Abstract
In two brothers born to consanguineous parents, we identified an unusual neurological disease that manifested with ataxia, psychomotor retardation, cerebellar and cerebral atrophy, and leukodystrophy. Via linkage analysis and exome sequencing, we identified homozygous c.2801C>T (p.(Ser934Leu)) in POLR1A (encoding RPA194, largest subunit of RNA polymerase I) and c.511C>T (p.(Arg171Trp)) in OSBPL11 (encoding oxysterol-binding protein-like protein 11). Although in silico analysis, histopathologic evidence and functional verification indicated that both variants were deleterious, segregation with the patient phenotype established that the POLR1A defect underlies the disease, as a clinically unaffected sister also was homozygous for the OSBPL11 variant. Decreased nucleolar RPA194 was observed in the skin fibroblasts of only the affected brothers, whereas intracellular cholesterol accumulation was observed in the skin biopsies of the patients and the sister homozygous for the OSBPL11 variant. Our findings provide the first report showing a complex leukodystrophy associated with POLR1A. Variants in three other RNA polymerase subunits, POLR1C, POLR3A and POLR3B, are known to cause recessive leukodystrophy similar to the disease afflicting the present family but with a later onset. Of those, POLR1C is also implicated in a mandibulofacial dysostosis syndrome without leukodystrophy as POLR1A is. This syndrome is absent in the family we present.
Introduction
Three mammalian polymerases, RNA polymerases (Pol) I, II and III, transcribe the major classes of RNAs: the ribosomal, messenger and transfer RNAs. Of these, Pol I transcribes the 47S precursor rRNA that is processed to the mature 28S, 18S and 5.8S rRNAs. Pol I transcription is the critical rate-limiting step in ribosome biogenesis. The transcription is compartmentalized to the nucleolus and attunes to the critical needs of protein synthesis during cell growth, division and differentiation.1, 2, 3 Defects in three subunits of Pol III, namely, POLR1C (shared by Pol I), POLR3A and POLR3B, have been associated with recessive hypomyelinating leukodystrophy.4, 5, 6 Defects in Pol I complex subunits POLR1A, POLR1C and POLR1D cause craniofacial anomalies consistent with perturbed ribosome biogenesis.7, 8
Leukodystrophies are heterogeneous disorders that primarily affect the white matter and are associated with anomalies of glial cells and myelin sheath.9 They are classified as hypomyelinating and demyelinating according to MRI results. The two brothers born to consanguineous Turkish parents we present here have cerebellar ataxia associated with spasticity, intellectual disability, cerebellar and cerebral atrophy, and demyelinating leukodystrophy. Candidate loci were found by linkage analysis, and homozygous variants in POLR1A and OSBPL11 were identified by exome sequencing. An unaffected sister was homozygous for the OSBPL11 variant but did not carry the POLR1A variant. The POLR1A defect was verified to be pathogenic by molecular modelling studies and functional assays. These findings constitute the first report showing a leukodystrophy syndrome associated with POLR1A, encoding RPA194, catalytic subunit of RNA polymerase I.
Subjects and methods
Family
The parents were half-first cousins once removed, with two affected sons and two healthy daughters (Figure 1). The study was conducted in accordance with the Declaration of Helsinki and national guidelines. Informed consent was obtained from/for participants in accordance with the regulations of the Boğaziçi University Institutional Review Board for Research with Human Participants that approved the study protocol.
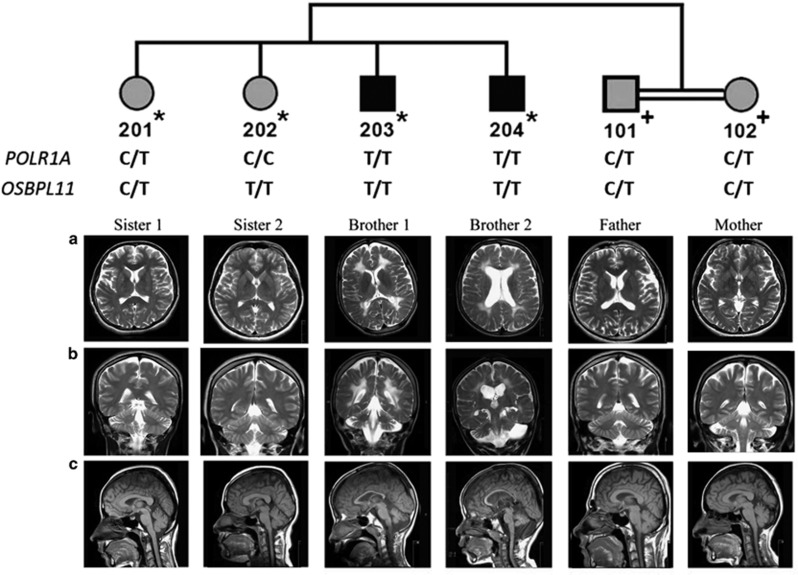
Figure 1.
The pedigree and cranial MR images. DNA samples subjected to SNP genotyping are indicated by * on the pedigree, and those for variant testing are indicated by +. POLR1A and OSBPL11 variant genotypes are given. MR images of the brothers revealed enlarged ventricles, cortical sulci and subarachnoid spaces indicative of cerebral atrophy, diffuse hyperintense involvement of periventricular white matter extending to subcortical white matter, atrophy of the cerebellar hemispheres and the vermis, and thin corpus callosum. Brother 2 has additionally a subarachnoid cyst in the left posterior fossa. The sisters have normal MRI findings. Parents have normal white matter and very mild cerebral atrophy. (a) Axial T2 weighted. (b) Coronal T2 weighted. (c) Sagittal T1 weighted.
Genetic analyses
Single-nucleotide polymorphism (SNP) genome scan was performed for the siblings, and multipoint LOD scores were calculated assuming autosomal recessive inheritance, initially with full penetrance and later with 70% penetrance. All loci >200 kb and yielding LOD scores of >2 were investigated for homozygosity possibly identical by descent. In addition, regions of homozygosity shared by the brothers only were searched using Homozygosity Mapper. Exome of brother 1 was sequenced and data were evaluated as described in Supplementary Methods. Hg19 map was used throughout the study.
RPA194 homologue structure, domains, and tertiary structure prediction
The full-length human RPA194 (O95602) and S. cerevisiae A190 (P10964) sequences were identified and divided into three sections, based on their predicted domain architecture, for submission to tertiary structure predictions. Yeast A190 was used as a control for the accuracy of the tertiary structure prediction programs as compared with the solved structure.10, 11 Domain architecture predictions were made via Pfam, PROSITE, SMART and the CDD. The residue of interest, Ser934, locates to the middle section of RPA194 that we will refer to as RPA194_Segment2. The sequences of RPA194_Segment2 and A190_Segment2 were submitted for tertiary structure prediction via Phyre2.12 All tertiary structure predictions were subjected to evaluation using ProSAweb and Verify3D.13, 14 Models were then ranked by these results, and the best model for RPA194_Segment2 was selected for further analysis. Solved structures of yeast A190 were submitted for model evaluation as control. PyMOL was used to visualize the best model and the solved structure 4c2m. Using PyMOL, the human RPA194_Segment2 was superimposed with the solved structures of yeast A190. Ser934 was then mutated to Leu using the PyMOL mutagenesis tool. PyMOL distance measurement tool was used to assess possible interactions between RPA194 and RPA135 (A127 in yeast) based on the prediction of interactions described in 4c2m.
Cell culture
Fibroblast cultures were established from skin biopsies of the father and sons. Control fibroblast cell lines AG08498 and GM0323 were obtained from Coriell Institute for Medical Research (Camden, NJ, USA), and F92-99 has been described previously.15 Fibroblast cell cultures were maintained in Eagle's minimum essential medium supplemented with 15% FBS, 2 mM L-glutamine, 100 IU/ml penicillin and 100 μg/ml streptomycin.
Immunofluorescence microscopy and image analysis
Fibroblasts grown on coverslips were fixed in 3.5% PFA, permeabilized with 0.5% NP-40 and blocked in 3% BSA. The primary antibodies used were RPA194 (C-1, Santa Cruz Biotechnology, Santa Cruz, CA, USA) and FBL (ab582, Abcam, Cambridge, MA, USA). Secondary Alexa488 and Alexa594-conjugated anti-mouse and anti-rabbit antibodies were from Invitrogen (Carlsbad, CA, USA). DNA was stained using DAPI. Images were captured using Leica DM6000B fluorescence wide-field microscope equipped with Hamamatsu Orca-Flash sCMOS camera, 20 × objective (20 × /0.7 HC PL APO CS, Leica, Wetzlar, Germany) and LasX software (Leica). Image analysis was conducted using FrIDA software as described previously.16 An average of 100 cells were quantified from two fields for each sample.
Filipin staining of skin biopsy samples
Skin samples were obtained from the two patients, sister 2 (202 in Figure 1) and the parents via punch biopsy, and 5-μm thick cryosections were fixed in 2.5% glutaraldehyde and 2% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) solution. Skin cryosections provided by the Medical Faculty of Istanbul Pathology Department were used as controls. Sections were stained for 2 h at room temperature in a mixture that contained 1 ml of fetal bovine serum (FBS), 9 ml of PBS and 20 μl of filipin (Sigma, St Louis, MO, USA F9765) solution (5 mg/ml in DMSO). Slides washed in PBS were viewed under a fluorescent microscope (Leitz Wetzlar) using ultraviolet light with excitation of 340–380 nm and emission of 385–470 nm to observe free (unesterified) cholesterol. Photographs were obtained using 4 s exposures at a magnification of × 320.
Results
Clinical findings
According to their parents, the affected brothers had uneventful prenatal and natal histories and were born at term. They were small for gestational age and with relative macrocephaly. They had psychomotor retardation. For brother 1 (203 in Figure 1) ataxic gait was evident at age 5 years, and he became non-ambulatory at age 9.5 years. Brother 2 (204) never achieved head control, sat unsupported, walked or spoke. He began having seizures at the age of 6 years.
The brothers were referred together to the medical school with complaints of abnormal gait/inability to walk and intellectual disability. At presentation, they had relative macrocephaly and normal weight, but younger brother 2 was short (<3rd p). The clinical findings were similar in both and are compiled in Table 1. The brothers had intellectual disability but no autistic features. Physical examination showed truncal ataxia, head titubation and spasticity. Muscle tonus was increased, especially in the lower extremities. Deep tendon reflexes were hyperactive. Achilles clonus and Babinski sign were positive. Achilles tendons were tight, and bilateral equine deformity was evident. Horizontal and vertical eye movements were normal at the initial visit but became limited in the 5-year follow-up, more prominent for brother 2. The metacarpophalangeal joints, elbows and distal phalanges of the fingers were hyperextensible. In addition, brother 1 had developed thenar atrophy of the hands, flexion deformity of the left hand and bilateral pes cavus deformity. Brother 2 could not control his head, walk or speak but could sit with support. Whole blood and urine analyses were normal in both brothers. Both had bilateral optic atrophy, but hearing was normal. Motor nerve conduction studies were normal for brother 2, but in brother 1 distal latencies were prolonged in the peroneal and tibial nerves, with normal compound muscle action potential amplitudes. In both brothers, sensory nerve conduction studies were normal. Cranial MRI indicated cerebral atrophy, diffuse white matter hypointensity at T1-weighted images and hyperintensity at T2-weighted images compatible with demyelinating leukodystrophy, severe atrophy of the inferior cerebellar vermis and moderate atrophy of both the superior cerebellar vermis and cerebellar hemispheres, mega cisterna magna and a thin corpus callosum (Figure 1). Brother 2 had in addition a posterior fossa arachnoid cyst on the left side, without any signs of pressure; thus, the cyst was considered an incidental finding. 3D cranial CT scans performed recently did not show any sign of mandibulofacial dysostosis (Supplementary Figure 1). The clinical findings in full are in Supplementary Information. Neurologic examination was normal in the sisters and parents, but cranial MRI results showed very mild cerebral atrophy in the parents (Figure 1).
Table 1. Characteristics of the family members.
| Features | Brother 1 (203) | Brother 2 (204) | Sister 1 (201) | Sister 2 (202) | Father (101) | Mother (102) |
|---|---|---|---|---|---|---|
| Age (years) | 11 | 6.5 | 24 | 19 | 45 | 45 |
| Postnatal history | ||||||
| Low birth weight | + | + | − | − | − | − |
| Hypoglycaemia in newborn | − | + | − | − | − | − |
| Relative macrocephaly | + | + | − | − | − | − |
| Neurologic development | ||||||
| Head control | + | − | + | + | + | + |
| Sitting | + | − | + | + | + | + |
| Walking | − | − | + | + | + | + |
| Speech | Single word | − | + | + | + | + |
| Mental retardation | Severe | Severe | − | − | − | − |
| Normal development | − | − | + | + | + | + |
| Developmental regression | + | − | − | − | − | − |
| Seizures | − | + | − | − | − | − |
| Physical examination | ||||||
| Truncal ataxia | + | + | − | − | − | − |
| Head titubation | + | + | − | − | − | − |
| Pyramidal signs | + | + | − | − | − | − |
| Joint hyperextensibility | + | + | − | − | − | − |
| Pes cavus | + | − | − | − | − | − |
| Equinism | + | − | − | − | − | − |
| Skin findings | − | − | − | − | − | − |
| Short stature | − | + | − | − | − | − |
| Eye examination | ||||||
| Bilateral optic atrophy | + | + | NA | NA | NA | NA |
| Gaze palsy | + | + | − | − | − | − |
| Nerve conduction studies | ||||||
| Motor nerves | Prolonged distal latencies; normal CMAP | N | NA | NA | NA | NA |
| Sensory nerves | N | N | NA | NA | NA | NA |
| Cranial MRI findings | ||||||
| Cerebral atrophy | + | + | − | − | + | − |
| White matter involvement | + | + | − | − | − | − |
| Cerebellar atrophy | + | + | − | − | + | + |
| Posterior fossa arachnoid cyst | − | + | − | − | − | − |
| Thin corpus callosum | + | + | − | − | − | − |
| Thick corpus callosum | − | − | + | + | + | + |
| 3D cranial CT | N | N | NA | NA | NA | NA |
| Skin biopsy | ||||||
| Cholesterol storage | + + | + + | NA | + + | + | + |
Abbreviations: N, normal; NA, not analysed.
Genetic findings
In linkage analysis assuming either full or 70% penetrance not to miss any possible candidate variant with reduced penetrance, the maximal LOD scores were 2.65 and 2.05, respectively, both lower than the critical value 3, because of small family size (Supplementary Figure 2). No additional candidate locus was found by homozygosity mapping. At each locus haplotype segregation analysis was applied to investigate whether the homozygosity that the patients shared was possibly due to identity by descent. A maximal LOD score of 0.3 excluded X-linked inheritance.
As no clinical phenotype similar to that in our patients was reported on the OMIM Phenotype Map (NCBI Map Viewer) at the candidate loci (Supplementary Table 1), we proceeded with exome sequencing (Supplementary Methods). Those loci together harboured two novel or rare variants that were predicted to affect protein structure (Supplementary Table 1; see Supplementary Figure 3 for Sanger validation). POLR1A NM_015425.3 c.2801C>T (p.(Ser934Leu)) was novel, whereas dbSNP138 listed OSBPL11 NM_022776.4 c.511C>T (p.(Arg171Trp)) as rs370760880. The variants were not found in the tested 398 or 278 Turkish control samples, respectively. OSBPL11 variant was recently found in a new individual in the Turkish Exome Database (Bayram Yüksel, personal communication), raising the population frequency to ∼0.0005, and is reported in ExAC database in only Europeans and South Asians, with a frequency of 0.00004.
Three prediction algorithms were utilized to investigate the effects of the variants on protein function. Mutation Taster predicted both variants as damaging.17 PolyPhen-2 predicted OSBPL11 c.511C>T (p.(Arg171Trp)) as damaging and POLR1A c.2801C>T (p.(Ser934Leu)) as benign; in contrast, SIFT predicted the former variant as tolerated and the latter as damaging.18, 19 In both variants the altered residues were conserved across species (Supplementary Figure 4). POLR1A c.2801C>T (p.(Ser934Leu)) results in the substitution of polar serine by nonpolar leucine at position 934 in RPA194. The residue is within a region of 104 amino acids that are conserved in full among all mammals. In addition, the region adjacent to the residue is highly conserved even in lower eukaryotes. OSBPL11 c.511C>T (p.(Arg171Trp)) leads to the substitution of positively charged, hydrophilic arginine with nonpolar, hydrophobic tryptophan at position 171 in the 747 amino-acid protein. Arg171 is located in a stretch of 116 amino acids that are fully conserved in mammals and highly conserved among vertebrates. The substitution could limit the protein's function because of a change in the three-dimensional structure, the loss of a hydrophilic functional residue and/or the lack of phosphorylation at the adjacent serine residue.20 Together, these findings suggest that both variants are potentially deleterious. Of note, the affected brothers were homozygous for both variants, whereas sister 2 (202) was homozygous for the OSBPL11 variant but did not carry the POLR1A variant (Figure 1). Thus, POLR1A c.2801C>T (p.(Ser934Leu)) is identified as the cause of the severe neurologic phenotype in the two brothers. Whether the deleterious OSBPL11 variant contributes to the complex clinical manifestations in the family can be discerned when new families with OSBPL11 mutations are detected.
Dissection of the role of POLR1A p.Ser934Leu
Predicted effects on protein structure
POLR1A encodes RPA194, the largest, catalytic subunit of the Pol I core complex. Two high-resolution crystal structures have provided detailed information on the interactions and functional roles of the subunits.10, 11 RPA194 interfaces with RPA135 (encoded by POLR1B), forming the composite active site of the enzyme. These subunits also form the horseshoe-shaped framework conserved among Pols I, II and III. Based on the Pol I crystal structure in yeast, the region around the variant forms the enzyme funnel and one of the interaction faces with RPA135.10, 11
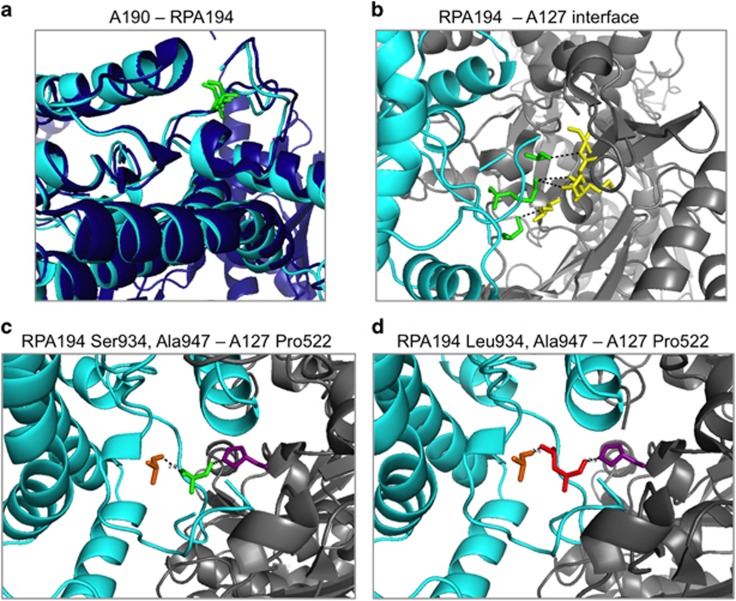
To assess the potential impact of the variant on the protein–protein (RPA194-RPA135) interaction, we first superpositioned human RPA194 on the yeast A190 crystal structure11 and used several molecular modelling tools for the prediction of the folding of the human RPA194, and conducted several tertiary structure predictions. We modelled RPA194_Segment2 containing residues 616–1287 that consists of the pore domain, the funnel domain in which the residue of interest lies, the cleft domains containing both the trigger loop and the bridge helix and the foot domain. Superimposition of RPA194_Segment2 with 4c2m solved structure of yeast A190 produced a RMSD score of 0.744, indicating that there is a high degree of similarity in the folding and secondary structure of both the structured and unstructured regions in the RPA194 interaction domain with RPA135 (Figure 2a). Based on the sequence alignment of yeast A190 to human RPA194, amino acids Ser931, Lys933 and Pro936 in the vicinity of the altered residue Ser934Leu are predicted to interact with A127 (the yeast RPA135 subunit; Figure 2b). In this model, Ser934 is located in the intramolecular side of RPA194. Two residues were identified to be within range to interact with the residue at 934. Pro522 in A127 forms a nonbonding contact with Ser934, and this interaction does not change upon substitution with a leucine (Figure 2c). The Ser934Leu substitution brings the leucine methyl groups in close vicinity (1.6 Å) of RPA194 Ala947, raising the potential for its intramolecular interaction (Figure 2d).
Figure 2.
Molecular modelling of human RPA194 and its Ser934Leu variant. (a) Modelling of RPA194 based on yeast A190. The ribbon structures of RPA194 (light blue) and A190 (dark blue) and the conserved Ser934 and Thr965 residues (both green) are shown. (b) The interface between RPA194 (light blue) and A127 (grey) is shown and residues with predicted intermolecular interactions based on the crystal structure are shown in green and yellow. (c, d) RPA194 (light blue) interface with A127 (grey) is shown alternatively with wild-type Ser934 (green, c) and mutant Leu934 (red, d). RPA194 Ala947 (orange) and A127 Pro522 (purple) predicted to interact with residue at 934 are shown. Note the shortened distance (1.6 Å) between the mutant Leu934 and RPA194 Ala947.
Decreased amount of nucleolar RPA194 in patient cells
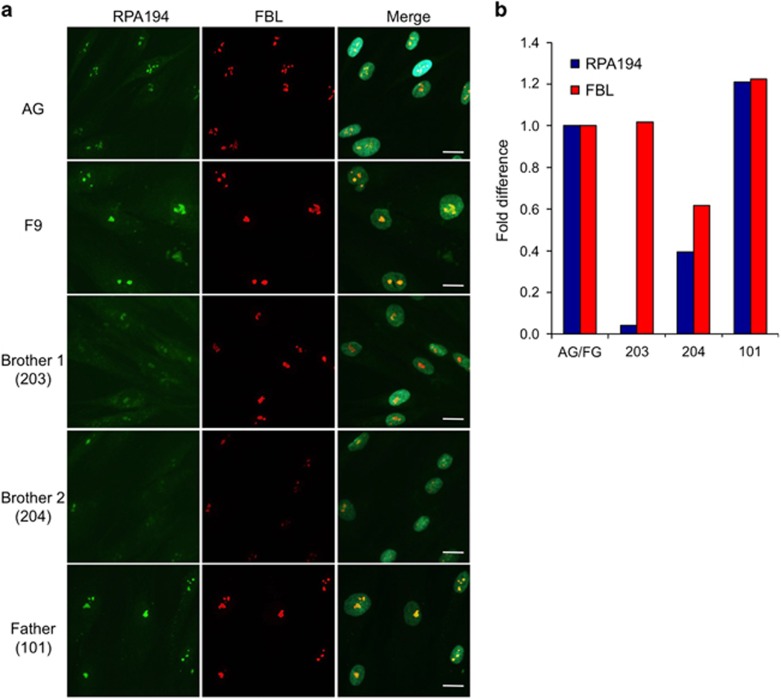
We used fibroblasts isolated from the two affected brothers, the father and unrelated controls to assess the subcellular distribution of RPA194. The cells were stained for RPA194 and fibrillarin (FBL), a nucleolar protein that modifies the newly synthesized rRNA, and conducted quantitative image analysis for the staining intensities of the proteins. Nucleolar RPA194 was found to be markedly reduced in both patients compared with the controls and the father. Nucleolar FBL was also decreased in brother 2 (Figure 3). These results suggested that the substitution compromises the ability of RPA194 to engage in its nucleolar activities.
Figure 3.
Expression of RPA194 in patient and control fibroblasts. (a) Immunostaining of RPA194 and fibrillarin (FBL) in the patients, father and control (AG for AG08498 and F9 for F92-99) fibroblasts. Cells were fixed and stained for RPA194 (green) and FBL (red), and counterstained for DNA (DAPI). Merged images are shown to the right. Scale bar, 20 μm. (b) Image quantification. Image analysis was conducted using FrIDA image analysis software for the expression of RPA194 and FBL and normalized to DNA. The specimens were then compared with the average of the control fibroblasts.
Putative role of OSBPL11 c.511C>T (p.(Arg171Trp)) in increased cholesterol deposition
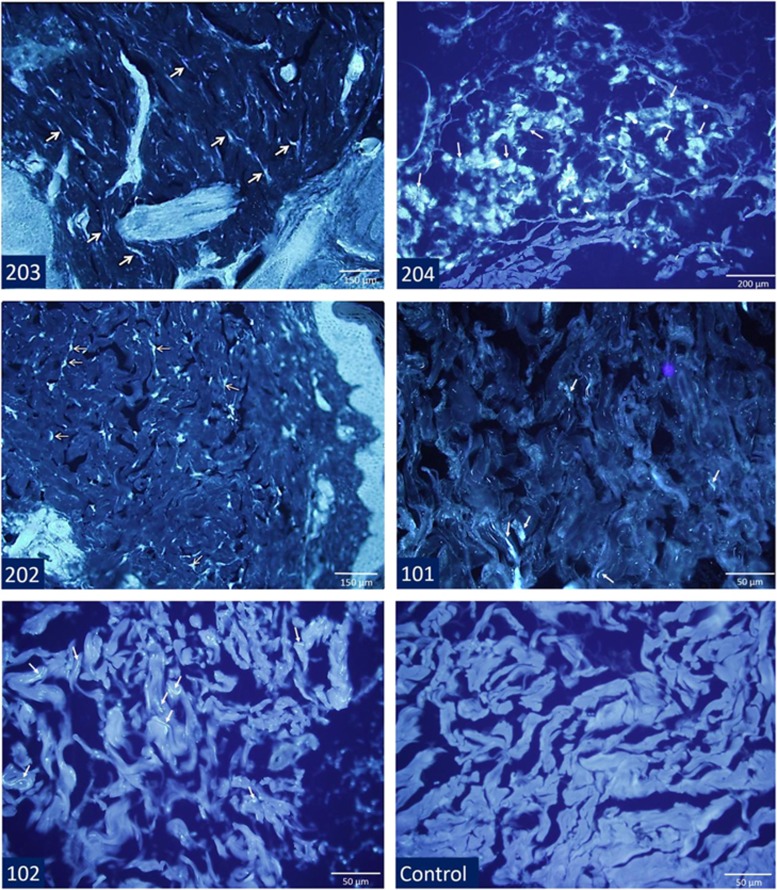
OSBPL11 was suggested to have a role an intracellular lipid transport,21 and OSBPL proteins may be cooperating with the NPC1 protein.22 We therefore hypothesized that an amino acid-altering variant in OSBPL11 might result in a cellular pathology similar to that in Niemann–Pick disease type C (NPC) disease and could increase the severity of the disease in the family under study. We stained dermal tissue specimens with the fluorescent dye filipin that binds free cholesterol. This histopathologic analysis revealed bright fluorescent droplets in the cytoplasm of the cells, predominantly in dermal fibroblasts, indicative of free cholesterol accumulation in affected brothers and sister 2 homozygous for OSBPL11 c.511C>T (p.(Arg171Trp)), and mildly in heterozygous parents (Figure 4). Such fluorescently stained inclusions were absent in the control preparations. However, no filipin-stainable cholesterol deposition was observed in fibroblasts cultured from dermal biopsies of the brothers (Supplementary Figure 5). This suggests that the cholesterol accumulation phenotype is distinct from the cholesterol accumulation caused by loss of NPC1 function. A putative functional impact of the OSBPL11 c.511C>T variant was further pursued in RNA interference/phenotypic rescue experiments (Supplementary Figure 6,Supplementary Methods and Supplementary Data). The results demonstrated that a cholesterol deposition resulted specifically from OSBPL11 silencing and transfection of OSBPL11-silenced cells with the c.511C>T mutant 'but not with the wildtype' failed to rescue the cholesterol deposition, suggesting that the variant protein is functionally defective and unable to facilitate normal cholesterol trafficking.
Figure 4.
Filipin staining of dermal tissue from the brothers, the sister homozygous for the OSBPL11 c.511C>T (p.(Arg171Trp)) variant (202), the parents and a control sample. The samples of the sibs have free cholesterol-related fluorescent staining in granular pattern (arrows) in dermal fibroblasts interspersed among collagen bundles and ground substance, whereas staining in parents' tissues was less prominent. Granular staining of cholesterol was not observed in the control dermal tissue.
Discussion
We showed by functional studies that the homozygous POLR1A variant we identified in the two brothers underlies the new demyelinating leukodystrophy associated with atrophy of the cerebellum, cerebrum and corpus callosum. We thus define a novel complex leukodystrophy and a new phenotype for POLR1A defect. Recently, heterozygous variants in the gene were shown to cause dominant acrofacial dysostosis type Cincinnati;7 however, we found that the zygomatic, maxillary and mandibular bones of the boys were normal. Neither the two sisters nor the parents have any neurological signs, but the parents display very mild cerebral atrophy.
With the recent advances in genotyping arrays and exome sequencing, identification of novel monogenic disease genes in small families has become feasible. Although the currently most commonly applied strategy for this task is exome sequencing of all siblings, we did not use this approach, because we wanted to detect all loci possibly harbouring fully penetrant candidate variants as well as those with low penetrance, via linkage analysis. By evaluating exome data of one affected sib at candidate disease loci followed by Sanger sequencing validation of candidate variants, we found two missense homozygous candidate variants, namely, POLR1A c.2801C>T and OSBPL11 c.511C>T, in the two affected siblings of the study family. In functional analysis we focussed on both variants and showed that both are possibly deleterious. However, only the former segregated with the severe phenotype. Neither of these genes has previously been associated with a neurological disease.
Our data, molecular modelling and prediction models suggested that the POLR1A Ser934Leu change leads to an alteration in the conformation of the encoded RPA194 protein. The modelling showed that Ser934 was located in an unstructured flexible domain that interacts with A127. This interaction is mediated by amino acids in the immediate vicinity of Ser934, namely, Ser931, Lys933 and Pro936, that protrude into the RPA135 domain. However, the modelling suggested that the Leu934 substitution may lead to a change in the intramolecular association of RPA194, raising the potential that this would decrease the flexibility of the linker domain interacting with RPA135. Subsequently, this could pose a challenge in the stability of the RPA194–RPA135 interaction and localization, compromising the Pol I transcription activity.
We further observed that the amount of RPA194 in the nucleolus was decreased in skin fibroblasts derived from the patients. This is consistent with the modelling studies predicting that the protein–protein interactions of mutant RPA194 may be compromised. We speculate that this defect becomes a vulnerability during times of development and growth when highly active Pol I transcription is critically needed. POLR1C and POLR1D have been implicated in recessive and dominant Treacher Collins syndrome, respectively.8 This rare disease manifests at birth with mandibulofacial dysostosis but normal intelligence. Recently, three heterozygous variants in POLR1A, namely, c.1777G>C (p.(Glu593Gln)), c.3649delC (p.(Gln1217Argfs*)) and c.3895G>T (p.(Val1299Phe)), were found associated with acrofacial dysostosis type Cincinnati.7 The deduced changes in the protein would locate to the RPA194 activation domain, trigger loop and the jaw and cause severe, moderate and mild dysostosis, respectively. Hypomorphic polr1a mutations introduced to zebrafish caused developmental defects including small, misshapen heads, microphthalmia, cerebral hypoplasia, jaw agenesis and changes in pigmentation and morphology of the heart. These findings and our results here are suggestive that specific POLR1A variants potentially cause broad multiorgan developmental defects, including the brain and cerebellum. Importantly, the clinical phenotypes of our patients show striking resemblance, except for the earlier onset, to Pol III-related leukodystrophies resulting from variants in POLR1C, POLR3A or POLR3B.4, 5, 6 Given that Pol III transcribes several noncoding RNAs, most notably tRNAs but also ribosomal 5S RNAs and other small nuclear, cytoplasmic and mitochondrial RNAs, the present findings suggest that defects in noncoding RNA synthesis have the potential to compromise development and cause complex cognitive and motor disease syndromes. Considering also our findings, variants in only POLR1A and POLR1C are so far known to cause either leukodystrophy or mandibulofacial dysostosis. As the presented family is the first one with POLR1A-associated leukodystrophy, detection of new families with POLR1A mutations will clarify the severity of the phenotype.
Genetic defects of lipid metabolism often associate with neurodegenerative diseases. For an example, NPC is a rare, autosomal recessive lysosomal cholesterol storage disease that causes progressive neurodegeneration and visceral involvement with variable clinical manifestations. The NPC1 and NCP2 proteins mutated in this disease mediate the egress of cholesterol from endo-lysosomal compartments.23 It has been proposed that NPC1 links to cytoplasmic sterol transport by members of the oxysterol-binding protein (OSBP) and its homologues (OSBP-like proteins (OSBPL))22 that constitute a large family of cytoplasmic lipid binding/transport proteins.24, 25 OSBPL11 was found to localize at the Golgi–late endosome interface, and the OSBPL11–OSBPL9 dimer was suggested to act as an intracellular lipid sensor or transporter.21, 26 Moreover, OSBPL11 was reported to function in adipogenesis,21, 27 and the gene has been associated with cardiovascular risk factors in obesity.28 NPC1 deficit causes intracellular lipid accumulation and leukodystrophy,29, 30 and we propose that the moderate accumulation of free cholesterol in the dermal biopsies of the patients and of sister 2, all homozygous for the OSBPL11 variant, and the mild accumulation in heterozygous parents could be because of OSBPL11 deficit. Moreover, knockdown of OSBPL11 in A431 cells resulted in an increase of filipin-stainable free cholesterol, an effect reversed by overexpression of wild-type OSBPL11 but not the c.511C>T variant. In some other diseases that develop as a result of intracellular lipid deposition, such as certain variants of neuronal ceroid lipofuscinoses (NCLs), similar cerebellar ataxia and diffuse demyelinating leukodystrophy as in the present patients are observed. Thus, in the light of all the available evidence, we find it possible that OSBPL11 c.511C>T may act as a modifier of the patient phenotype.
In conclusion, the present findings expand the spectrum of inherited human leukodystrophies by reporting association of a homozygous POLR1A variant with such a disease phenotype. Our genetic, histopathologic, in silico and experimental cell biological analyses together suggest POLR1A as the gene responsible for the novel disease in the two, severely affected male patients. We expect that our findings will facilitate the detection of new patients with POLR1A mutations. In particular, families afflicted with initial symptoms of ataxia, spasticity, psychomotor retardation and leukodystrophy could benefit from testing for mutations in this gene. Our findings also demonstrate the challenge in gene identification in a novel disease when variants damaging to protein function in different genes could be exerting their effects additively or epistatic.
Acknowledgments
We thank the family members for their cooperation and TÜBITAK Advanced Genomics and Bioinformatics Group (IGBAM) for sharing with us the Turkish Exome Database. This work was supported by the Boğaziçi University Research Fund (Grant 7695 to AT), Academy of Finland (Grants 263841 and 272130 to EI; 285223 to VMO; and 288364 to ML), the Sigrid Juselius Foundation (to VMO and EI., the Magnus Ehrnrooth Foundation (to VMO) and the National Institutes of Health (R01 CA172069 to ML).
Author contributions
Bülent Kara contributed to patient acquisition, analysis and interpretation of clinical data, study supervision and drafting/revising the manuscript. Çiğdem Köroğlu analysed the genetic data and drafted/revised the manuscript. Karita Peltonen carried out the RPA194 stainings and their quantification. Ruchama C Steinberg performed the molecular modelling, variant analysis and revising the manuscript. Maarit Hölttä-Vuori, Kristiina Kanerva and You Zhou carried out the filipin staining/quantification experiments in cultured cells, analysis of the imaging data and drafting/revising the manuscript. Hülya Maraş Genç participated in acquisition, analysis and interpretation of clinical data and drafting/revising the manuscript. Ayşe Güven generated and analysed the genetic data and drafted the manuscript. Tuğba Kotil contributed to acquisition, analysis and interpretation of histologic data and drafting/revising the manuscript. Seyhun Solakoglu contributed to acquisition, analysis and interpretation of histologic data, study supervision and drafting/revising the manuscript. Vesa M Olkkonen designed cultured cell experiments and drafted/revised the manuscript. Elina Ikonen and Marikki Laiho contributed to the design of the cell experiments, interpretation of their results and drafting/revising the manuscript. Aslıhan Tolun created study concept and design, interpreted the data, supervised the study and drafted/revised the manuscript.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Russell J, Zomerdijk JC: The RNA polymerase I transcription machinery. Biochem Soc Symp 2006; 73: 203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I: Wisely chosen paths—regulation of rRNA synthesis. FEBS J 2010; 277: 4626–4639. [DOI] [PubMed] [Google Scholar]
- Pederson T: The nucleolus. Cold Spring Harb Perspect Biol 2011; 3: a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard G, Chouery E, Putorti ML et al: Mutations of POLR3A encoding a catalytic subunit of RNA polymerase pol III cause a recessive hypomyelinating leukodystrophy. Am J Hum Genet 2011; 89: 415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitsu H, Osaka H, Sasaki M et al: Mutations in POLR3A and POLR3B encoding RNA polymerase III subunits cause an autosomal-recessive hypomyelinating leukoencephalopathy. Am J Hum Genet 2011; 89: 644–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiffault I, Wolf NI, Forget D et al: Recessive mutations in POLR1C cause a leukodystrophy by impairing biogenesis of RNA polymerase III. Nat Commun 2015; 6: 7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver KN, Watt KE, Hufnagel RB et al: Acrofacial dysostosis, Cincinnati type, a mandibulofacial dysostosis syndrome with limb anomalies, is caused by POLR1A dysfunction. Am J Hum Genet 2015; 96: 765–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauwerse JG, Dixon J, Seland J et al: Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat Genet 2011; 43: 20–22. [DOI] [PubMed] [Google Scholar]
- Vanderver A, Prust M, Tonduti D et al: Cas definition and classification of leukodystrophies and leukoencephalopathies. Mol Genet Metab 2015; 114: 494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel C, Sainsbury S, Cheung AC, Kostrewa D, Cramer P: RNA polymerase I structure and transcription regulation. Nature 2013; 502: 650–655. [DOI] [PubMed] [Google Scholar]
- Fernández-Tornero C, Moreno-Morcillo M, Rashid UJ et al: Crystal structure of the 14-subunit RNA polymerase I. Nature 2013; 502: 644–649. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ: The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 2015; 10: 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederstein M, Sippl MJ: ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 2007; 35: W407–W410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Lüthy R, Bowie JU: VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol 1997; 277: 396–404. [DOI] [PubMed] [Google Scholar]
- Hölttä-Vuori M, Määttä J, Ullrich O, Kuismanen E, Ikonen E: Mobilization of late-endosomal cholesterol is inhibited by Rab guanine nucleotide dissociation inhibitor. Curr Biol 2000; 10: 95–98. [PubMed] [Google Scholar]
- Peltonen K, Colis L, Liu H et al: A targeting modality for destruction of RNA polymerase I that possesses anticancer activity. Cancer Cell 2014; 25: 77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, Seelow D: MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 2014; 11: 361–362. [DOI] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L et al: A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC: Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- Cantin GT, Yi W, Lu B et al: Combining protein-based IMAC, peptide-based IMAC, and MudPIT for efficient phosphoproteomic analysis. J Proteome Res 2008; 7: 1346–1351. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Li S, Mäyränpää MI et al: OSBP-related protein 11 (ORP11) dimerizes with ORP9 and localizes at the golgi-late endosome interface. Exp Cell Res 2010; 316: 3304–3316. [DOI] [PubMed] [Google Scholar]
- Yang H: Nonvesicular sterol transport: two protein families and a sterol sensor? Trends Cell Biol 2006; 16: 427–432. [DOI] [PubMed] [Google Scholar]
- Vanier MT: Niemann-Pick disease type C. Orphanet J Rare Dis 2010; 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehto M, Laitinen S, Chinetti G et al: The OSBP-related protein family in humans. J Lipid Res 2001; 42: 1203–1213. [PubMed] [Google Scholar]
- Jaworski CJ, Moreira E, Li A, Lee R, Rodriguez IR: A family of 12 human genes containing oxysterol-binding domains. Genomics 2001; 78: 185–196. [DOI] [PubMed] [Google Scholar]
- Vihervaara T, Jansen M, Uronen RL, Ohsaki Y, Ikonen E, Olkkonen VM: Cytoplasmic oxysterol-binding proteins: sterol sensors or transporters? Chem Phys Lipids 2011; 164: 443–450. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Robciuc MR, Wabitsch M et al: OSBP related proteins (ORPs) in human adipose depots and cultured adipocytes: evidence for impacts on the adipocyte phenotype. PLoS One 2012; 7: e45352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard L, Faucher G, Tchernof A et al: Association of OSBPL11 gene polymorphisms with cardiovascular disease risk factors in obesity. Obesity (Silver Spring) 2009; 17: 1466–1472. [DOI] [PubMed] [Google Scholar]
- Soccio RE, Breslow JL: Intracellular cholesterol transport. Arterioscler Thromb Vasc Biol 2004; 24: 1150–1160. [DOI] [PubMed] [Google Scholar]
- Ikonen E, Hölttä-Vuori M: Cellular pathology of Niemann–Pick type C disease. Semin Cell Dev Biol 2004; 15: 445–454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.