Abstract
Accurate human leukocyte antigen (HLA) genotyping is critical in studies involving the immune system. Several algorithms to estimate HLA genotypes from whole-exome data were developed. We compared the accuracy of seven algorithms, including Optitype, Polysolver and PHLAT, as well as investigated patterns and possible causes of miscalls using 12 clinical samples and 961 individuals from the 1000 Genomes Project. Optitype showed the highest accuracy of 97.2% for HLA class I alleles at the second field resolution, followed by 94.0% in Polysolver and 85.6% in PHLAT. In Optitype, 34 (21.1%) of 161 miscalls were across different serological types, and common miscalls were HLA-A*26:01 to HLA-A*25:01, HLA-B*45:01 to HLA-B*44:15 and HLA-C*08:02 to HLA-C*05:01 with error rates of 4.1%, 10.0% and 4.1%, respectively. In Polysolver, 193 (55.9%) of 345 miscalls occurred across different serological alleles, and a specific pattern of genotyping error from HLA-A*25:01 to HLA-A*26:01 was observed in 93.3% of HLA-A*25:01 carriers, due to dropping of HLA-A*25:01 sequence reads during the extraction process of HLA reads. In PHLAT, 147 (59.8%) of 246 miscalls in HLA-A were due to erroneous assignment of multiple alleles to either HLA-A*01:22 or HLA-A*01:81. These results suggest that careful considerations needed to be taken when using exome-based HLA class I genotyping data and applying these results in clinical settings.
Introduction
The human leukocyte antigen (HLA) gene cluster, located on the short arm of chromosome 6, is among the most polymorphic regions in human genome with thousands of documented alleles.1, 2 This cluster includes several genes involved in functions of the immune system, including major histocompatibility complex (MHC) classes I and II.3 Both MHC classes are encoded by three major loci (MHC class I: HLA-A, HLA-B and HLA-C; MHC class II: HLA-DP, HLA-DQ and HLA-DR), which are co-dominantly expressed. The polymorphisms in the HLA region have been reported to play critical roles in rejection and graft-versus-host disease of hematopoietic stem cell transplants and the risks of several diseases, including autoimmune diseases.4, 5 Certain HLA genotypes were shown to link with increased risk of drug-induced skin hypersensitivity and liver inquiry.6, 7 In cancer research, HLA information is very important because HLA class I molecules are critical mediators of the cytotoxic T-cell response, presenting antigen peptides on the cell surface to be recognized by the T cell receptor.8, 9 The recognition of HLA-peptide complexes on cancer cells by T-cell receptor of the cytotoxic CD8+ T cells is crucial for anti-tumor immune responses. Indeed, HLA dysfunction caused by genetic and epigenetic alteration in the HLA genes or β2-microglobulin has implicated as a possible mechanism of immune evasion during the development and progression processes of cancer.10, 11, 12, 13, 14 Because of the success of immune checkpoint inhibitors, there is no skepticism against immune-based therapies for cancer and it is almost certain that cancer cells present cancer-specific antigens on HLA molecules. With advances in genome sequencing, it is possible to predict a new class of tumor-specific antigens (‘neoantigens') derived from somatically mutated proteins that are present uniquely in tumor cells. Immune checkpoint antibody therapies, such as anti-CTLA-4 and anti-PD-1 antibodies, have been used for the treatment of melanoma, non-small cell lung cancer and kidney cancer.15, 16, 17, 18 In fact, accumulative evidence has supported that higher somatic mutation burden and predicted-neoantigen load were strongly associated with better clinical outcome in patients treated with CTLA-4 and PD-1 blockades.19, 20 These lines of evidence support the significance of neoantigen prediction in cancer immunotherapy. Since cancer genome information is usually readily available in these studies, it is important that HLA class I genotype information can be accurately extracted so that possible neoantigens can be predicted, and neoantigen-specific T cells are in a scope for development of next-generation cancer immunotherapies.
HLA allele definition comprises the gene name indicating the locus (that is, A, B or C) followed by successive sets of digits separated by colons.21 While the first two digits (field 1) specify the allele groups by serological activity (allele level resolution, for example, A*01 or A*02), the second field indicates the protein sequence (protein level resolution, for example, A*02:01 or A*02:02). The remaining two sets distinguish synonymous polymorphisms and non-coding variations. HLA typing can be done with different degrees of resolution.22 Conventional HLA typing is performed using serology- and/or PCR-based methods, such as sequence-specific oligonucleotide and sequencing-based typing techniques. These techniques are labor-intensive, time-consuming, and often lead to ambiguous genotyping results because of limitation of oligonucleotide probe design or phase ambiguity for HLA allele assignment.23, 24 Several protocols have recently been established for HLA-targeted multiplexed PCR or long-range PCR methods coupled with next-generation sequencing that enable us to obtain accurate and super high resolution of HLA information up to fourth field level,25, 26, 27 although errors introduced by PCR artifacts, sequencing procedures, bioinformatics or inadequately genotyped sequence references are still inherent problems.28, 29 Recently, several tools to extract HLA allele information from genome-wide sequencing data, including whole-exome, whole-genome and transcriptome data, but it is still challenging to improve the accuracy of these tools.29
Materials and methods
Samples
We used genomic DNA from 12 patients with malignant methothelioma. All samples were obtained under Institutional Review Board approval in the University of Chicago (No. IRB15-0128) and with written informed consent.
HLA typing
Genomic DNA was extracted from peripheral blood mononuclear cells using Qiagen DNA Mini Kit (Qiagen, Valencia, CA, USA). PCR amplicon-based high-resolution typing on MiSeq (Illumina, San Diego, CA, USA) was performed for HLA-A, HLA-B and HLA-C in Scisco Genetics Inc. (Seattle, WA, USA).25
Whole-exome sequencing
DNA libraries were prepared using SureSelect XT Human All Exon V5 (Agilent Technologies, Santa Clara, CA, USA) and whole-exome sequencing was performed by 100-bp paired-end reads on HiSeq2500 (Illumina) according to the manufacturers' protocol.
Fastq data of the 1000 Genomes Project samples
To compare with PCR-based approaches, we selected a total of 961 samples across 13 different populations in the 1000 Genomes Project database (http://www.1000genomes.org/) that had both exome data, which were sequenced by paired-end reads on Illumina sequencer (Genome Analyzer II or HiSeq 2000), and HLA type information experimentally determined by Sanger sequencing (Supplementary Table 1).30 The fastq files were downloaded from the 1000 Genomes Project database.
Read mapping to hg19 reference genome
After the exclusion of low-quality reads (base quality of<20 for more than 80% of bases) using FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/), sequence reads were mapped to the human reference genome GRCh37/hg19 using Burrows-Wheeler Aligner (v0.7.10).31 BAM files were generated using SAMTools (v0.1.19),32 and possible PCR duplicated reads were removed using Picard v1.117 (http://broadinstitute.github.io/picard/).
HLA typing tools
We have compared publically available algorithms for HLA typing, including OptiType,33 Polysolver,34 PHLAT,35 HLAreporter,36 HLAforest,37 HLAminer38 and seq2HLA.39 All algorithms were run according to respective instructions. For Polysolver analysis, we used BAM files generated as described above as an input. For the other algorithms, we used fastq files after filtering low-quality reads (base quality of<20 for more than 80% of bases) as an input. Multiple predictions for an allele at a locus detected in HLAreporter were considered as ambiguous results, and only first field information was used.
Data analysis
Sanger sequencing-based HLA genotype data were used as a reference.30 The accuracy was simply calculated as the ratio between the number of correctly called alleles and the total number of the alleles. The total number of the alleles were 2 alleles × number of samples for the accuracy calculation in each of the HLA-A, HLA-B or HLA-C gene, or 2 alleles × 3 loci × number of samples to calculate the accuracy among all three HLA class I genes (HLA-A,B,C). Ambiguous results were considered as incorrect predictions. Error rate for each miscall was calculated by dividing the number of miscalls by total number of the alleles in the 961 samples. The significance of the error rate was evaluated by Fisher's exact test. Since ambiguous HLA alleles existed in Sanger sequencing results,30 we compared all possibilities of ambiguous HLA alleles to test concordance of identified alleles, while we merged ambiguous HLA alleles into the most common HLA allele among ambiguous HLA alleles to calculate allele frequency or error rates. Allelic frequency of HLA in 961 samples from the 1000 Genomes Project was summarized in Supplementary Table 2. HLA coding DNA and genomic nucleotide sequences and feature annotation were obtained from the IMGT/HLA (https://www.ebi.ac.uk/ipd/imgt/hla/)1, 2 or dbMHC database (http://www.ncbi.nlm.nih.gov/projects/gv/mhc/). To visualize the mapping data, we used IGV software.40, 41
Results
As a preliminary screening, we evaluated seven reported algorithms for HLA typing: Optitype,33 Polysolver,34 PHLAT,35 HLAreporter,36 HLAforest,37 HLAminer38 and seq2HLA,39 using exome data from 12 clinical samples. Default parameters were applied for all the algorithms, with BAM files mapped to hg19 reference genome as an input for Polysolver analysis, while with fastq files as an input for the other algorithms according to the manual of each tool. As summarized in Supplementary Table 3, the results were discordant among these algorithms. To evaluate the accuracy of each exome-based software, we genotyped HLA of these 12 samples using PCR amplicon-based high-resolution HLA typing on MiSeq.25 We calculated the accuracies in each of HLA-A, HLA-B and HLA-C genes, and the accuracy of all three HLA class I genes (HLA-A,B,C), which was a percentage of correctly identified alleles among the total 72 alleles (2 alleles × 3 loci × 12 samples; see Materials and methods). The accuracies for HLA-A,B,C in each software varied from 36.1 to 100% at the resolution level of the second field (Supplementary Table 4). Based on this result, we focused on three algorithms, Optitype, Polysolver and PHLAT, which showed more than 90% accuracy, for further analyses to investigate patterns and possible causes of miscalls. These algorithms are most recently reported three, and Figure 1 briefly summarizes the workflow for these software. These three used different aligners and different statistical analysis to determine most-likely HLA alleles, and only two of them, Optitype and Polysolver adopted pre-selection step of HLA reads.
Figure 1.
Workflows of Optitype, Polysolver and PHLAT. The parts enclosed by dotted lines are the steps included in each software.
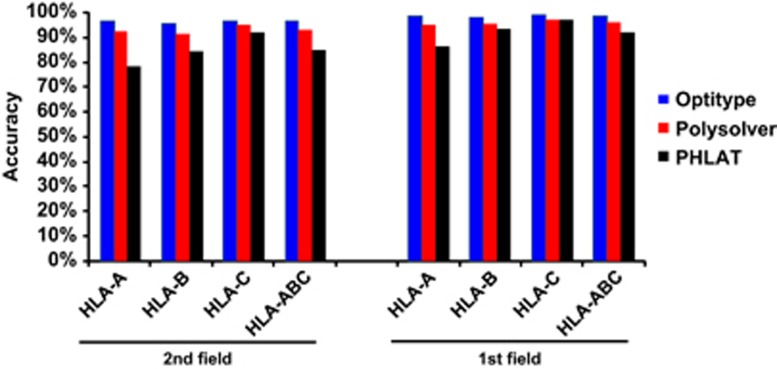
We selected 961 samples in the 1000 Genomes Project database that had both fastq files of paired-end Illumina exome data (N=1142) and experimentally determined HLA genotype information by Sanger sequencing technique (N=1274) (Supplementary Table 1),30 then ran exome-based HLA typing algorithms to determine HLA class I genotype (Supplementary Table 5). Among these three algorithms, Optitype showed the highest accuracy of 97.2% for HLA class I alleles (HLA-A,B,C) at the second field level, followed by 94.0% in Polysolver and 85.6% in PHLAT (Figure 2 and Supplementary Table 6). The allele estimation accuracies for each of HLA-A, HLA-B and HLA-C by Optitype were as high as 97.3%, 96.6% and 97.7%, respectively. Polysolver achieved the accuracies as high as 93.4% for HLA-A, 92.5% for HLA-B and 96.1% for HLA-C. However, in PHLAT, the accuracies were significantly lower than the other two methods, with the accuracies of 79.1%, 85.1% and 92.8% for HLA-A, HLA-B and HLA-C, respectively. Importantly, nearly 20% of HLA-A alleles were incorrectly assigned by PHLAT.
Figure 2.
Performance comparison of three exome-based HLA-typing algorithms. Accuracy of HLA alleles typed at the second field (4-digit) and first field (2-digit) resolution in 961 individuals from the 1000 Genomes Project. Accuracy was calculated by the fraction of total number of alleles that were correctly called.
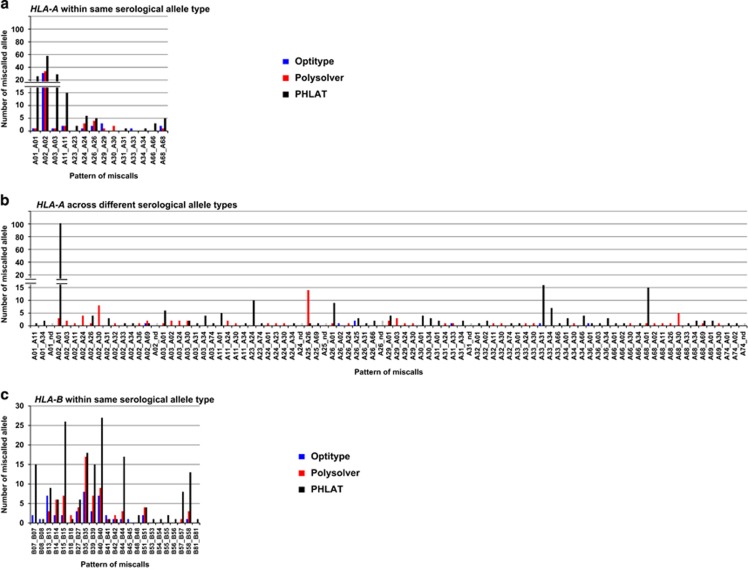
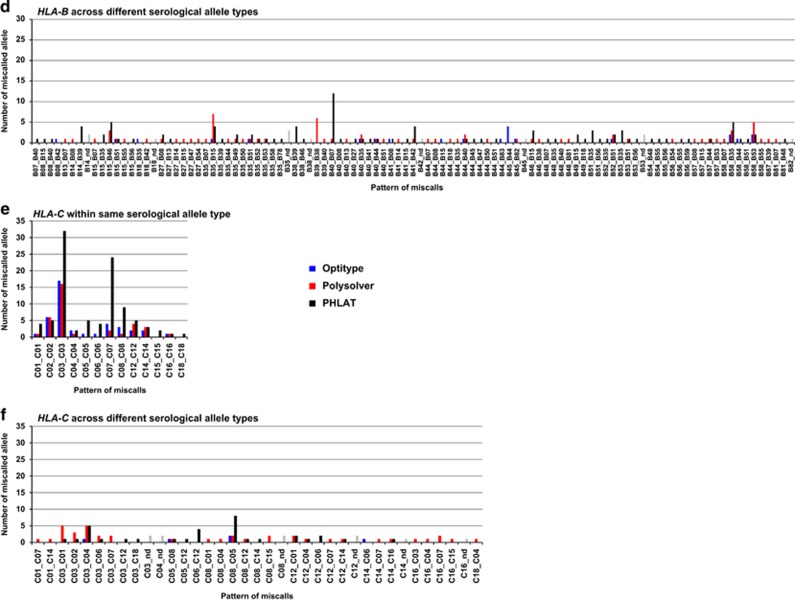
We next investigated specific patterns of discordance in each algorithm. In Optitype, 44 (86.3%) of 51 errors observed in HLA-A were within the same serological allele group, especially in the HLA-A*02 group (60.8%) (Figures 3a and b and Supplementary Table 7). Only 7 (13.7%) of the 51 errors were across different serological types. Similar to HLA-A, most miscalls found in Optitype were within the same serological allele type in HLA-B (43 of 65 (66.2%)) and in HLA-C (40 of 45 (89.9%)) (Figures 3c–f and Supplementary Tables 8, 9). The discordances across serological allele groups detected in multiple samples were HLA-B*45:01 to HLA-B*44:15 and HLA-C*08:02 to HLA-C*05:01, with the error rate of 10.0% and 4.1%, respectively. HLA-C*08:02 to HLA-C*05:01 was also detected in Polysolver and PHLAT with error rates of 4.1% and 6.1%, respectively (Table 1). Although these alleles showed high sequence similarity (Supplementary Figure 1), exact causes for these miscalls are still unclear.
Figure 3.
Distribution of the pattern of genotyping errors in HLA-A, HLA-B and HLA-C. (a, b) The pattern of discordant HLA-A alleles within the same serological allele (a) and across different serological alleles (b). (c, d) The pattern of discordant HLA-B alleles within the same serological allele (c) and across different serological alleles (d). (e, f) The pattern of discordant HLA-C alleles within the same serological allele (e) and across different serological alleles (f). nd; not determined (as shown as gray bars).
Table 1. Summary of miscalls observed in multiple software and at least twice in one software.
|
Experimental typing |
Predicted data |
Optitype |
Polysolver |
PHLAT |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | 2nd field | 1st field | 2nd field | 1st field | No of miscalls | Error rate | P | No of miscalls | Error rate | P | No of miscalls | Error rate | P |
| HLA-A | |||||||||||||
| A*02:01 | A*02 | A*02:06 | A*02 | 3 | 0.8% | 0.25 | 1 | 0.3% | 1.0 | 4 | 1.1% | 0.12 | |
| A*02:01 | A*02 | A*02:07 | A*02 | 10 | 2.7% | 0.0018 | 9 | 2.4% | 0.0037 | 4 | 1.1% | 0.12 | |
| A*02:06 | A*02 | A*02:01 | A*02 | 7 | 18.4% | 0.012 | 5 | 13.2% | 0.054 | 4 | 10.5% | 0.12 | |
| A*02:01 | A*02 | A*26:01 | A*26 | 0 | 0% | − | 1 | 0.3% | 1.0 | 3 | 0.8% | 0.25 | |
| A*02:01 | A*02 | A*69:01 | A*69 | 1 | 0.3% | 1.0 | 2 | 0.5% | 0.50 | 0 | 0% | − | |
| A*03:01 | A*03 | A*01:01 | A*01 | 0 | 0% | − | 1 | 0.5% | 1.0 | 3 | 1.6% | 0.25 | |
| A*11:01 | A*11 | A*11:02 | A*11 | 1 | 0.6% | 1.0 | 1 | 0.6% | 1.0 | 2 | 1.2% | 0.50 | |
| A*24:04 | A*24 | A*24:02 | A*24 | 0 | 0% | − | 2 | 100.0% | 0.33 | 1 | 50.0% | 1.0 | |
| A*25:01 | A*25 | A*26:01 | A*26 | 0 | 0% | − | 14 | 93.3% | 2.1E-07 | 1 | 6.7% | 1.0 | |
| A*26:01 | A*26 | A*25:01 | A*25 | 2 | 4.1% | 0.49 | 0 | 0% | − | 1 | 2.0% | 1.0 | |
| A*26:01 | A*26 | A*26:02 | A*26 | 1 | 2.0% | 1.0 | 2 | 4.1% | 0.49 | 1 | 2.0% | 1.0 | |
| A*26:08 | A*26 | A*26:01 | A*26 | 0 | 0% | − | 2 | 66.7% | 0.40 | 3 | 100.0% | 0.10 | |
| A*29:02 | A*29 | A*29:01 | A*29 | 3 | 5.7% | 0.24 | 1 | 1.9% | 1.0 | 0 | 0% | − | |
| A*33:03 | A*33 | A*31:01 | A*31 | 1 | 1.6% | 1.0 | 0 | 0% | − | 16 | 25.4% | 1.0E-05 | |
| A*68:01 | A*68 | A*69:01 | A*69 | 0 | 0% | − | 1 | 1.9% | 1.0 | 2 | 3.8% | 0.50 | |
| HLA-B | |||||||||||||
| B*13:01 | B*13 | B*13:02 | B*13 | 1 | 4.3% | 1.0 | 1 | 4.3% | 1.0 | 8 | 34.8% | 0.0038 | |
| B*13:02 | B*13 | B*13:01 | B*13 | 6 | 12.8% | 0.026 | 2 | 4.3% | 0.49 | 0 | 0% | − | |
| B*14:01 | B*14 | B*14:02 | B*14 | 2 | 22.2% | 0.47 | 1 | 11.1% | 1.0 | 4 | 44.4% | 0.082 | |
| B*15:01 | B*15 | B*15:07 | B*15 | 0 | 0% | − | 1 | 1.4% | 1.0 | 3 | 4.1% | 0.24 | |
| B*15:15 | B*15 | B*15:01 | B*15 | 0 | 0% | − | 1 | 33.3% | 1.0 | 2 | 66.7% | 0.40 | |
| B*15:01 | B*15 | B*46:01 | B*46 | 0 | 0% | − | 2 | 2.7% | 0.50 | 2 | 2.7% | 0.50 | |
| B*27:05 | B*27 | B*08:01 | B*08 | 0 | 0% | − | 1 | 2.9% | 1.0 | 2 | 5.9% | 0.49 | |
| B*27:03 | B*27 | B*27:05 | B*27 | 3 | 75.0% | 0.14 | 1 | 25.0% | 1.0 | 1 | 75.0% | 0.14 | |
| B*35:03 | B*35 | B*35:01 | B*35 | 0 | 0% | − | 1 | 4.5% | 1.0 | 2 | 9.1% | 0.49 | |
| B*35:11 | B*35 | B*35:01 | B*35 | 0 | 0% | − | 2 | 100.0% | 0.33 | 2 | 100.0% | 0.33 | |
| B*35:12 | B*35 | B*35:02 | B*35 | 0 | 0% | − | 4 | 80.0% | 0.048 | 1 | 20.0% | 1.0 | |
| B*35:17 | B*35 | B*35:01 | B*35 | 1 | 14.3% | 1.0 | 6 | 85.7% | 0.0047 | 2 | 28.6% | 0.46 | |
| B*35:43 | B*35 | B*35:14 | B*35 | 2 | 20.0% | 0.47 | 0 | 0% | − | 2 | 20.0% | 0.47 | |
| B*35:43 | B*35 | B*49:01 | B*49 | 0 | 0% | − | 1 | 10.0% | 1.0 | 2 | 20.0% | 0.47 | |
| B*39:06 | B*39 | B*39:01 | B*39 | 2 | 12.5% | 0.48 | 1 | 6.3% | 1.0 | 11 | 68.8% | 6.8E-05 | |
| B*40:01 | B*40 | B*07:02 | B*07 | 0 | 0% | − | 1 | 0.9% | 1.0 | 12 | 10.3% | 0.00036 | |
| B*40:04 | B*40 | B*40:02 | B*40 | 1 | 25.0% | 1.0 | 3 | 75.0% | 0.14 | 3 | 75.0% | 0.14 | |
| B*40:06 | B*40 | B*40:02 | B*40 | 5 | 31.3% | 0.043 | 4 | 25.0% | 0.10 | 4 | 25.0% | 0.10 | |
| B*42:02 | B*42 | B*42:01 | B*42 | 1 | 33.3% | 1.0 | 2 | 66.7% | 0.40 | 0 | 0% | − | |
| B*44:03 | B*44 | B*44:02 | B*44 | 1 | 1.2% | 1.0 | 1 | 1.2% | 1.0 | 2 | 2.4% | 0.50 | |
| B*52:01 | B*52 | B*51:01 | B*51 | 1 | 2.1% | 1.0 | 2 | 4.2% | 0.49 | 2 | 4.2% | 0.49 | |
| B*58:01 | B*58 | B*35:01 | B*35 | 1 | 1.8% | 1.0 | 2 | 3.5% | 0.50 | 3 | 5.3% | 0.24 | |
| B*58:01 | B*58 | B*53:01 | B*53 | 1 | 1.8% | 1.0 | 3 | 5.3% | 0.24 | 1 | 1.8% | 1.0 | |
| B*58:02 | B*58 | B*53:01 | B*53 | 1 | 5.3% | 1.0 | 2 | 10.5% | 0.49 | 1 | 5.3% | 1.0 | |
| B*58:02 | B*58 | B*58:01 | B*58 | 1 | 5.3% | 1.0 | 0 | 0% | − | 10 | 52.6% | 0.00039 | |
| HLA-C | |||||||||||||
| C*02:02 | C*02 | C*02:10 | C*02 | 5 | 11.6% | 0.055 | 5 | 11.6% | 0.055 | 5 | 11.6% | 0.055 | |
| C*03:03 | C*03 | C*03:04 | C*03 | 5 | 4.9% | 0.059 | 1 | 1.0% | 1.0 | 1 | 1.0% | 1.0 | |
| C*03:04 | C*03 | C*03:02 | C*03 | 1 | 0.6% | 1.0 | 0 | 0% | − | 3 | 5.9% | 0.029 | |
| C*03:04 | C*03 | C*03:03 | C*03 | 6 | 3.7% | 0.030 | 6 | 3.7% | 0.030 | 13 | 3.9% | 0.12 | |
| C*03:03 | C*03 | C*04:01 | C*04 | 0 | 0% | − | 1 | 1.0% | 1.0 | 4 | 1.0% | 1.0 | |
| C*03:04 | C*03 | C*04:01 | C*04 | 1 | 0.6% | 1.0 | 2 | 1.2% | 0.50 | 1 | 1.8% | 0.25 | |
| C*07:01 | C*07 | C*07:02 | C*07 | 0 | 0% | − | 1 | 0.7% | 1.0 | 7 | 4.8% | 0.015 | |
| C*08:02 | C*08 | C*05:01 | C*05 | 2 | 4.1% | 0.49 | 2 | 4.1% | 0.49 | 5 | 6.1% | 0.24 | |
| C*08:01 | C*08 | C*08:03 | C*08 | 2 | 4.1% | 0.49 | 1 | 2.0% | 1.0 | 3 | 2.0% | 1.0 | |
| C*12:03 | C*12 | C*12:02 | C*12 | 1 | 1.6% | 1.0 | 4 | 6.3% | 0.12 | 4 | 6.3% | 0.12 | |
| C*14:03 | C*14 | C*14:02 | C*14 | 2 | 13.3% | 0.48 | 3 | 20.0% | 0.22 | 3 | 20.0% | 0.22 | |
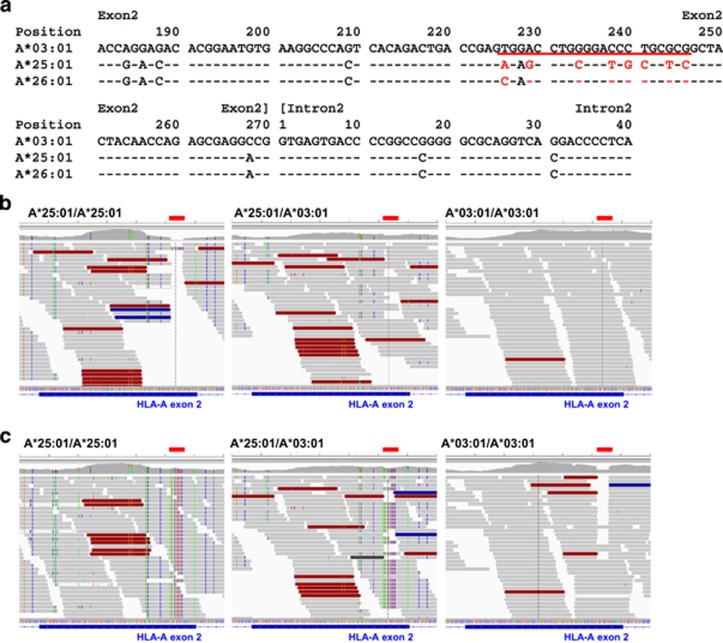
In Polysolver, 78 (61.9%) of 126 discordances in HLA-A were across different serological alleles, and the most common (14 out of 126) miscalls were HLA-A*25:01 to HLA-A*26:01 with an error rate of 93.3% (P=2.1 × 10−7, Figures 3a and b and Supplementary Table 10). This error was observed in one sample using PHLAT and an opposite miscall of HLA-A*26:01 to HLA-A*25:01 was observed in two samples analyzed with Optitype (Table 1). The HLA-A*25:01 to HLA-A* 26:01 error was also observed in two of the 12 samples in our preliminary screening samples (Supplementary Table 3). This miscall was observed regardless to the allele combination; that is, heterozygous with HLA-A*01:01 (N=3), HLA-A*02:01 (N=3), HLA-A*03:01 (N=3), HLA-A*11:01 (N=2) and HLA-A*24:02 (N=3). As shown in Figure 4a, HLA-A*25:01 and HLA-A*26:01 showed 99.2% similarity in 1098 nucleotides in its coding sequence, with only eight nucleotide differences at the 3′ part (226th to 246th bases) of exon 2. This 3′ part of exon 2 in HLA-A*25:01 also has nine nucleotide differences from HLA-A*03:01, which is the HLA-A allele in the hg19 reference. During the first step of the Polysolver pipeline,34 all exome sequence reads were aligned to the human hg19 reference genome (Figure 1), and then the reads mapped to HLA gene loci were extracted as HLA reads. IGV view of a homozygous sample for HLA-A*25:01 after mapping to the hg19 reference showed that there was only one read mapped to the 3′ part of exon 2 of the HLA-A gene (Figure 4b and Table 2). Similarly, in the heterozygous sample for HLA-A*25:01 and HLA-A*03:01, all reads mapped to this 3′ part of exon 2 matched to the HLA-A*03:01 allele (average 23.7 ×) but not to the HLA-A*25:01 allele. Since the sequence reads in HLA-A*03:01 homozygous sample were correctly mapped with an average of 59.9 × depth, we assumed that the HLA-A*25:01 reads were not efficiently mapped to the hg19 reference genome because of the mismatches between HLA-A*25:01 and HLA-A*03:01 sequences. To examine this possibility, we attempted to replace the 3′ part of HLA-A exon 2 of the hg19 reference genome to the corresponding sequences of HLA-A*25:01 allele, and analyzed the same data using this modified genome sequence (Figure 4c). As a result, we could successfully map the sequence reads of the HLA-A*25:01/A25:01 sample to the 3′ part of exon 2 with an average depth of 37.9. In the heterozygous sample with HLA-A*03:01/A25:01, sequence reads mapped to this region were in average 28.2 depth; however, 90% (25.1 ×) of the reads corresponded to the HLA-A*25:01, and the numbers (in average 3.1 ×) of reads corresponding to HLA-A*03:01 were significantly decreased from 23.7 × when we used the standard hg19 reference sequence with HLA-A*03:01. These results clearly indicated that when reference genome sequence with only a single HLA allele was used, serious bias in the mapping of HLA reads occurred, and the initial extracting process of HLA reads using the hg19 reference genome with only HLA-A03:01 allele was the main cause of miscalls in Polysolver. The second most common error in Polysolver was HLA-A*02 to HLA-A*30 with the error rate as low as 1.5% (P=0.0076). In HLA-B and HLA-C genes, no specific discordant pattern was observed for Polysolver, although there were several miscall patterns observed in multiple samples (Figures 3c–f and Supplementary Tables 11, 12).
Figure 4.
Bias among the HLA sequences affecting exome-based HLA-typing results. (a) Sequence alignment of exon 2 region of HLA-A*03:01 (a coordinate with the hg19 reference genome), HLA-A*25:01 and HLA-A*26:01. The sequence underlined in red is the region where nucleotides are different between HLA-A*25:01 and HLA-A*26:01, and between HLA-A*25:01 and HLA-A*03:01, which is corresponding to the red lines in (b) and (c). (b) IGV views of HLA-A exon 2 region after mapping to hg19 reference genome (corresponding to HLA-A*03:01 allele) in the samples with HLA-A*25:01/HLA-A*25:01, HLA-A*25:01/HLA-A*03:01 and HLA-A*03:01/HLA-A*03:01. Red lines represent nucleotides different between HLA-A*25:01 and HLA-A*26:01 as shown in (a). (c) IGV views of HLA-A exon 2 region after mapping to modified hg19 reference genome, which was replaced by HLA-A*25:01 exon 2 sequences, in the samples with HLA-A*25:01/HLA-A*25:01, HLA-A*25:01/HLA-A*03:01 and HLA-A*03:01/HLA-A*03:01. In IGV viewing, standard hg19 was used as a reference sequence. Red lines represent nucleotides different between HLA-A*25:01 and HLA-A*26:01 as shown in (a).
Table 2. Sequence depth and proportion of each allele around the 3′ part of exon 2 region in HLA-A gene.
|
A*25:01/A*25:01 |
A*25:01/A*03:01 |
A*03:01/A*03:01 |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Depth |
Depth |
Depth |
|||||||||||||
| No | Position in HLA-Aa | Chromosome location | Total | A*03:01 | A*25:01 | Proportion of A*25:01 | Total | A*03:01 | A*25:01 | Proportion of A*25:01 | Total | A*03:01 | A*25:01 | Proportion of A*25:01 | |
| Mapped to hg19 reference (A*03:01) | |||||||||||||||
| 1 | exon 2 | 184 | 29 910 716 | 31 | 0 | 31 | 100% | 34 | 20 | 14 | 41% | 69 | 69 | 0 | 0% |
| 2 | exon 2 | 186 | 29 910 717 | 31 | 0 | 31 | 100% | 33 | 19 | 14 | 42% | 59 | 59 | 0 | 0% |
| 3 | exon 2 | 188 | 29 910 730 | 30 | 0 | 30 | 100% | 33 | 19 | 14 | 42% | 55 | 55 | 0 | 0% |
| 4 | exon 2 | 209 | 29 910 742 | 26 | 0 | 26 | 100% | 30 | 20 | 10 | 33% | 63 | 63 | 0 | 0% |
| 5 | exon 2 | 226 | 29 910 759 | 1 | 0 | 1 | 100% | 23 | 23 | 0 | 0% | 60 | 60 | 0 | 0% |
| 6 | exon 2 | 228 | 29 910 761 | 1 | 0 | 1 | 100% | 23 | 23 | 0 | 0% | 60 | 60 | 0 | 0% |
| 7 | exon 2 | 229 | 29 910 762 | 1 | 0 | 1 | 100% | 23 | 23 | 0 | 0% | 60 | 60 | 0 | 0% |
| 8 | exon 2 | 234 | 29 910 767 | 1 | 0 | 1 | 100% | 25 | 25 | 0 | 0% | 60 | 60 | 0 | 0% |
| 9 | exon 2 | 238 | 29 910 771 | 1 | 0 | 1 | 100% | 24 | 24 | 0 | 0% | 60 | 60 | 0 | 0% |
| 10 | exon 2 | 240 | 29 910 773 | 1 | 0 | 1 | 100% | 23 | 23 | 0 | 0% | 60 | 60 | 0 | 0% |
| 11 | exon 2 | 241 | 29 910 774 | 1 | 0 | 1 | 100% | 23 | 23 | 0 | 0% | 60 | 60 | 0 | 0% |
| 12 | exon 2 | 244 | 29 910 777 | 1 | 0 | 1 | 100% | 24 | 24 | 0 | 0% | 60 | 60 | 0 | 0% |
| 13 | exon 2 | 246 | 29 910 779 | 1 | 0 | 1 | 100% | 25 | 25 | 0 | 0% | 59 | 59 | 0 | 0% |
| 14 | exon 2 | 268 | 29 910 801 | 14 | 0 | 14 | 100% | 31 | 26 | 5 | 16% | 60 | 60 | 0 | 0% |
| 15 | Intron 2 | 17 | 29 910 820 | 18 | 0 | 18 | 100% | 30 | 22 | 8 | 27% | 56 | 56 | 0 | 0% |
| 16 | Intron 2 | 31 | 29 910 834 | 21 | 0 | 21 | 100% | 29 | 20 | 9 | 31% | 48 | 48 | 0 | 0% |
| Mapped to modified hg19 reference (A*25:01) | |||||||||||||||
| 1 | exon 2 | 184 | 29 910 716 | 53 | 0 | 53 | 100% | 37 | 14 | 23 | 62% | 47 | 47 | 0 | 0% |
| 2 | exon 2 | 186 | 29 910 717 | 53 | 0 | 53 | 100% | 35 | 12 | 23 | 66% | 43 | 43 | 0 | 0% |
| 3 | exon 2 | 188 | 29 910 730 | 53 | 0 | 53 | 100% | 36 | 13 | 23 | 64% | 44 | 44 | 0 | 0% |
| 4 | exon 2 | 209 | 29 910 742 | 41 | 0 | 41 | 100% | 35 | 12 | 23 | 66% | 50 | 50 | 0 | 0% |
| 5 | exon 2 | 226 | 29 910 759 | 36 | 0 | 36 | 100% | 29 | 3 | 26 | 90% | 21 | 21 | 0 | 0% |
| 6 | exon 2 | 228 | 29 910 761 | 39 | 0 | 39 | 100% | 29 | 3 | 26 | 90% | 19 | 19 | 0 | 0% |
| 7 | exon 2 | 229 | 29 910 762 | 39 | 0 | 39 | 100% | 29 | 3 | 26 | 90% | 19 | 19 | 0 | 0% |
| 8 | exon 2 | 234 | 29 910 767 | 41 | 0 | 41 | 100% | 27 | 3 | 24 | 89% | 19 | 19 | 0 | 0% |
| 9 | exon 2 | 238 | 29 910 771 | 40 | 0 | 40 | 100% | 27 | 3 | 24 | 89% | 19 | 19 | 0 | 0% |
| 10 | exon 2 | 240 | 29 910 773 | 39 | 0 | 39 | 100% | 28 | 3 | 25 | 89% | 19 | 19 | 0 | 0% |
| 11 | exon 2 | 241 | 29 910 774 | 38 | 0 | 38 | 100% | 28 | 3 | 25 | 89% | 19 | 19 | 0 | 0% |
| 12 | exon 2 | 244 | 29 910 777 | 35 | 0 | 35 | 100% | 28 | 3 | 25 | 89% | 19 | 19 | 0 | 0% |
| 13 | exon 2 | 246 | 29 910 779 | 34 | 0 | 34 | 100% | 29 | 4 | 25 | 86% | 19 | 19 | 0 | 0% |
| 14 | exon 2 | 268 | 29 910 801 | 36 | 0 | 36 | 100% | 38 | 14 | 24 | 63% | 49 | 49 | 0 | 0% |
| 15 | Intron 2 | 17 | 29 910 820 | 29 | 0 | 29 | 100% | 35 | 14 | 21 | 60% | 48 | 48 | 0 | 0% |
| 16 | Intron 2 | 31 | 29 910 834 | 30 | 0 | 30 | 100% | 34 | 14 | 20 | 59% | 45 | 45 | 0 | 0% |
In PHLAT, 246 (60.7%) of 405 miscalls were across different serological types when predicting HLA-A genotypes (Figures 3a and b and Supplementary Table 13). Notably, PHLAT erroneously assigned various types of allele to either HLA-A*01:22 or HLA-A*01:81 (that is, the error rates of HLA-A*02:01 to HLA-A*01:22, HLA-A*02:01 to HLA-A*01:81, HLA-A*02:06 to HLA-A*01:81, HLA-A*26:01 to HLA-A*01:22 and HLA-A*68:01 to HLA-A*01:22 were 6.1%, 9.5%, 26.3%, 16.3% and 17.0% with P-values of 4.0 × 10−8, 3.8 × 10−15, 0.0010, 0.0057 and 0.0027, respectively), despite relatively low sequence similarities between HLA-A*01:22 or HLA-A*01:81 and these alleles (Supplementary Figure 2a). These errors were PHLAT platform-specific errors, and not found in Optitype or Polysolver (Supplementary Tables 7, 10). The second frequent miscalls in PHLAT were between HLA-A*33:03 and HLA-A*31:01, which showed high sequence similarities (error rate of 25.4%, P=1.0 × 10−5; Supplementary Figure 2b). Proportion of miscalls across different serological types were 100 (34.7%) of 288 for HLA-B and 33 (23.6%) of 140 for HLA-C. A few specific patterns were observed such as HLA-B*40:01 to HLA-B*07:02 (error rate of 10.3%, P=3.6 × 10−4) and HLA-C*08 to HLA-C*05:01 (error rate of 6.9%, P=0.0069), although HLA-C*08 to HLA-C*05:01 miscalls were also found in Optitype and Polysolver as described above (Figures 3c–f and Supplementary Tables 14, 15).
Discussion
In this study, we evaluated seven algorithms for HLA typing using whole-exome data: Optitype,33 Polysolver,34 PHLAT,35 HLAreporter,36 HLAforest,37 HLAminer38 and seq2HLA,39 using 12 clinical samples, and further evaluated three of them, Optitype, Polysolver and PHLAT, which displayed the highest accuracy (>90%) in the preliminary screening, using 961 samples from the 1000 Genomes Project database.
One of the common platform-specific miscalls is from HLA-A*25:01 to HLA-A*26:01 that was found in Polysolver. This miscall is caused by the lacking of HLA-A*25:01 reads during the extraction of HLA reads using the human hg19 reference genome, which has a single HLA allele of HLA-A*03:01 at HLA-A locus. On the other hand, Optitype used reference sequences that are a collection of all the HLA allele sequences when extracting HLA reads. Although extraction of HLA reads is included in the script of Polysolver, we may be able to improve the accuracy of Polysolver if we apply HLA read extraction using a collection of all the HLA allele sequences.
Among the software we tested, Optitype showed the highest performance with 97.2% accuracy at the second field resolution. However, this value was still lower than the accuracy in HLA typing using PCR-based next-generation sequencing methods, which is nearly 100% accuracy.25, 28 One of the causes of lower accuracy in exome-based HLA typing may be because of the low number of HLA reads. The lower accuracy was found in the samples with lower number of HLA reads (accuracy of 82.0%, 95.4% and 98.0% in the groups with HLA reads of <1000 (N=25), 1000–2000 (N=193) and >2000 (N=743), respectively). Although this explains only a part of the causes of miscalls in Optitype, input number of reads into the analysis is critical for accurate HLA genotype prediction.
In this study, we analyzed the potential problems that contributed to prediction errors in computational analysis, including the mapping process to the reference genome sequence with a single HLA allele. Currently, Optitype showed the highest performance; however, certain patterns of miscalls still occurred and needed to be addressed. In order to apply these techniques into clinical settings, further studies and validations are required to solve these problems to improve the accuracy of HLA genotype estimation.
Acknowledgments
The super-computing resource was provided by Human Genome Center, the Institute of Medical Science, the University of Tokyo (http://sc.hgc.jp/shirokane.html).
Footnotes
Supplementary Information accompanies the paper on Journal of Human Genetics website (http://www.nature.com/jhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Robinson, J., Mistry, K., McWilliam, H., Lopez, R., Parham, P. & Marsh, S.G. The IMGT/HLA database. Nucleic Acids Res 39, D1171–D1176 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J., Halliwell, J.A., Hayhurst, J.D., Flicek, P., Parham, P. & Marsh, S.G. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res 43, D423–D431 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The MHC sequencing consortium Complete sequence and gene map of a human major histocompatibility complex. Nature 401, 921–923 (1999). [DOI] [PubMed] [Google Scholar]
- Sasazuki, T., Juji, T., Morishima, Y., Kinukawa, N., Kashiwabara, H., Inoko, H. et al. Effect of matching of class I HLA alleles on clinical outcome after transplantation of hematopoietic stem cells from an unrelated donor. Japan Marrow Donor Program. N Engl J Med 339, 1177–1185 (1998). [DOI] [PubMed] [Google Scholar]
- International MHC and Autoimmunity Genetics Network, Rioux, J.D., Goyette, P., Vyse, T.J., Hammarstrom, L., Fernando, M.M. et al. Mapping of multiple susceptibility variants within the MHC region for 7 immune-mediated diseases. Proc Natl Acad Sci USA 106, 18680–18685 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniwa, N. & Saito, Y. Pharmacogenomics of severe cutaneous adverse reactions and drug-induced liver injury. J Hum Genet 58, 317–326 (2013). [DOI] [PubMed] [Google Scholar]
- Kiyotani, K. Prediction of drug-induced adverse reactions: skin hypersensitivity and liver toxicity. in Immunopharmacogenomics (ed. Nakamura, Y.) 47–61 (Springer, Tokyo, Japan, 2016). [Google Scholar]
- Townsend, A. & Bodmer, H. Antigen recognition by class I-restricted T lymphocytes. Annu Rev Immunol 7, 601–624 (1989). [DOI] [PubMed] [Google Scholar]
- Bjorkman, P.J. & Parham, P. Structure, function, and diversity of class I major histocompatibility complex molecules. Annu Rev Biochem 59, 253–288 (1990). [DOI] [PubMed] [Google Scholar]
- Stransky, N., Egloff, A.M., Tward, A.D., Kostic, A.D., Cibulskis, K., Sivachenko, A. et al. The mutational landscape of head and neck squamous cell carcinoma. Science 333, 1157–1160 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, M.S., Stojanov, P., Mermel, C.H., Robinson, J.T., Garraway, L.A., Golub, T.R. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney, M.S., Shukla, S.A., Wu, C.J., Getz, G. & Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghaei, H., Paz-Ares, L., Horn, L., Spigel, D.R., Steins, M., Ready, N.E. et al. Nivolumab versus docetaxel in advanced nonsquamous non-samall-cell lung cancer. N Engl J Med 373, 1627–1639 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motzer, R.J., Escudier, B., McDermott, D.F., George, S., Hammers, H.J., Srinivas, S. et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med 373, 1803–1813 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmer, J., Reckamp, K.L., Baas, P., Crino, L., Eberhardt, W.E., Poddubskaya, E. et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med 373, 123–135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, J., Chiarion-Sileni, V., Gonzalez, R., Grob, J.J., Cowey, C.L., Lao, C.D. et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med 373, 23–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder, A., Makarov, V., Merghoub, T., Yuan, J., Zaretsky, J.M., Desrichard, A. et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 371, 2189–2199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi, N.A., Hellmann, M.D., Snyder, A., Kvistborg, P., Makarov, V., Havel, J.J. et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh, S.G., Albert, E.D., Bodmer, W.F., Bontrop, R.E., Dupont, B., Erlich, H.A. et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens 75, 291–455 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth, R., Hurley, C.K., Marsh, S.G., Lau, M., Noreen, H.J., Kempenich, J.H. et al. The HLA dictionary 2008: a summary of HLA-A, -B, -C, -DRB1/3/4/5, and -DQB1 alleles and their association with serologically defined HLA-A, -B, -C, -DR, and -DQ antigens. Tissue Antigens 73, 95–170 (2009). [DOI] [PubMed] [Google Scholar]
- Adams, S.D., Barracchini, K.C., Simonis, T.B., Stroncek, D. & Marincola, F.M. High throughput HLA sequence-based typing (SBT) utilizing the ABI Prism 3700 DNA Analyzer. Tumori 87, S40–S43 (2001). [PubMed] [Google Scholar]
- Itoh, Y., Mizuki, N., Shimada, T., Azuma, F., Itakura, M., Kashiwase, K. et al. High-throughput DNA typing of HLA-A, -B, -C, and -DRB1 loci by a PCR-SSOP-Luminex method in the Japanese population. Immunogenetics 57, 717–729 (2005). [DOI] [PubMed] [Google Scholar]
- Nelson, W.C., Pyo, C.W., Vogan, D., Wang, R., Pyon, Y.S., Hennessey, C. et al. An integrated genotyping approach for HLA and other complex genetic systems. Hum Immunol 76, 928–938 (2015). [DOI] [PubMed] [Google Scholar]
- Bentley, G., Higuchi, R., Hoglund, B., Goodridge, D., Sayer, D., Trachtenberg, E.A. et al. High-resolution, high-throughput HLA genotyping by next-generation sequencing. Tissue Antigens 74, 393–403 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina, T., Suzuki, S., Ozaki, Y., Taira, H., Kikkawa, E., Shigenari, A. et al. Super high resolution for single molecule-sequence-based typing of classical HLA loci at the 8-digit level using next generation sequencers. Tissue Antigens 80, 305–316 (2012). [DOI] [PubMed] [Google Scholar]
- Shiina, T., Suzuki, S., Kulski, J.K. MHC genotyping in human and nonhuman species by PCR-based next-generation sequencing in Next Generation Sequencing - Advances, Applications and Challenges (ed. Kulski, J.K.) 82–109 (InTech, Rijeka, Croatia, 2016). [Google Scholar]
- Juhos, S., Rigó, K., Horváth, G. On genotyping polymorphic HLA genes—ambiguities and quality measures using NGS in Next Generation Sequencing—Advances, Applications and Challenges (ed. Kulski, J.K.) 370–386 (InTech, Rijeka, Croatia, 2016). [Google Scholar]
- Gourraud, P.A., Khankhanian, P., Cereb, N., Yang, S.Y., Feolo, M., Maiers, M. et al. HLA diversity in the 1000 genomes dataset. PLoS ONE 9, e97282 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szolek, A., Schubert, B., Mohr, C., Sturm, M., Feldhahn, M. & Kohlbacher, O. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics 30, 3310–3316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla, S.A., Rooney, M.S., Rajasagi, M., Tiao, G., Dixon, P.M., Lawrence, M.S. et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat Biotechnol 33, 1152–1158 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, Y., Ni, M., Cooper, B., Wei, Y. & Fury, W. Inference of high resolution HLA types using genome-wide RNA or DNA sequencing reads. BMC Genomics 15, 325 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y., Yang, J., Ying, D., Zhang, Y., Shotelersuk, V., Hirankarn, N. et al. HLAreporter: a tool for HLA typing from next generation sequencing data. Genome Med 7, 25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.J. & Pourmand, N. HLA typing from RNA-seq data using hierarchical read weighting. PLoS ONE 8, e67885 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, R.L., Choe, G., Freeman, D.J., Castellarin, M., Munro, S., Moore, R. et al. Derivation of HLA types from shotgun sequence datasets. Genome Med 4, 95 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boegel, S., Lower, M., Schafer, M., Bukur, T., de Graaf, J., Boisguerin, V. et al. HLA typing from RNA-Seq sequence reads. Genome Med 4, 102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J.T., Thorvaldsdottir, H., Winckler, W., Guttman, M., Lander, E.S., Getz, G. et al. Integrative genomics viewer. Nat Biotechnol 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir, H., Robinson, J.T. & Mesirov, J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.