Johnson SA, Campbell IG. Genetic and epigenetic analysis of CHEK2 in sporadic breast, colon, and ovarian cancers. Clinical Cancer Research. 2006;12:6967–6972. doi: 10.1158/1078-0432.CCR-06-1770. [DOI] [PubMed] [Google Scholar]
- 45.Okamoto K, Kashima K, Pereg Y, et al. DNA damage-induced phosphorylation of MdmX at Serine 367 activates p53 by targeting MdmX for Mdm2-dependent degradation. Molecular and Cellular Biology. 2005;25:9608–9620. doi: 10.1128/MCB.25.21.9608-9620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siddik ZH. Mechanism of action of cancer chemotherapeutic agents: DNA-interactive alkylating agents and antitumor platinum-based drugs. In: Alison MR, editor. The Cancer Handbook. London: Macmillan Publishers; 2002. pp. 1295–1311. [Google Scholar]