Summary
Oxygen (O2) homeostasis is important for all aerobic animals. However, the manner by which O2 sensing and homeostasis contribute to lifespan regulation is poorly understood. Here, we use the nematode Caenorhabditis elegans to address this question. We demonstrate that a loss‐of‐function mutation in the neuropeptide receptor gene npr‐1 and a deletion mutation in the atypical soluble guanylate cyclase gcy‐35 O2 sensor interact synergistically to extend worm lifespan. The function of npr‐1 and gcy‐35 in the O2‐sensing neurons AQR, PQR, and URX shortens the lifespan of the worm. By contrast, the activity of the atypical soluble guanylate cyclase O2 sensor gcy‐33 in these neurons is crucial for lifespan extension. In addition to AQR, PQR, and URX, we show that the O2‐sensing neuron BAG and the interneuron RIA are also important for the lifespan lengthening. Neuropeptide processing by the proprotein convertase EGL‐3 is essential for lifespan extension, suggesting that the synergistic effect of joint loss of function of gcy‐35 and npr‐1 is mediated through neuropeptide signal transduction. The extended lifespan is regulated by hypoxia and insulin signaling pathways, mediated by the transcription factors HIF‐1 and DAF‐16. Moreover, reactive oxygen species (ROS) appear to play an important function in lifespan lengthening. As HIF‐1 and DAF‐16 activities are modulated by ROS, we speculate that joint loss of function of gcy‐35 and npr‐1 extends lifespan through ROS signaling.
Keywords: Caenorhabditis elegans, lifespan, NPR‐1, oxygen sensing, reactive oxygen species, soluble guanylate cyclase
Introduction
To survive, animals must sense key environmental factors, including temperature, food, and oxygen (O2). Sensory information then regulates homeostatic responses to maintain physiological parameters within a narrow range (Zimmer et al., 2009). O2 and reactive oxygen species (ROS) are important for animal viability but may be toxic when misregulated (Halliwell & Gutteridge, 2006).
The Caenorhabditis elegans (C. elegans) nematode uses atypical soluble guanylate cyclases (sGCs) to sense changes in O2 levels. The sGCs GCY‐35 and GCY‐36 are expressed in the O2‐sensing neurons AQR, PQR, and URX and appear to be activated by hyperoxia ([O2]>12%) (Gray et al., 2004; Abergel et al., 2016). Upon activation, cGMP production is increased and triggers the opening of cyclic nucleotide‐gated channels, such as TAX‐2 and TAX‐4, which depolarize the cells (Gray et al., 2004). A decrease in ambient O2 concentration inhibits AQR, PQR, and URX activity but activates the BAG sensory neurons. In BAG, the atypical sGC GCY‐31 and GCY‐33 are activated by low O2 concentration (Zimmer et al., 2009). Therefore, GCY‐31/GCY‐33 and GCY‐35/GCY‐36 display reciprocal responses to O2.
Previous studies showed that, in addition to sGC signaling, neuropeptide signaling plays a critical role in C. elegans O2 sensing (Gray et al., 2004). N2 worms that express the gain‐of‐function allele of neuropeptide receptor 1, npr‐1(215V), do not avoid hyperoxia on food. However, animals bearing the weaker natural allele npr‐1(215F) or the loss‐of‐function npr‐1(ad609) allele show strong 21% O2 avoidance on food and aggregate on the bacterial lawn border, where there is a thicker growth of bacteria and, therefore, less O2 (~13%) (Gray et al., 2004). The aggregation of worms may further decrease the O2 concentration inside the clump and so create a preferable O2 concentration of around 8%. Therefore, N2 worms may experience higher O2 levels compared with npr‐1(ad609) animals under standard laboratory growth conditions. In this study, we explored the function of O2 sensing and neuropeptide signaling in lifespan regulation.
Results
NPR‐1 activity reduces Caenorhabditis elegans lifespan
The gain‐of‐function npr‐1(215V) allele appears to have arisen during domestication of the N2 strain to laboratory conditions (McGrath et al., 2009). As npr‐1(215V) activity suppresses the natural hyperoxia avoidance response on bacteria, we asked whether N2 worms suffer from misregulated O2‐homeostatic responses and therefore have a shorter lifespan than the aggregating strain npr‐1(ad609). We measured the lifespans of N2 and npr‐1(ad609) worms on food. The lifespan of N2 worms was slightly but significantly shorter than that of npr‐1(ad609) animals (Fig. 1A; P = 0.0023), suggesting that NPR‐1(215V) activity is harmful to C. elegans longevity.
Figure 1.
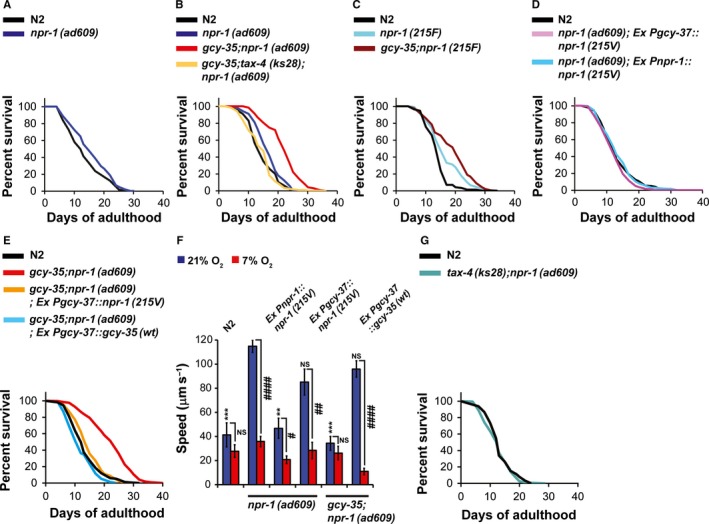
NPR‐1 and GCY‐35 regulate Caenorhabditis elegans lifespan. (A–E, G) Survival curves comparing the lifespans of worm strains. These experiments were performed at 21°C on live OP50. (F) Speed measurements. The speed of worms was measured at 21% and 7% O2 in the presence of OP50. Asterisks indicate significance for comparisons with npr‐1 animals’ speed at 21% O2, Kruskal–Wallis test with Dunn's post‐test. Number sign indicates significance between speeds, within a strain. Unpaired t‐test with Welch's correction. n = 6 or more assays performed over at least 3 days. ** P < 0. 01, *** P < 0.001, ##P < 0. 01, ####P < 0.0001, NS, nonsignificant, Error bars represent SEM.
Deletion of gcy‐35 significantly increases npr‐1(ad609) worms’ lifespan
GCY‐35 activity is important for the O2 responses of npr‐1(ad609) animals (Gray et al., 2004). To explore whether GCY‐35 is also important for the extended lifespan of npr‐1(ad609) worms, we created an npr‐1(ad609) strain bearing the gcy‐35(ok769) deletion allele of gcy‐35 and compared its lifespan to N2 and npr‐1(ad609) animals (Fig. 1B). The lifespan of gcy‐35;npr‐1(ad609) mutants was significantly longer than both N2 and npr‐1(ad609) worms, suggesting that the catalytic activity of GCY‐35 shortens the lifespan of npr‐1(ad609) worms.
We explored the lifespan of N2 worms bearing the npr‐1(215F) ancestral allele (this strain, QX1155 (McGrath et al., 2009), was kindly provided by the Bargmann laboratory) and of the gcy‐35(ok769);npr‐1(215F) double mutant. The lifespan of npr‐1(215F) worms was significantly longer than N2 controls (Fig. 1C, P < 0.0001), further supporting the conclusion that npr‐1(215V) activity shortens the lifespan of N2 worms. The deletion of gcy‐35 significantly lengthened the lifespan of npr‐1(215F) worms (P = 0.0006), further supporting our conclusion that GCY‐35 activity shortens the lifespan of worms bearing either a nonfunctional or a weak allele of NPR‐1.
GCY‐35 and GCY‐36 are thought to form a functional heterodimer in AQR, PQR, and URX (Cheung et al., 2004; Zimmer et al., 2009). Therefore, npr‐1(ad609) gcy‐36 mutants should have extended lifespan compared with N2 and npr‐1(ad609) worms. To test this, we crossed npr‐1(ad609) worms with worms bearing the gcy‐36(ok2208) loss‐of‐function allele and compared the lifespan of npr‐1(ad609) gcy‐36 mutants to N2 and npr‐1(ad609) worms. Joint loss of function of npr‐1 and gcy‐36 significantly extended lifespan compared with both N2 and npr‐1(ad609) controls (Fig. S1A, Supporting information, P < 0.0001 and P = 0.0034, respectively). As loss of function of either gcy‐35 or gcy‐36 extended the lifespan of npr‐1(ad609) worms and tracking the gcy‐35 deletion was easier than tracking the gcy‐36 deletion, we focused our studies on how combined loss of function of gcy‐35 and npr‐1 increases lifespan.
NPR‐1 and GCY‐35 function in the AQR, PQR, and URX neurons to regulate lifespan
As both npr‐1 and gcy‐35 are expressed in the O2‐sensing neurons AQR, PQR, and URX, we hypothesized that the function of npr‐1 and gcy‐35 in these neurons decreases lifespan. To test this, we generated npr‐1(ad609) transgenes expressing genomic npr‐1(215V) only in AQR, PQR, and URX (using the gcy‐37 promoter), or under its own promoter. In addition, we generated gcy‐35;npr‐1(ad609) transgenic animals expressing genomic npr‐1(215V) only in the AQR, PQR, and URX neurons, as well as gcy‐35;npr‐1(ad609) transgenes expressing gcy‐35 cDNA in AQR, PQR, and URX. npr‐1(ad609) animals expressing npr‐1(215V) under its own promoter or the gcy‐37 promoter had similar lifespans to N2 worms (Fig. 1D), indicating that npr‐1(215V) activity in the AQR, PQR, and URX neurons is essential and sufficient for shortening the extended lifespan of npr‐1(ad609) worms. Similarly, driving npr‐1(215V) expression only in the AQR, PQR, and URX neurons significantly shortened the lifespan of gcy‐35;npr‐1(ad609) mutants (Fig. 1E, P < 0.0001). Finally, restoring gcy‐35 activity in AQR, PQR, and URX shortened the lifespan of gcy‐35;npr‐1(ad609) animals (Fig. 1E). Notably, a previous study by Powell‐Coffman and colleagues observed that gcy‐37 is also expressed in AVM neurons and in two unidentified neurons in the head (Qin et al., 2006). However, we did not observe mCherry staining in AVM or any other neurons in the head beside AQR, and URX (see Fig. S1B, Supporting information). We performed two control experiments in which we used the gcy‐34 promoter region to rescue the activity of gcy‐35 and npr‐1(215V) in the AQR, PQR, and URX neurons of gcy‐35;npr‐1(ad609) mutants (gcy‐34 is specifically expressed in AQR, PQR, and URX (Qin et al., 2006)). Similar to our previous results, expressing either gcy‐35 or npr‐1(215V) under the promoter of gcy‐34 significantly shortened the lifespan of gcy‐35;npr‐1(ad609) mutants (Fig. S1C, Supporting information), supporting the conclusion that gcy‐35 and npr‐1(215V) act in AQR, PQR, and URX to shorten the lifespan of gcy‐35;npr‐1(ad609) mutants.
We verified the functionality of the npr‐1 and gcy‐35 expression constructs using speed measurements (Fig. 1F). Previous studies showed that npr‐1(ad609) worms move quickly on food at 21% O2 but sharply slow down at 7% O2 (Abergel et al., 2016), whereas N2 worms move slowly at both O2 concentrations (similar to npr‐1(ad609) worms at 7% O2). Moreover, the high foraging speed of npr‐1(ad609) worms on food requires gcy‐35 activity (Cheung et al., 2005). Restoring npr‐1(215V) activity using its own promoter in npr‐1(ad609) worms reduced the speed of worms at 21% O2 to the low levels observed for N2 worms. npr‐1(215V) expression in AQR, PQR, and URX did not attenuate speed at 21% O2, but did suppress the extended lifespan of gcy‐35;npr‐1(ad609) worms (Fig. 1E), suggesting that npr‐1 function in aging is not mediated by the hub‐neuron RMG (Macosko et al., 2009). Conversely, restoring gcy‐35 activity in AQR, PQR, and URX in gcy‐35;npr‐1(ad609) worms significantly increased their speed at 21% O2 (Fig. 1F). Together, our data show that our rescuing constructs are functional and that the synergistic life extension of gcy‐35;npr‐1(ad609) worms depends on the combined loss of function of npr‐1 and gcy‐35 in AQR, PQR, and URX.
TAX‐4 activity is important for the extended lifespan of gcy‐35;npr‐1(ad609) mutants
The activity of TAX‐2/TAX‐4 is important for cGMP signaling in some or all O2‐sensing neurons (Coates & de Bono, 2002; Zimmer et al., 2009). Indeed, previous studies showed that the tax‐4(ks28) strong loss‐of‐function allele suppresses GCY‐35 activity in AQR, PQR, and URX (Gray et al., 2004). Therefore, we hypothesized that joint loss of function of tax‐4 and npr‐1 would recapitulate the synergistic effect of gcy‐35 and npr‐1 loss of function on lifespan. However, our results showed that the lifespan of tax‐4(ks28);npr‐1(ad609) double mutants is similar to N2 worms (Fig. 1G). Moreover, we found that activity of TAX‐4 is required for the extended lifespan of gcy‐35;npr‐1(ad609) mutants, as the gcy‐35;tax‐4(ks28);npr‐1(ad609) triple mutants and N2 worms had similar lifespans (Fig. 1B, P = 0.8745).
A previous study showed that the tax‐4 (ky89) and tax‐4(p678) loss‐of‐function alleles significantly lengthen the lifespan of N2 worms at 20°C (Apfeld & Kenyon, 1999). However, our experiments show that tax‐4(ks28) mutants and N2 worms have similar lifespans (P = 0.6112, Fig. S1D, Supporting information). Apart from using a different loss‐of‐function allele of tax‐4, a significant difference between our experiments and previous studies is that we did not use the thymidylate synthetase inhibitor FUDR in our lifespan assays. Therefore, we investigated whether the function of tax‐4 in lifespan regulation is modulated by FUDR. FUDR significantly increased the lifespan of tax‐4(ks28) mutants compared with control tax‐4(ks28) mutants that grew without FUDR (P < 0.0001, Fig. S1D, Supporting information). In addition, a significant lengthening of the lifespan of N2 worms was observed (Fig. S1E, Supporting information). However, the lifespans of tax‐4(ks28) mutants and N2 worms that grew on FUDR were not significantly different (P = 0.6091). Therefore, while our results show that FUDR significantly affects the function of tax‐4 in lifespan regulation, they do not recapitulate previous results. As FUDR inhibits both DNA synthesis in worms and bacterial proliferation (Portal‐Celhay et al., 2012), we explored whether the effect of tax‐4 on lifespan is modulated by bacterial viability. We repeated the lifespan experiments using UV‐killed bacteria as a food source. The lifespan of tax‐4(ks28) mutants that were grown on dead bacteria was similar to tax‐4(ks28) mutants that grew on live bacteria and FUDR (P = 0.0858), suggesting that FUDR extends the lifespan of tax‐4(ks28) mutants by suppressing bacterial toxicity. Finally, under this experimental condition the lifespan of N2 worms was significantly lengthened (P < 0.0001) and was similar to the lifespan of tax‐4(ks28) (Fig. S1F, Supporting information), suggesting that tax‐4 may be important in the defense mechanism against bacterial toxicity.
GCY‐33 activity is essential for the extended lifespan of gcy‐35;npr‐1(ad609) animals
Our results show that the function of tax‐4 is important for the extended lifespan of both npr‐1(ad609) and gcy‐35;npr‐1(ad609) worms. As the sGC gcy‐33 and tax‐4 function in URX and BAG (Gray et al., 2004; Zimmer et al., 2009), we next explored the function of gcy‐33 in lifespan regulation. To explore whether gcy‐33 regulates the lifespan of npr‐1(ad609) and gcy‐35;npr‐1(ad609) worms, we replaced the wild‐type allele of gcy‐33 with the gcy‐33(ok232) deletion allele. The deletion of gcy‐33 did not affect N2 worms’ lifespan (Fig. 2A). In contrast, gcy‐33 deletion significantly shortened the lifespan of npr‐1(ad609) and gcy‐35;npr‐1(ad609) worms (Fig. 2B). Notably, the mean lifespan of gcy‐35;gcy‐33;npr‐1(ad609) mutants was significantly shorter than npr‐1(ad609) and gcy‐33;npr‐1(ad609) mutants (P < 0.0001), suggesting a negative synergistic interaction between gcy‐33 and gcy‐35.
Figure 2.
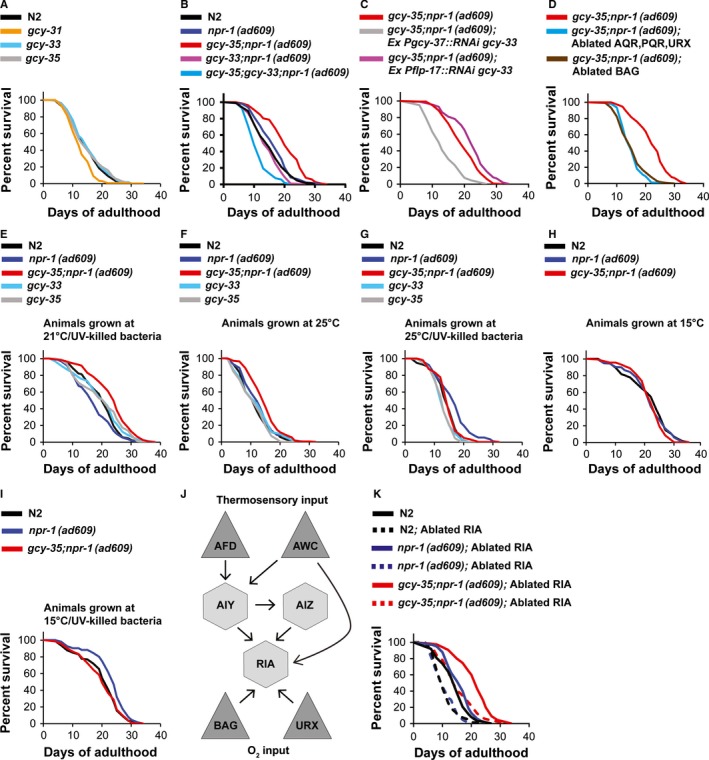
GCY‐31 and GCY‐33 are important for gcy‐35;npr‐1(ad609) animals’ extended lifespan. (A–D, K) Survival curves comparing the lifespans of worm strains at 21°C on live OP50. (C) Survival curves comparing the lifespan of gcy‐35;npr‐1(ad609) mutants to gcy‐35;npr‐1(ad609) transgenic worms expressing gcy‐33 RNAi in AQR, PQR, and URX (under the gcy‐37 promoter) or in BAG (under the flp‐17 promoter). (E–I) Survival curves comparing the lifespans of worm strains at different growth conditions, as indicated by the labels above the graphs. (J) Circuit diagram of neurons with synaptic connections to RIA.
Previous studies suggested that GCY‐31 and GCY‐33 form a functional O2‐sensor complex in BAG (Zimmer et al., 2009). To see whether deletion of gcy‐31 and gcy‐33 has a similar effect, we measured the lifespan of N2 worms bearing the gcy‐31(ok296) deletion allele. Unlike gcy‐33 mutants, gcy‐31 mutants had a significantly shorter lifespan than N2 animals (Fig. 2A). Next, we generated gcy‐35;gcy‐31 npr‐1(ad609) mutants and compared their lifespan to gcy‐35;npr‐1(ad609) worms. The lifespan of gcy‐35;gcy‐31 npr‐1(ad609) mutants was significantly shorter than gcy‐35;npr‐1(ad609) worms but significantly longer than gcy‐35;gcy‐33;npr‐1(ad609) mutants (P < 0.0001, and P = 0.0137, respectively) (Fig. S2A, Supporting information), suggesting that the function of gcy‐33 in lifespan regulation is not restricted to the GCY‐31/GCY‐33 complex.
GCY‐33 activity in AQR, PQR, and URX is essential for lifespan extension of gcy‐35;npr‐1(ad609) animals
Unlike gcy‐31, the expression pattern of gcy‐33 is not limited to BAG and includes the AQR, PQR, and URX neurons (Zimmer et al., 2009). To explore where gcy‐33 function is important for lifespan regulation, we performed cell‐specific RNAi experiments. To knock down gcy‐33 activity in BAG, we generated transgenes expressing gcy‐33 RNAi under the flp‐17 promoter. To knock down gcy‐33 activity in AQR, PQR, and URX, we generated transgenic worms expressing gcy‐33 RNAi under the gcy‐37 promoter. Knocking down gcy‐33 expression in the AQR, PQR, and URX neurons of gcy‐35;npr‐1(ad609) worms shortened the lifespan of these worms to below the level of N2 worms (P = 0.0153; Fig. 2C), recapitulating the results obtained for the gcy‐35;gcy‐33;npr‐1(ad609) mutant (Fig. 2B). By contrast, gcy‐35;npr‐1(ad609) worms expressing gcy‐33 RNAi only in BAG had a significantly longer lifespan than gcy‐35;npr‐1(ad609) controls (Fig. 2C, P = 0.0002). Knocking down gcy‐33 expression in N2 worms, either in BAG or AQR, PQR, and URX did not have a significant effect on worm lifespan (Fig. S2B, Supporting information). Knocking down gcy‐33 in AQR, PQR, and URX significantly shortened the lifespan of npr‐1(ad609) worms (Fig. 2SC, Supporting information); however, gcy‐33 RNAi in BAG did not affect their lifespan. Together, our results suggest that the effect of gcy‐33 activity on lifespan is cell specific and is dependent on the activity of gcy‐35 and npr‐1.
The O2‐sensing neurons AQR, PQR, URX, and BAG are important for the extended lifespan of gcy‐35;npr‐1(ad609) animals
Our results show that both gcy‐31 and gcy‐33 are essential for the extended lifespan of gcy‐35;npr‐1(ad609) mutants (Figs 2B and S2A, Supporting information). As gcy‐31 is exclusively expressed in BAG, and the function of gcy‐33 in AQR, PQR, and URX is essential for the lifespan extension of gcy‐35;npr‐1(ad609) worms, we hypothesized that all four O2‐sensing neurons are required for the lifespan extension of gcy‐35;npr‐1(ad609) mutants. To test this, we made strains expressing the death activator egl‐1 only in AQR, PQR, and URX and strains expressing caspase 3 only in BAG (the parental strains for these crosses, CX7102 and CX11697, were kindly provided by the Bargmann laboratory). Ablation of AQR, PQR, and URX significantly shortened the lifespan of N2, npr‐1(ad609), and gcy‐35;npr‐1(ad609) worms (Figs 2D and S2D,E, Supporting information). BAG ablation significantly shortened the lifespan of gcy‐35;npr‐1(ad609) and npr‐1(ad609) worms (Figs 2D and S2D, Supporting information, respectively) and, however, significantly increased the lifespan of N2 worms (Fig. S2E, Supporting information). Therefore, our experiments suggest that AQR, PQR, and URX are generally important for life extension and that BAG regulates lifespan in an npr‐1‐dependent manner.
Temperature and food quality affect the function of npr‐1 and gcy‐35 in lifespan regulation
A previous study, conducted on the N2 background, showed that gcy‐33 deletion extends lifespan, whereas deletion of gcy‐35 shortens lifespan (Liu & Cai, 2013). However, in our experiments, the same gcy‐33 and gcy‐35 deletion alleles did not affect N2 worm lifespan significantly (Fig. 2A). As Liu & Cai used different experimental conditions from ours (our assays were performed on live OP50 bacteria at 21°C, whereas Liu & Cai's experiments used UV‐killed OP50 bacteria at 25°C), we asked whether temperature and bacterial viability could explain the difference between the results. We measured lifespan in three growth conditions: 21°C/UV‐killed OP50; 25°C/live OP50; and 25°C/UV‐killed OP50. All strains lived longest on UV‐killed OP50 at 21°C (Fig. 2E). These results support previous studies that showed that live OP50 shortens C. elegans lifespan (Garigan et al., 2002). Importantly, under this experimental condition, gcy‐33 and gcy‐35 mutants had similar lifespans to N2 controls. However, N2 worms lived significantly longer compared with npr‐1(ad609) worms (P = 0.0098), suggesting that thriving on dead OP50 bacteria requires NPR‐1(215V) activity. Growing the worms on live OP50 at 25°C shortened the lifespan of all strains (Fig. 2F). The lifespans of gcy‐33, gcy‐35, and npr‐1(ad609) mutants grown in this environment were similar to N2 controls. Finally, gcy‐33 and gcy‐35 mutants had shorter lifespans than N2 controls on dead OP50 at 25°C (Fig. 2G). Notably, gcy‐35;npr‐1(ad609) worms lived longer (compared with the other strains) on live and dead OP50 at 21°C (Fig. 2B,E) and on live OP50 at 25°C (Fig. 2F). However, when grown at 25°C on dead OP50, their lifespan was similar to N2 animals (Fig. 2G). Intriguingly, in this growth condition, npr‐1(ad609) animals lived significantly longer compared with the other strains, indicating that gcy‐35 activity is essential to thriving in this environment. Taken together, our results show that the synergistic effect of npr‐1 and gcy‐35 loss of function on lifespan is modulated by both temperature and bacterial viability. Moreover, the discrepancy between our data and that of and Liu & Cai could be attributed to experimental conditions in the case of gcy‐35 mutants, as our findings at 25°C on dead OP50 are in agreement with theirs. However, the discrepancy between the gcy‐33 mutant lifespan data could not be explained by differences in temperature or bacteria viability, as the gcy‐33 deletion did not lengthen the lifespan of N2 worms (nor npr‐1(ad609) or gcy‐35;npr‐1(ad609) mutants) in any condition we tested.
Joint loss of function of gcy‐35 and npr‐1 does not extend lifespan at 15°C
Our results show that joint loss of function of gcy‐35 and npr‐1 affects lifespan in a temperature‐ and food quality‐dependent way. To further explore this observation, we measured the lifespan of N2, npr‐1(ad609), and gcy‐35;npr‐1(ad609) worms while feeding on either live or UV‐killed OP50 at 15°C. The three strains had similar lifespans at 15°C when grown on live OP50 (Fig. 2H). Notably, at 15°C the lifespans of N2 and npr‐1(ad609) worms were significantly increased (compared with 21°C, P < 0.0001), but that of gcy‐35;npr‐1(ad609) animals was not affected, suggesting that low temperature may recapitulate the effect of gcy‐35 and npr‐1 joint loss of function on lifespan. The lifespan of npr‐1(ad609) worms on UV‐killed OP50 at 15°C was significantly longer than both N2 and gcy‐35;npr‐1(ad609) worms (Fig. 2I, P = 0.0002 and P = 0.0003, respectively), further supporting our conclusion that gcy‐35 and npr‐1 function in lifespan regulation is modulated by temperature and bacterial viability.
RIA is important for the extended lifespan of gcy‐35;npr‐1(ad609) mutants
Previous studies suggested that the RIA interneuron integrates information about temperature from the AFD and AWC neurons, and information about O2 from the URX and BAG neurons ((Kimata et al., 2012; Luo et al., 2014) for illustration, see Fig. 2J). Therefore, we hypothesized that RIA is important for the extended lifespan of npr‐1(ad609) and gcy‐35;npr‐1(ad609) mutants. To explore this, we genetically ablated RIA by expressing the death activator egl‐1 with the promoter region of the RIA‐specific gene glr‐3 (Brockie et al., 2001). The ablation of RIA shortened the lifespan of gcy‐35;npr‐1(ad609) mutants so it was similar to that of npr‐1(ad609) worms (P = 0.6392, Fig. 2K), suggesting that RIA function is essential for the synergistic effect of gcy‐35 and npr‐1 loss of function on lifespan. Moreover, the lifespans of RIA genetically ablated N2 and npr‐1(ad609) transgenic worms were similar (P = 0.6906) and significantly shorter than control worms (P < 0.0001), suggesting that RIA is also important for the individual effect of npr‐1 loss of function on lifespan. Notably, the lack of difference between the lifespans of N2 and npr‐1(ad609) worms with ablated RIA cannot be attributed to suppression of behavioral O2 responses, because the ablation of RIA did not reduce the accumulation of npr‐1(ad609) worms on the bacterial lawn border (Fig. S2F, Supporting information).
These results show that the RIA interneurons play an important role in lengthening the lifespan of npr‐1(ad609) and gcy‐35;npr‐1(ad609) worms and so suggest that the effect of joint loss of function of npr‐1 and gcy‐35 on lifespan regulation is mediated by neuronal communication.
GCY‐35 and NPR‐1(215V) function in lifespan regulation is mediated by neurotransmitter/neuropeptide signaling
UNC‐31 regulates the release of neuropeptides from dense core vesicles (DCV) (Taylor & Dillin, 2013), and UNC‐13 and UNC‐64 are important for both neuropeptide and neurotransmitter release from DCV and small clear vesicles (Saifee et al., 1998; Sieburth et al., 2007). To explore whether neurotransmitter and/or neuropeptide signaling are important for the extended lifespan of gcy‐35;npr‐1(ad609) mutants, we made strains bearing the unc‐13(e450), unc‐31(e928), and unc‐64(e246) loss‐of‐function mutations. The unc‐13 gcy‐35;npr‐1(ad609) and gcy‐35;unc‐31;npr‐1(ad609) mutants had significantly shorter lifespans than gcy‐35;npr‐1(ad609) worms (Fig. S3A, Supporting information). However, the unc‐64 mutation did not significantly reduce the lifespan of gcy‐35;npr‐1(ad609) mutants. Therefore, although these results suggest that neuropeptide and/or neurotransmitter release is important for the lengthened lifespan of gcy‐35;npr‐1(ad609) mutants, they should be interpreted with caution. The unc‐13 and unc‐31 mutants had significantly longer lifespans than N2 worms (Fig. S3B, Supporting information). However, these mutations did not affect the lifespan of npr‐1(ad609) mutants (Fig. S3C, Supporting information), suggesting that the extended lifespan of unc‐13 and unc‐31 mutants requires the activity of NPR‐1(215V). The lifespan of unc‐64 mutants was similar to N2 controls (Fig. S3B, Supporting information). However, unc‐64;npr‐1(ad609) mutants had a significantly shorter lifespan than both N2 and npr‐1(ad609) animals (P < 0.0001, Fig. S3C, Supporting information), suggesting that the beneficial effect of npr‐1(215V) loss of function on worm lifespan is mediated through neuropeptide and/or neurotransmitter signaling.
EGL‐3 is required for the extended lifespan of gcy‐35;npr‐1(ad609) mutants
Our results suggest that joint loss of function of npr‐1 and gcy‐35 lengthens lifespan through neuropeptide signaling (Fig. S3A, Supporting information). The activity of the proprotein convertase EGL‐3 is important for transforming proneuropeptides into functional neuropeptides in C. elegans (Edwards et al., 2009). Therefore, we hypothesized that EGL‐3 is essential for the lengthened lifespan of gcy‐35;npr‐1(ad609) worms. To explore this, we created a gcy‐35;npr‐1(ad609) strain bearing the missense mutation allele egl‐3(n150). The lifespan of gcy‐35;egl‐3;npr‐1(ad609) mutants was significantly shorter than gcy‐35;npr‐1(ad609) controls (Fig. 3A), suggesting that neuropeptide signaling is important for the lengthening of gcy‐35;npr‐1(ad609) mutant lifespan.
Figure 3.
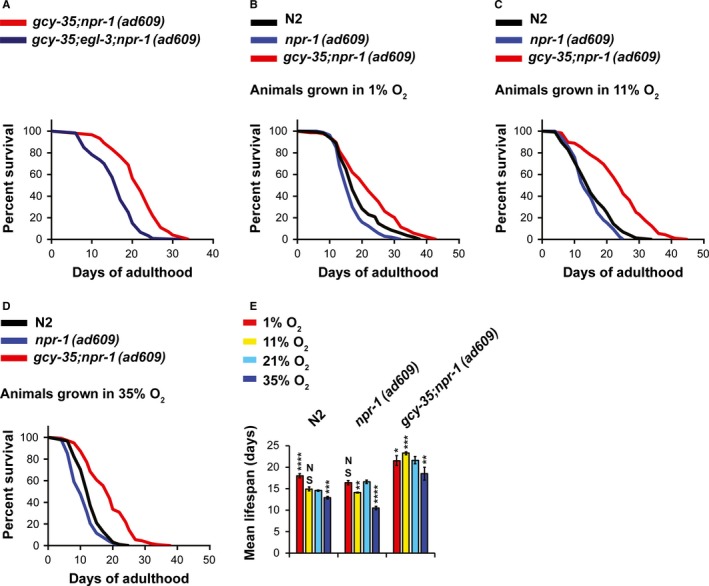
Neurotransmitter/neuropeptide signaling regulate the lifespan of gcy‐35;npr‐1(ad609) mutants. (A) Survival curves comparing the lifespans of worm strains at 21°C on live OP50 at 21% O2. (B–D) Survival curves comparing the lifespans of worm strains at different O2 levels, as indicated by the labels above the graphs. (E) Bar graph comparing mean lifespan at different O2 concentrations. The mean lifespan data for 1%, 11%, and 35% O2 were taken from C–E, respectively. Data for 21% O2 lifespan measurements were taken from Fig. 1B. Asterisks indicate significance for comparisons with N2's lifespan at each O2 level. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, NS (not significant). Error bars represent SEM.
NPR‐1(215V) regulates lifespan in an O2‐dependent manner
O2 regulates C. elegans lifespan. For example, exposure to 60% and 1% O2, respectively, shortens and lengthens the lifespan of N2 worms (Adachi et al., 1998). To explore the effect of O2 level on the lifespan of N2, npr‐1(ad609), and gcy‐35;npr‐1(ad609) animals, we grew the worms at three O2 concentrations: 1%, 11%, and 35%. We chose these concentrations because 1% O2 induces hypoxic responses in C. elegans and was found to increase N2 lifespan (Adachi et al., 1998); 11% O2 inhibits high foraging speed and aggregation behavior of npr‐1(ad609) worms on food and is in the preferred O2 range for C. elegans (Gray et al., 2004); and 35% O2 may trigger oxidative stress but is still within the physiological range that nematodes experienced in the carboniferous period (~300 million years ago) (Halliwell & Gutteridge, 2006). In general, gcy‐35;npr‐1(ad609) mutants had a significantly longer lifespan at all tested O2 conditions than both N2 and npr‐1(ad609) animals (Fig. 3B–E), suggesting that the synergistic effect of gcy‐35 and npr‐1 loss of function on lifespan is O2 independent. By contrast, the effect of npr‐1(215V) on lifespan was O2 dependent (Fig. 3B–E). At 1% O2, N2 worms lived significantly longer than npr‐1(ad609) worms (Fig. 3B); at 11% O2, the lifespan of N2 and npr‐1(ad609) animals was similar (Fig. 3C); the lifespan of N2 worms at 35% was significantly longer than npr‐1(ad609) worms (Fig. 3D). In fact, only at 21% O2 was the lifespan of npr‐1(ad609) mutants longer than N2 worms. Therefore, we conclude that npr‐1(215V) effects lifespan in an O2‐dependent way.
The extended lifespan of npr‐1(ad609) and gcy‐35;npr‐1(ad609) mutants cannot be explained by decreased metabolism
Some longed‐lived mutants, such as clk‐1 and daf‐2 (mutants with reduced mitochondrial activity and insulin signaling, respectively), have reduced metabolic rates compared with N2 worms (Van Voorhies & Ward, 1999). Therefore, we asked whether the extended lifespan of gcy‐35;npr‐1(ad609) mutants is associated with decreased metabolic activity. We measured the locomotory activity of worms on both solid medium and liquid medium (speed and thrashing assays respectively), feeding (pumping assays), development, egg laying rate, O2 consumption, and ATP levels (Fig. 4). These parameters (apart from development) were measured on the first and fifth days of adulthood, which precede the rapid decline of N2 worm viability. The locomotory activity and pharyngeal pumping of N2, npr‐1(ad609), and gcy‐35;npr‐1(ad609) worms were similar on days 1 and 5 (Fig. 4A–C). By contrast, N2 worms developed slightly faster compared with the other strains (Fig. 4D) and had a higher egg laying rate on day 1, but not on day 5 (Fig. 4E). Notably, npr‐1(ad609) worms developed slightly faster than gcy‐35;npr‐1(ad609) worms, but laid fewer eggs on day 1. Therefore, although our results suggest that NPR‐1(215V) and GCY‐35 signaling is important for both development and egg‐laying regulation, the small differences between the strains probably cannot explain the lengthened lifespan of gcy‐35;npr‐1(ad609) mutants. To directly compare the metabolic activity of N2, npr‐1(ad609), and gcy‐35;npr‐1(ad609) animals, we measured O2 consumption and ATP levels in the three strains (Fig. 4F,G). Consumption of O2 and ATP levels of the N2, npr‐1(ad609), and gcy‐35;npr‐1(ad609) were similar on both days 1 and 5, indicating that the metabolic activity of these strains is similar.
Figure 4.
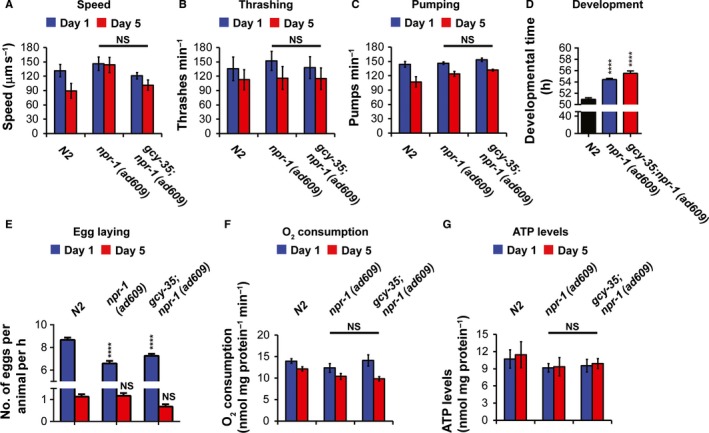
NPR‐1 and GCY‐35 effects on behavior, development, O2 consumption, and ATP levels. All experiments (apart from D) were performed on the first and fifth days of adulthood. (A, B) The speed and thrashing measurement were performed in the absence of bacteria. (C) Pharyngeal pumping behavior. (D) The development of worms was monitored from the L1 to the adult stage. Worms that had at least one egg in their bodies were counted as adults. Asterisks indicate significance for comparisons with N2 worms (one‐way ANOVA with Bonferroni post‐test). (E) Egg‐laying measurements. Asterisks indicate significance for comparisons with N2 worms at days 1 and 5 (two‐way ANOVA with Bonferroni post‐test). (F, G) O2 consumption and ATP measurements. One‐way ANOVA with Tukey's multiple comparisons post‐test. All assays were performed over the course of 3 days. The number of assays and the number of worms tested in each assay are indicated in Table S11 (Supporting information). Error bars represents SEM. ****P < 0.0001, NS (not significant).
The canonical daf‐2/daf‐16 pathway modulates npr‐1/gcy‐35 function in lifespan regulation
Our results suggest that reduction in metabolic rates cannot explain the extended lifespan of gcy‐35;npr‐1(ad609) mutants. Therefore, we set out to find the signaling pathway that lengthens the lifespan of gcy‐35;npr‐1(ad609) mutants. Insulin and insulin‐like growth factor signaling (IIS) plays a critical role in lifespan regulation (Murphy & Hu, 2013). In C. elegans, the activity of the insulin receptor DAF‐2 stops DAF‐16 (the C. elegans ortholog of the mammalian FOXO transcription factor) entering the nucleus. Therefore, when daf‐2 is inhibited, DAF‐16 translocates to the nucleus and activates genes involved in stress resistance and longevity. In addition, DAF‐16 translocation to the nucleus is triggered by various environmental stimuli, including oxidative stress (Murphy & Hu, 2013). To explore whether IIS is important for npr‐1/gcy‐35 signaling, we generated N2, npr‐1(ad609), and gcy‐35;npr‐1(ad609) strains bearing the daf‐16(mu86) deletion allele and the daf‐2(e1370) loss‐of‐function allele and performed lifespan experiments. As expected, daf‐16 significantly shortened the lifespan of the three strains (Fig. 5A). However, daf‐16;npr‐1(ad609) mutants lived significantly longer than daf‐16 mutants (P < 0.0001), indicating that the beneficial effect of NPR‐1(215V) inhibition is not mediated by daf‐16 signaling. By contrast, the lifespan of daf‐16;npr‐1(ad609) mutants was similar to that of daf‐16 gcy‐35;npr‐1(ad609) mutants, indicating that the extended lifespan of gcy‐35;npr‐1(ad609) worms is dependent on DAF‐16 activity. The lifespan of daf‐2(e1370) mutants was not further extended by either npr‐1 loss of function or the joint loss of function of npr‐1/gcy‐35 (Fig. 5B), suggesting that lifespan lengthening by inhibition of daf‐2 and npr‐1/gcy‐35 takes place through overlapping mechanisms.
Figure 5.
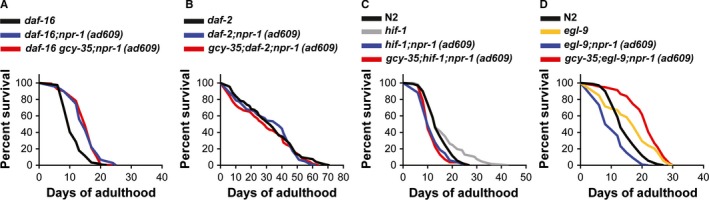
IIS and HIF‐1 signaling interact genetically with npr‐1 and gcy‐35. (A–D) Survival curves comparing the lifespans of worm strains at 21°C on live OP50. One‐way ANOVA with Tukey's multiple comparisons post‐test. NS (not significant). Error bars represents SEM.
HIF‐1 is essential for the extended lifespan of gcy‐35;npr‐1(ad609) mutants
Our results suggest that npr‐1 and gcy‐35 interact with the canonical IIS pathway. As the IIS pathway is important for life span extension in hypoxia (Leiser et al., 2013), we next explored whether the hypoxia‐inducible factor 1 (HIF‐1) is needed for the lifespan lengthening of gcy‐35;npr‐1(ad609) mutants. The transcription factor hif‐1 plays a critical role in hypoxia signaling (Halliwell & Gutteridge, 2006). N2 worms bearing the hif‐1(ia4) deletion allele lived significantly longer than N2 controls (Fig. 5C). By contrast, the deletion of hif‐1 shortened the lifespans of npr‐1(ad609) and gcy‐35;npr‐1(ad609) mutants to less than that of N2 (P < 0.0001), suggesting that the beneficial effect of HIF‐1 on worm lifespan is mediated by NPR‐1(215V) activity. Moreover, our data suggest that the synergistic effect of npr‐1/gcy‐35 loss of function on lifespan is mediated by HIF‐1.
HIF‐1 levels are regulated by the prolyl hydroxylase EGL‐9, which targets HIF‐1 for degradation. Therefore, inhibition of EGL‐9 results in accumulation of HIF‐1 even at 21% O2 (Zhang et al., 2009). To examine the effect of HIF‐1 accumulation on worm lifespan, we introduced the egl‐9(sa307) loss‐of‐function allele to N2, npr‐1(ad609), and gcy‐35;npr‐1(ad609) worms and measured lifespan (Fig. 5D). egl‐9 loss of function significantly increased the lifespan of N2 worms, but significantly shortened the lifespan of npr‐1(ad609) animals (Fig. 5D, P < 0.0001), indicating that the function of egl‐9 in lifespan is regulated by npr‐1. Intriguingly, egl‐9(sa307) did not shorten the lifespan of gcy‐35;npr‐1(ad609) mutants, suggesting that the synergistic effect of npr‐1/gcy‐35 on lifespan is not mediated by egl‐9 function. In conclusion, our studies show that HIF‐1 function is required for the extended lifespan of gcy‐35;npr‐1(ad609) mutants (Fig. 5C). However, constitutive stabilization of HIF‐1 at 21% O2 is not required for the synergistic effect of npr‐1/gcy‐35 loss of function on lifespan. npr‐1 activity is essential for the beneficial effects of both hif‐1 deletion and stabilization on lifespan.
Are gcy‐35;npr‐1(ad609) mutants more resistant to stress?
The extended lifespan of gcy‐35;npr‐1(ad609) mutants is modulated by the activity of DAF‐16 and HIF‐1. As these transcription factors are important for protection against unfolded protein toxicity, heat, oxidative damage, and pathogenic bacteria (Back et al., 2012), we asked whether these animals live longer because they are more stress resistant. To address this question, we exposed N2, npr‐1(ad609), and gcy‐35;npr‐1(ad609) worms on their first and fifth days of adulthood to various stresses and measured survival. Exposure to Pseudomonas aeruginosa PA14 bacteria induced rapid death in all three strains. However, the survival of gcy‐35;npr‐1(ad609) mutants was higher in both day 1 and day 5 of adulthood compared with N2 and npr‐1(ad609) worms (Fig. 6A), indicating that joint loss of function of gcy‐35 and npr‐1 provides protection against PA14 toxicity. Similarly, gcy‐35;npr‐1(ad609) mutants were more resistant to UV stress than N2 and npr‐1(ad609) worms (Fig. 6B). Interestingly, a previous study showed a striking similarity between the transcription of genes involved in DNA damage and innate immunity (Ermolaeva et al., 2013), suggesting that joint loss of function of gcy‐35 and npr‐1 induces an innate immunity response that extends the lifespan of gcy‐35;npr‐1(ad609) mutants. The increased resistance of gcy‐35;npr‐1(ad609) mutants to stress was specific to PA14 and UV. gcy‐35;npr‐1(ad609) mutants were significantly more sensitive to heat stress than N2 and npr‐1(ad609) worms on the first day of adulthood (Fig. S4A, Supporting information). However, on day 5 of adulthood the three strains showed similar survival rates, which were significantly higher than day 1, suggesting that older worms are generally more resistant to heat stress than young worms. Tunicamycin prevents the first step of N‐linked glycosylation of proteins in the ER so inducing accumulation of unfolded proteins and activating the unfolded protein response of the endoplasmic reticulum (UPRER) (Taylor & Dillin, 2013). N2, npr‐1(ad609), and gcy‐35;npr‐1(ad609) worms showed similar survival in response to tunicamycin (both on days 1 and 5 of adulthood, Fig. S4B, Supporting information). Finally, to explore whether gcy‐35;npr‐1(ad609) mutants are more resistant to mitochondrial oxidative stress, we exposed the three strains to 200 mM paraquat, a superoxide generator (Halliwell & Gutteridge, 2006), and measured their survival over 24 h. The survival of N2 and npr‐1(ad609) worms (on days 1 and 5) was similar to gcy‐35;npr‐1(ad609) mutants (Fig. S4C, Supporting information). Therefore, our results show that gcy‐35;npr‐1(ad609) mutants are not generally resistant to stress, but rather show specific resistance to both PA14 and UV toxicity.
Figure 6.
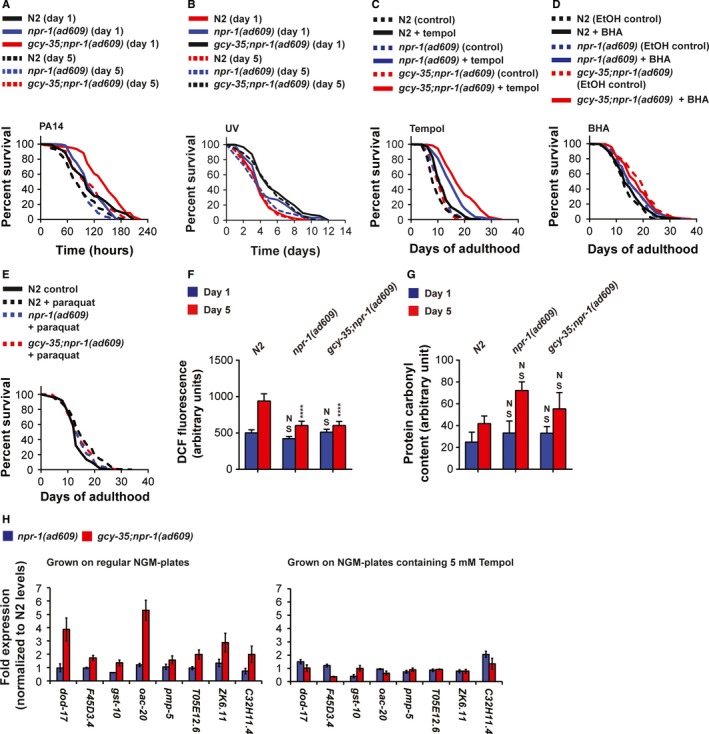
Survival assays of N2, npr‐1(ad609), and gcy‐35;npr‐1(ad609) animals. (A) PA14 killing assays (B) Survival after UV exposure. The assays were performed on days 1 and 5 of adulthood. (C, D) Lifespan experiments in the presence of the antioxidants tempol (5 mM) and BHA (25 μm). (E) Lifespan experiments in the presence of 0.1 mm paraquat. (F) Bar graph comparing the levels of ROS in N2, npr‐1(ad609), and gcy‐35;npr‐1(ad609) worms at days 1 and 5. Asterisks indicate significance for comparisons with N2 worms at days 1 and 5 (two‐way ANOVA with Bonferroni post‐test). (G) Protein oxidation measurements by Oxyblot. Quantification of Western blot analysis from at least four independent biological repeats, for each condition. Two‐way ANOVA with Bonferroni post‐test. (H) Gene expression measurements in N2, npr‐1(ad609), and gcy‐35;npr‐1(ad609) worms that were either grown on regular NGM plates (left panel) or on NGM plates containing 5 mm Tempol (right panel). Gene expression was measured by qPCR. Each measurement represents, at least, three biological repeats.
ROS is required for the extended lifespan of gcy‐35;npr‐1(ad609) mutants
As ROS signaling appears to be important for tolerance to UV and pathogenic bacteria (Hideg et al., 2013; Schieber & Chandel, 2014), we explored whether ROS is needed for the extended lifespan of gcy‐35;npr‐1(ad609) mutants, by performing lifespan experiments in the presence of antioxidants. We used the superoxide scavenger tempol (5 mm), and butylated hydroxyanisole (BHA). Tempol treatment significantly reduced the lifespan of N2, npr‐1(ad609), and gcy‐35;npr‐1(ad609) worms (Fig. 6C), suggesting that either excessive scavenging of superoxide is generally damaging to C. elegans or long exposure to 5 mM tempol induces a toxic effect that is not associated with ROS. That being said, tempol treatment affected the three strains differently. The lifespans of npr‐1(ad609) and gcy‐35;npr‐1(ad609) mutants were similar, suggesting that ROS is required for the positive synergistic effect of npr‐1 and gcy‐35 loss of function on lifespan. However, npr‐1(ad609) mutants lived longer than N2 worms in the presence of tempol (Fig. 6C), suggesting that the effect of npr‐1 on lifespan is not mediated by ROS. BHA significantly increased the lifespan of N2 worms (Fig. 6D, P = 0.0394) compared with N2 worms living on NGM plates with ethanol (the concentration of ethanol was similar in all plates). However, npr‐1(ad609) worms growing on BHA lived longer than N2 worms with BHA (P = 0.0272). These results are in agreement with the tempol experiments that suggested that the effect of npr‐1 on lifespan is not mediated by ROS. The lifespan of gcy‐35;npr‐1(ad609) animals growing with BHA was longer than the lifespan of npr‐1(ad609) worms with BHA. However, the difference was much smaller and barely reached significance (P = 0.0438). Therefore, although the BHA results are not as conclusive as the tempol results, they do suggest that the synergistic effect of gcy‐35/npr‐1 loss of function on lifespan in regulated by ROS.
Paraquat extends the lifespan of N2 worms but reduces the lifespan of both npr‐1(ad609) and gcy‐35;npr‐1(ad609) mutants
Previous studies showed that low levels of the superoxide generator paraquat lengthen the lifespan of N2 worms (Yang & Hekimi, 2010). Intriguingly, either a small increase or decrease in paraquat concentration can diminish the beneficial effect, suggesting that ROS act within a narrow range of concentrations to lengthen lifespan. Our studies suggest that the extended lifespan of gcy‐35;npr‐1(ad609) mutants is mediated by ROS. Therefore, we hypothesized that low levels of paraquat will not further extend the lifespan of gcy‐35;npr‐1(ad609) mutants and may even induce a toxic effect. We measured the lifespan of N2, npr‐1(ad609), and gcy‐35;npr‐1(ad609) worms grown on NGM plates supplemented with 0.1 mM paraquat, a concentration shown to maximize the lifespan extension of N2 worms (Yang & Hekimi, 2010). As expected, paraquat treatment lengthened the lifespan of N2 worms significantly (Fig. 6E, P < 0.0001), supporting the results from previous studies. By contrast, it shortened the lifespans of npr‐1(ad609) and gcy‐35;npr‐1(ad609) mutants to N2 control level, suggesting that the level of ROS in these worms exceeded the beneficial level and elicited a toxic effect. Together, these results further support the hypothesis that ROS act within a narrow range of concentrations to lengthen worm lifespan and that excessive ROS is damaging.
Accumulation of ROS and protein oxidation are not correlated with the extended lifespan of gcy‐35;npr‐1(ad609) mutants
To explore the metabolism of ROS and its consequences, we measured the levels of ROS and protein carbonyl oxidation in N2, npr‐1(ad609), and gcy‐35;npr‐1(ad609) worms. To measure the overall level of ROS, we used the 2′,7′dichlorofluorescin diacetate (DCFDA) fluorescent dye. We measured ROS levels in N2, and npr‐1(ad609), and gcy‐35;npr‐1(ad609) worms on the first and fifth days adulthood. The levels of ROS in all three trains were similar on day 1 (Fig. 6F), and also similar in npr‐1(ad609) and gcy‐35;npr‐1(ad609) mutants on day 5. However, the levels of ROS on day 5 were significantly higher in N2 worms compared with both npr‐1(ad609) and gcy‐35;npr‐1(ad609) mutants (P < 0.001). Nevertheless, the conclusion from these studies is that ROS levels do not correlate with extended lifespan of gcy‐35;npr‐1(ad609) mutants. As degradation of ROS could be different in N2, npr‐1(ad609), and gcy‐35;npr‐1(ad609) worms, we measured the oxidation of proteins on the first and fifth days of adulthood. Protein oxidation, as monitored by carbonyl accumulation, was similar in all strains on days 1 and 5 (Figs 6G and S4D, Supporting information), suggesting that ROS do not accumulate to levels that cause this commonly used gauge of protein oxidation.
In conclusion, our experiments failed to detect any correlation between the extended lifespan of gcy‐35;npr‐1(ad609) mutants and ROS levels or protein oxidation. However, our antioxidant, paraquat, DAF‐16, and HIF‐1 experiments strongly suggest that ROS is important for the extended lifespan of gcy‐35;npr‐1(ad609) mutants. A potential explanation for this discrepancy is that ROS are needed at small quantities in specific cells (e.g., the O2‐sensing neurons). However, future studies are needed to explore this hypothesis.
The transcription of innate immunity genes in gcy‐35;npr‐1(ad609) mutants is ROS dependent
ROS controls the function of DAF‐16 and HIF‐1 and the activity of these transcription factors is important for both gcy‐35;npr‐1(ad609) life extension (Fig. 5) and innate immunity (Singh & Aballay, 2009; Hwang et al., 2014). Moreover, a previous study showed that innate immunity genes are differentially expressed in gcy‐35;npr‐1(ad609) mutants and npr‐1(ad609) mutants (Styer et al., 2008). Therefore, we asked whether ROS regulates the expression of innate immunity genes in gcy‐35;npr‐1(ad609) mutants. We extracted RNA from N2, npr‐1(ad609), and gcy‐35;npr‐1(ad609) worms that were grown throughout their life cycle on either regular NGM plates (as controls) or NGM plates containing 5 mm tempol (experimental group) and measured the expression of eight genes known to be involved in the innate immunity (Engelmann et al., 2011; Gaglia et al., 2012; Twumasi‐Boateng & Shapira, 2012), using reverse‐transcription quantitative PCR (RT–qPCR). As shown in Fig. 6H (left panel), all eight genes (in the control group) were upregulated in gcy‐35;npr‐1(ad609) mutants compared with N2 and npr‐1(ad609) worms. However, in the presence of tempol the expression level of these genes was substantially downregulated in gcy‐35;npr‐1(ad609) mutants (Fig. 6H, right panel), indicating that the induction of these genes in gcy‐35;npr‐1(ad609) mutants is regulated by ROS.
Discussion
ROS signaling is essential for the extended lifespan of gcy‐35;npr‐1(ad609) animals
Harman's theory of aging (Harman, 1956) suggested that the creation of ROS via aerobic respiration underlies the development and progression of aging. However, studies from the last decade have challenged this theory. For example, C. elegans lacking all superoxide dismutases (sod‐1‐sod‐5) live longer than N2 animals, and chemical agents that enhance mitochondrial ROS increase lifespan (Van Raamsdonk & Hekimi, 2012). Our results suggest that tight regulation of ROS level is essential for the extended lifespan of gcy‐35;npr‐1(ad609) mutants (Fig. 6). Both ROS scavenging by antioxidants and increased ROS by sublethal level of paraquat shorten the lifespan of gcy‐35;npr‐1|(ad609) worms. This hypothesis is further supported by the requirement of DAF‐16 and HIF‐1 for the extended lifespan of gcy‐35;npr‐1(ad609) animals (Fig. 5), as these transcription factors are regulated by ROS (Hwang & Lee, 2011; Zou et al., 2013). Notably, the interpretation of our results should be made with caution, because we failed to detect any significant difference in either ROS accumulation or oxidative damage in gcy‐35;npr‐1(ad609) worms compared with both N2 and npr‐1(ad609) animals. That being said, we suspect that ROS act in specific neurons to regulate lifespan. Therefore, more specific and sensitive methods are needed to detect these localized changes.
Despite the above caveats, we suggest the following working model. The function of NPR‐1(215V) and GCY‐35/GCY‐36 in the O2‐sensing neurons AQR, PQR, and URX inhibits lifespan extension. By contrast, GCY‐33 signaling through TAX‐2/TAX‐4 triggers the secretion of neuropeptide/neurotransmitter that increases the level of ROS in either the O2‐sensing neurons themselves, downstream neurons, or even non‐neuronal tissues. The increase in ROS levels activates the HIF‐1 and DAF‐16 transcription factors which induce innate immunity. This induction of innate immunity increases lifespan. Although much of the details in this model are still not known, for example, the identity of its interacting partner of GCY‐33 in AQR, PQR, and URX, and where DAF‐16 and HIF‐1 function is needed for the life extension, this model provides a framework for future studies to identify the ‘anti‐aging’ neurotransmitter/neuropeptide and to elucidate how innate immunity genes prolong lifespan. In this respect, it is interesting to note that a previous study showed that the expression of the innate immunity genes dod‐17, C32H11.4, and ZK6.11 is substantially downregulated in daf‐10(m79) worms (Gaglia et al., 2012). Moreover, the high sensitivity of daf‐10(m79) animals to PA14 is rescued by the gcy‐35(ok769) mutation. DAF‐10 is important for the activity of ciliated neurons, such as AQR, PQR, and BAG. Therefore, in the future it will be intriguing to explore whether daf‐10 function in AQR, PQR, and BAG is important for the extended lifespan of gcy‐35;npr‐1(ad609) mutants and whether this function is ROS dependent.
Experimental procedures
We used standard molecular biology and genetic protocols. A detailed description of strains and oligonucleotides used is found in Table S10 (Supporting information) and Supplemental Experimental Procedures.
Lifespan analysis
All lifespan assays were conducted at 21°C and started with synchronized young adults, unless otherwise mentioned. Statistical analysis on lifespan survival curves was performed using the log‐rank (Mantel–Cox) test with Prism 6 software, and the results are displayed in Tables S1–S9 (Supporting information). Further details are found in Supplemental Experimental Procedures.
Funding info
Research leading to these results received funding from the Marie Curie Career Integration Grants (CIG), agreement no. PCIG11‐GA‐2012‐322003. (L.L and R.A).
Conflict of interest
The authors declare that they have no conflicts of interest.
Author contributions
R.A., and L.L., and E.G. designed research; R.A., L.L., M.S., and A.K.C. performed research; R.A., L.L., and E.G. analyzed data; and R.A., L.L., and E.G. wrote the paper.
Supporting information
Fig. S1 The effect of GCY‐36 and TAX‐4 on N2 and npr‐1(ad609) worms’ lifespan (related to Figure 1).
Fig. S2 GCY‐33 is important for npr‐1(ad609) animals’ extended lifespan (related to Figure 2).
Fig. S3 The function of NPR‐1 in lifespan regulation is modulated by neuropeptide/neurotransmitter signaling (related to Figure 3).
Fig. S4 Joint loss‐of‐function of npr‐1 and gcy‐35 does not increase tolerance to heat, ER UPR, or mitochondrial UPR stress (related to Figure 6).
Table S1 Numerical values for data plotted in Fig. 1.
Table S2 Numerical values for data plotted in Fig. S1.
Table S3 Numerical values for data plotted in Fig. 2.
Table S4 Numerical values for data plotted in Fig. S2.
Table S5 Numerical values for data plotted in Fig. S3.
Table S6 Numerical values for data plotted in Fig. 3.
Table S7 Numerical values for data plotted in Fig. 5.
Table S8 Numerical values for data plotted in Fig. 6.
Table S9 Numerical values for data plotted in Fig. S4.
Table 10 List of primers used for npr‐1, gcy‐35 and egl‐1 sequencing, primers for cell specific RNAi by PCR fusion, and quantitative RT‐PCR experiments.
Table S11 Number of animals and experimental data.
Appendix S1 Supplemental Experimental Procedures.
Acknowledgments
We thank C. Bargmann and E. Cohen for strains. We thank E. Cohen, D. Fass, and G. Kay for critical reading of the manuscript, and members of Gross laboratory for comments and advice. We obtained some strains from the Caenorhabditis elegans Genetics Stock Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
References
- Abergel Z, Chatterjee AK, Zuckerman B, Gross E (2016) Regulation of neuronal oxygen responses in C. elegans is mediated through interactions between globin 5 and the H‐NOX domains of soluble guanylate cyclases. J. Neurosci. 36, 963–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi H, Fujiwara Y, Ishii N (1998) Effects of oxygen on protein carbonyl and aging in Caenorhabditis elegans mutants with long (age‐1) and short (mev‐1) life spans. J. Gerontol. A Biol. Sci. Med. Sci. 53, B240–B244. [DOI] [PubMed] [Google Scholar]
- Apfeld J, Kenyon C (1999) Regulation of lifespan by sensory perception in Caenorhabditis elegans . Nature 402, 804–809. [DOI] [PubMed] [Google Scholar]
- Back P, Braeckman BP, Matthijssens F (2012) ROS in aging Caenorhabditis elegans: damage or signaling? Oxid. Med. Cell. Longev. 2012, 608478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockie PJ, Madsen DM, Zheng Y, Mellem J, Maricq AV (2001) Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC‐42. J. Neurosci. 21, 1510–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung BH, Arellano‐Carbajal F, Rybicki I, de Bono M (2004) Soluble guanylate cyclases act in neurons exposed to the body fluid to promote C. elegans aggregation behavior. Curr. Biol. 14, 1105–1111. [DOI] [PubMed] [Google Scholar]
- Cheung BH, Cohen M, Rogers C, Albayram O, de Bono M (2005) Experience‐dependent modulation of C. elegans behavior by ambient oxygen. Curr. Biol. 15, 905–917. [DOI] [PubMed] [Google Scholar]
- Coates JC, de Bono M (2002) Antagonistic pathways in neurons exposed to body fluid regulate social feeding in Caenorhabditis elegans . Nature 419, 925–929. [DOI] [PubMed] [Google Scholar]
- Edwards SL, Charlie NK, Richmond JE, Hegermann J, Eimer S, Miller KG (2009) Impaired dense core vesicle maturation in Caenorhabditis elegans mutants lacking Rab2. J. Cell Biol. 186, 881–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann I, Griffon A, Tichit L, Montanana‐Sanchis F, Wang G, Reinke V, Waterston RH, Hillier LW, Ewbank JJ (2011) A comprehensive analysis of gene expression changes provoked by bacterial and fungal infection in C. elegans . PLoS ONE 6, e19055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva MA, Segref A, Dakhovnik A, Ou HL, Schneider JI, Utermohlen O, Hoppe T, Schumacher B (2013) DNA damage in germ cells induces an innate immune response that triggers systemic stress resistance. Nature 501, 416–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglia MM, Jeong DE, Ryu EA, Lee D, Kenyon C, Lee SJ (2012) Genes that act downstream of sensory neurons to influence longevity, dauer formation, and pathogen responses in Caenorhabditis elegans . PLoS Genet. 8, e1003133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C (2002) Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat‐shock factor and bacterial proliferation. Genetics 161, 1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JM, Karow DS, Lu H, Chang AJ, Chang JS, Ellis RE, Marletta MA, Bargmann CI (2004) Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature 430, 317–322. [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC (2006) Free Radicals in Biology and Medicine. Oxford: Oxford University Press. [Google Scholar]
- Harman D (1956) Aging: a theory based on free radical and radiation chemistry. J. Gerontol. 11, 298–300. [DOI] [PubMed] [Google Scholar]
- Hideg E, Jansen MA, Strid A (2013) UV‐B exposure, ROS, and stress: inseparable companions or loosely linked associates? Trends Plant Sci. 18, 107–115. [DOI] [PubMed] [Google Scholar]
- Hwang AB, Lee SJ (2011) Regulation of life span by mitochondrial respiration: the HIF‐1 and ROS connection. Aging 3, 304–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang AB, Ryu EA, Artan M, Chang HW, Kabir MH, Nam HJ, Lee D, Yang JS, Kim S, Mair WB, Lee C, Lee SS, Lee SJ (2014) Feedback regulation via AMPK and HIF‐1 mediates ROS‐dependent longevity in Caenorhabditis elegans . Proc. Natl Acad. Sci. USA 111, E4458–E4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata T, Sasakura H, Ohnishi N, Nishio N, Mori I (2012) Thermotaxis of C. elegans as a model for temperature perception, neural information processing and neural plasticity. Worm 1, 31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiser SF, Fletcher M, Begun A, Kaeberlein M (2013) Life‐span extension from hypoxia in Caenorhabditis elegans requires both HIF‐1 and DAF‐16 and is antagonized by SKN‐1. J. Gerontol. A Biol. Sci. Med. Sci. 68, 1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Cai D (2013) Counterbalance between BAG and URX neurons via guanylate cyclases controls lifespan homeostasis in C. elegans . EMBO J. 32, 1529–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Cook N, Venkatachalam V, Martinez‐Velazquez LA, Zhang X, Calvo AC, Hawk J, MacInnis BL, Frank M, Ng JH, Klein M, Gershow M, Hammarlund M, Goodman MB, Colon‐Ramos DA, Zhang Y, Samuel AD (2014) Bidirectional thermotaxis in Caenorhabditis elegans is mediated by distinct sensorimotor strategies driven by the AFD thermosensory neurons. Proc. Natl Acad. Sci. USA 111, 2776–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Pokala N, Feinberg EH, Chalasani SH, Butcher RA, Clardy J, Bargmann CI (2009) A hub‐and‐spoke circuit drives pheromone attraction and social behaviour in C. elegans . Nature 458, 1171–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Rockman MV, Zimmer M, Jang H, Macosko EZ, Kruglyak L, Bargmann CI (2009) Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron 61, 692–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, Hu PJ (2013) Insulin/insulin‐like growth factor signaling in C. elegans . (December 26, 2013), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.164.1, http://www.wormbook.org.. [DOI] [PMC free article] [PubMed]
- Portal‐Celhay C, Bradley ER, Blaser MJ (2012) Control of intestinal bacterial proliferation in regulation of lifespan in Caenorhabditis elegans . BMC Microbiol. 12, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Zhai Z, Powell‐Coffman JA (2006) The Caenorhabditis elegans AHR‐1 transcription complex controls expression of soluble guanylate cyclase genes in the URX neurons and regulates aggregation behavior. Dev. Biol. 298, 606–615. [DOI] [PubMed] [Google Scholar]
- Saifee O, Wei LP, Nonet ML (1998) The Caenorhabditis elegans unc‐64 locus encodes a syntaxin that interacts genetically with synaptobrevin. Mol. Biol. Cell 9, 1235–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieber M, Chandel NS (2014) ROS function in redox signaling and oxidative stress. Curr. Biol. 24, R453–R462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth D, Madison JM, Kaplan JM (2007) PKC‐1 regulates secretion of neuropeptides. Nat. Neurosci. 10, 49–57. [DOI] [PubMed] [Google Scholar]
- Singh V, Aballay A (2009) Regulation of DAF‐16‐mediated Innate Immunity in Caenorhabditis elegans . J. Biol. Chem. 284, 35580–35587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styer KL, Singh V, Macosko E, Steele SE, Bargmann CI, Aballay A (2008) Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR‐1/GPCR. Science 322, 460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Dillin A (2013) XBP‐1 is a cell‐nonautonomous regulator of stress resistance and longevity. Cell 153, 1435–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twumasi‐Boateng K, Shapira M (2012) Dissociation of immune responses from pathogen colonization supports pattern recognition in C. elegans . PLoS ONE 7, e35400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Hekimi S (2012) Superoxide dismutase is dispensable for normal animal lifespan. Proc. Natl Acad. Sci. USA 109, 5785–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Voorhies WA, Ward S (1999) Genetic and environmental conditions that increase longevity in Caenorhabditis elegans decrease metabolic rate. Proc. Natl Acad. Sci. USA 96, 11399–11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Hekimi S (2010) A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans . PLoS Biol. 8, e1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shao Z, Zhai Z, Shen C, Powell‐Coffman JA (2009) The HIF‐1 hypoxia‐inducible factor modulates lifespan in C. elegans . PLoS ONE 4, e6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer M, Gray JM, Pokala N, Chang AJ, Karow DS, Marletta MA, Hudson ML, Morton DB, Chronis N, Bargmann CI (2009) Neurons detect increases and decreases in oxygen levels using distinct guanylate cyclases. Neuron 61, 865–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou CG, Tu Q, Niu J, Ji XL, Zhang KQ (2013) The DAF‐16/FOXO transcription factor functions as a regulator of epidermal innate immunity. PLoS Pathog. 9, e1003660. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 The effect of GCY‐36 and TAX‐4 on N2 and npr‐1(ad609) worms’ lifespan (related to Figure 1).
Fig. S2 GCY‐33 is important for npr‐1(ad609) animals’ extended lifespan (related to Figure 2).
Fig. S3 The function of NPR‐1 in lifespan regulation is modulated by neuropeptide/neurotransmitter signaling (related to Figure 3).
Fig. S4 Joint loss‐of‐function of npr‐1 and gcy‐35 does not increase tolerance to heat, ER UPR, or mitochondrial UPR stress (related to Figure 6).
Table S1 Numerical values for data plotted in Fig. 1.
Table S2 Numerical values for data plotted in Fig. S1.
Table S3 Numerical values for data plotted in Fig. 2.
Table S4 Numerical values for data plotted in Fig. S2.
Table S5 Numerical values for data plotted in Fig. S3.
Table S6 Numerical values for data plotted in Fig. 3.
Table S7 Numerical values for data plotted in Fig. 5.
Table S8 Numerical values for data plotted in Fig. 6.
Table S9 Numerical values for data plotted in Fig. S4.
Table 10 List of primers used for npr‐1, gcy‐35 and egl‐1 sequencing, primers for cell specific RNAi by PCR fusion, and quantitative RT‐PCR experiments.
Table S11 Number of animals and experimental data.
Appendix S1 Supplemental Experimental Procedures.


