Abstract
Cell cultures are widely used model systems. Some immortalized cell lines can be grown in either two-dimensional (2D) adherent monolayers or in three-dimensional (3D) multicellular aggregates, or spheroids. Here, the quantitative proteome and phosphoproteome of colon carcinoma HT29 cells cultures in 2D monolayers and 3D spheroids were compared with a stable isotope labeling of amino acids (SILAC) labeling strategy. Two biological replicates from each sample were examined and notable differences in both the proteome and the phosphoproteome were determined by nano-liquid chromatography tandem mass spectrometry (nLC-MS/MS) to assess how growth configuration affects molecular expression. A total of 5867 protein groups, including 2523 phosphoprotein groups and 8733 phosphopeptides were identified in the samples. The Gene Ontology analysis revealed enriched GO terms in the 3D samples for RNA binding, nucleic acid binding, enzyme binding, cytoskeletal protein binding, and histone binding for their molecular functions (MF) and in the process of cell cycle, cytoskeleton organization, and DNA metabolic process for the biological process (BP). The KEGG pathway analysis indicated that 3D cultures are enriched for oxidative phosphorylation pathways, metabolic pathways, peroxisome pathways, and biosynthesis of amino acids. In contrast, analysis of the phosphoproteomes indicated that 3D cultures have decreased phosphorylation correlating with slower growth rates and lower cell-to extracellular matrix interactions. In sum, these results provide quantitative assessments of the effects on the proteome and phosphoproteome of culturing cells in 2D versus 3D cell culture configurations.
Keywords: Proteomic, phosphoproteomic, 2D culture, 3D culture, colon cancer
Graphical Abstract
INTRODUCTION
Immortalized cell culture models are valuable resources for in vitro research. Given the ethical limitations of working with human tissues, cell lines provide an essential in vitro alternative by which researchers may investigate mechanistic events in human and other types of cells. (1) Some of the positive attributes of cell culture include the ability to control culture conditions, ease of propagation, and relatively low cost. (2) While cell culture has enormous value, there are also some disadvantages, namely the high passage number of many commonly available cell lines, the effects of sustained growth in nonphysiological conditions, and the lack of matched normal cell lines for the large cohort of available cancerous cell lines. (1)
For adherent cells, the most common cell culture method for propagating cells is in two-dimensions (2D). In this approach, cells are seeded on a solid substrate in a volume of media. After a few hours, cells will attach to the surface and begin to propagate. For many adherent cell lines, the cellular population will form an adherent 2D monolayer covering the available surface area. They will be limited by contact inhibition and generally not grow in a vertical fashion. (3)
Many of the adherent cell lines can also be grown as 3D multicellular structures or spheroids. (4) To facilitate this configuration, adherent cells are grown instead in spinner flasks, (5) in hanging drops, (6, 7) or on non-adherent plates.(8) Without a substrate to attach to, the cells instead adhere to one another, forming a multicellular aggregate. Spheroids can be grown from a wide variety of sources including endometrial cancer, (9) ovarian cancer (2) (10), hepatocellular cancer,(11) and colon cancer (12). These models have proved to be valuable surrogates, especially for tumor studies. (13)
3D cell cultures have been employed experimentally to examine cellular function and phenotype. As an example, neuronal cells were found to more accurately model voltage gated calcium channel functionality in freshly dissected nerve tissue when they were cultured in 3D rather than 2D flat surfaces. (14) 3D cell cultures are also widely used to evaluate the penetration and metabolism of new drug compounds. (10, 15–18)
An important reason that 3D cell cultures are often selected as the culturing configuration is that they more closely mimic the chemical microenvironments in vivo. For example, ovarian cancer cells cultured as multicellular aggregates have mRNA gene expression patterns that more closely mimic in vivo tumors compared to 2D monolayers, especially if the 3D culture is allowed to adhere to a collagen surface. (6) There is also evidence that 3D cultures promote the viability of stem cells in culture. Human adipose-derived mesenchymal stem cells (MSCs) were cultured in 2D monolayers or in 3D spheroids. The MSCs cultured in 3D were found to secrete higher levels of antifibrotic factors like insulin growth factor 1 (IGF-1) and interleukin-6 (IL-6), which are helpful in treating fibrosis. (19) Similarly, MSCs cultured in 3D spheroids instead of 2D monolayer cultures were found to survive better and display improved therapeutic effects when injected in sites of acute kidney injury. (20) In addition, Gaedtke et al. performed proteome analysis on low-passage colon carcinoma cells cultured as multicellular spheroids versus monolayers, and identifies differences in protein expression levels relevant to tumor cell proliferation, survival, and chemoresistance. (21)
Given these intriguing differences between 2D and 3D cultured cells, previous studies have compared the global proteomes for the two types of cultures. The proteomes of 2D and 3D cultured gliomas cells were quantitatively compared and notable differences were observed in immune response pathways. (22) Hepatocellular carcinoma cells grown in 3D instead of 2D culture configurations showed differences in adhesion proteins and matrix metalloproteinases expression patterns. (11) Similarly, human parotid gland cells expressed higher levels of amylase and aquaporin proteins when cultured in 3D instead of 2D monolayers. (23)
Just as the global proteomes of 2D and 3D cultures have been examined, phosphoproteomic comparisons are also an area of interest. High-throughput phosphoproteome data could provide valuable targets for pharmaceutical therapies because many pathological changes occur at the phosphorylation level. Also, cellular processes such as DNA replication, cell cycle, apoptosis, and migration are mostly controlled through phosphorylation. The proteomes and phosphoproteomes of five glioma and six carcinoma cell lines grown in 2D and 3D were analyzed using reverse-phase protein arrays (24). As well as comparing the endogenous differences, the researchers also sought to determine the differences amongst the cell lines in normoxic versus hypoxic conditions as well as comparing the differences between 2D and 3D culture conditions. They determined that culture conditions (2D versus 3D) had a more striking effect on protein expression levels than the oxygen concentrations. They also observed that seven proteins were more highly expressed in 3D than 2D culture: FAK, AKT, SRC, GSK3AB, TSC2, MAPK14, and NFKBp65 and determined that seven other proteins are commonly decreased: ATRIP, ATR, CTNNB1, BCL2L1, CCNB1, EGR1, and HIF1A. Given these pronounced differences, there is value in examining the expression differences for 2D versus 3D cultures of commonly employed cell lines.
In this study, we examined the quantitative difference in the proteome and phosphoproteome of 2D and 3D cell cultures of the colon carcinoma cell line HT29. This cell line can be grown as an adherent monolayer or as 1mm spheroids. Using a stable isotope labeling of amino acids (SILAC) labeling strategy with liquid chromatography mass spectrometry (LC-MS), we quantified protein and phosphoprotein abundance differences in two biological replicates. We determined that there are notable differences in the proteomic and phosphoproteomic pathways in the two samples. In particular, pathways controlling growth rate in the 3D cells were more reduced in expression as compared to 2D.
MATERIALS AND METHODS
Chemicals and Materials
HPLC grade acetonitrile (ACN) and HPLC grade water were purchased from B & J Brand (Honeywell Burdick & Jackson, Muskegon, MI). The water used in all experiments except LC-MS/MS sample preparation was obtained from a Thermo Scientific Barnstead NANOpure™ Water Purification System (Thermo Scientific). Acetic acid (HAcO) was acquired from Sigma-Aldrich (St Louis, MO, USA). Trifluoroacetic acid (TFA) was obtained from Acros Organics (Belgium, NJ). Formic acid (FA, Optima LS/MS) and phosphoric acid (H3PO4) were acquired from Fisher Scientific (Fair Lawn, NJ). Cell lysis buffer reagents including urea, sodium chloride (NaCl), Trizma Base, sodium fluoride (NaF), β-glycerophosphate disodium salt hydrate (C3H7Na2O6P·xH2O), sodium orthovanadate (Na3O4V), sodium pyrophosphate tetra-basic decahydrate (Na4O7P2·10H2O), and phenylmethylsulfonyl fluoride (PMSF) were obtained from Sigma-Aldrich (St Louis, MO, USA). Complete protease inhibitors cocktail tablets mini (EDTA-free) were acquired from Roche Diagnostics (Mannheim, Germany). Reagents needed for tryptic digestion including dithiothreitol (DTT), iodoacetamide (IAA), calcium chloride (CaCl2) were acquired from Sigma-Aldrich (St Louis, MO, USA). Reagents used to make SCX and immobilized metal affinity chromatography (IMAC) buffers including potassium phosphate monobasic (KH2PO4), potassium chloride (KCl), potassium phosphate dibasic (K2HPO4·3H2O) were obtained from Sigma-Aldrich (St Louis, MO, USA). Ammonium bicarbonate (NH4HCO3) was ordered from Sigma-Aldrich (St Louis, MO, USA).
Cell Culture and Trypsin Digestion
The colorectal cancer cell line HT-29 was purchased from the American Type Culture Collection (ATCC, Manassas, VA). These cells were used within three months of resuscitation of frozen aliquots thawed from liquid nitrogen. The provider assured the authentication of this cell line by cytogenetic analysis.
HT-29 cells were cultured in McCoy’s 5A media supplemented with 10% fetal bovine serum and grown in 5% CO2 at 37 °C. For the 2D monolayer cultures, 2 × 105 cells were seeded in T-75 flask. For the 3D spheroid cultures, 3000 cells were seeded in 96-well ultra low attachment plates to form multicellular aggregates. In biological replicate 1, the 3D spheroids were cultured in “heavy” SILAC media while the 2D monolayer culture was grown in “light” media. The SILAC heavy media contained heavy arginine (13C6, 15N4) and lysine (13C6, 15N2) (Isotec, Sigma) while the light media contained the naturally occurring isotopes of arginine and lysine. In biological replicate 2, the 2D monolayer was cultured in heavy media and the 3D spheroids were grown in light media. For each biological replicate, the 3D culture is a pooled sample of 180 3D spheroids that were grown separately, and the 2D culture is pooled sample of 3 dishes of monolayer cells that were grown separately.
The 2D monolayer cells were grown to 80% confluence then rinsed twice with ice-cold phosphate buffered saline and lysed with 8M urea lysis buffer. Every 10 ml of lysis buffer contains 8 M urea, 75 mM NaCl, 50 mM Tris-HCl (pH8.2) (Sigma-Aldrich), 1 mM NaF, 1 mM β-glycerophosphate, 1 mM sodium orthovanadate, 10 mM sodium pyrophosphate, 1 mM PMSF, and 1 tablet of EDTA-free protease inhibitors cocktail. The 3D spheroids were grown for 10 days to a final diameter of 800 µm and harvested with 200 µl of lysis buffer per 60 spheroids. Cells were lysed with 1 min of sonication followed by 2 min rest intervals, repeated three times. The cell lysates were centrifuged for 10 min at 2500 g to remove cell debris and the total protein concentrations were determined with the bicinchoninic acid (BCA) protein assay kit (Thermo Scientific Pierce, Rockford, IL). After the BCA assay, proteins from the 2D monolayer and the 3D spheroids were mixed in a 1:1 (light: heavy) ratio. A 500 µg protein sample from each biological replicate was allocated for the proteomic profiling and 3 mg of proteins was used for phosphoproteomic analysis.
Western Blot
A 20 µg protein sample from each of the harvested cell lysates was run on a NuPAGE SDS-PAGE 4–12% gel (Life Technologies, Carlsbad, CA) for 1 hour. Proteins were transferred to a nitrocellulose membrane using transfer buffer (Life Technologies, Carlsbad, CA) containing 10% methanol. Transfer conditions were 25V, 500mA for 90 minutes after which the membrane was briefly washed with DI water. A buffer containing 5% Non-fat dry milk in 1× PBS was used for antibody incubation and washes. Primary antibodies against Rb1, MCM5, p-Rb1 (Ser 807/811) (Santa Cruz Biotechnology, Santa Cruz, CA), UQCRC1 (Abcam, Cambridge, MA) and COX IV (loading control) (Cell Signaling Technology, MA) were diluted between 1:250 to 1:1000 in milk buffer. The membrane was incubated in primary antibody at RT for 1 hour on a rocking platform. The membrane was rinsed 3 times for 10 minutes each in milk buffer. The secondary antibody, HRP anti-mouse or rabbit (Jackson ImmunoResearch, West Groove, PA), was diluted 1:10,000 in milk buffer and incubated with the membrane for 1 hour at RT on a rocking platform. Membrane was rinsed, as above, with a final rinse of DI water. Chemiluminescent substrate (Thermo Scientific, Rockford, IL) was incubated with membrane for 5 minutes. Solution was removed and membrane was wrapped in plastic wrap. In a dark room, the membrane was exposed to Kodak BioMax film (VWR, Radnor, PA) for 2 to 5 minutes. The film was then developed using GBX developer and fixer (VWR, Radnor, PA).
Preparation of Mass Spectrometric Samples
To reduce disulfide bonds, protein samples were treated with 5mM dithioreitol (DTT) for 25 min at 56°C. To alkylate the cysteines, iodoacetamide (IAA) was added to a final concentration of 14 mM and allowed to react in the dark for 30 min. To stop the alkylation reaction, another 5mM aliquot of DTT was added and reacted in the dark for another 15 min. To achieve a final urea concentration of 1.8 M, the protein mixtures were diluted with 25 mM Tris-HCl (pH 8.2). Protein samples were digested with trypsin from bovine pancreas (Sigma-Aldrich, St Louis, MO, USA) overnight at 37°C at a 50:1 protein to trypsin ratio in the presence of 1 mM CaCl2. To stop the digestion reaction, trifluoroacetic acid was added to a final concentration of 0.4% (vol/vol) and the samples were centrifuged at 2500 g for 10 min to remove precipitates. The peptides were then desalted by 500 mg reverse-phase C18 Sep-Pak solid-phase extraction cartridges (Waters Corporation, Milford, MA).
The two 500 µg peptide samples for proteomic were separated with high-pH reverse phase separation with OASIS HLB 1cc Vac Cartridges with 30 mg sorbent per cartridge and 30 µm of particle size from Waters Corporation (Milford, MA). In the HLB separation, the buffers contained 10 mM NH4HCO3 and were adjusted to pH 10 with NH4OH. The IMAC enriched samples were first washed three times with 3 mL of 80% acetonitrile/ NH4HCO3 and equilibrated with 3 mL of 1% acetonitrile/ NH4HCO3. The samples were then conditioned with 0.5 mL of 1% acetonitrile/ NH4HCO3 and loaded onto the equilibrated HLB columns. The samples were washed with 3 mL of 1% acetonitrile/ NH4HCO3 to remove salts. Stepwise serial elution was performed with 0.5 ml of each elution buffer in the order of 5%, 10%, 15%, 20%, 25%, 30%, 35%, and 80% acetonitrile/NH4HCO3. Eluates were collected in an Eppendorf tube containing 25 µl of 100% formic acid to acidify the phosphopeptides and stabilize them. Forced air was used to accelerate the solutions passing through the HLB cartridges. To minimize LC-MS/MS analysis time, fraction numbers were combined with 5% mixed with 25%, 10% mixed with 30%, 15% mixed with 35%, and 20% mixed with 80% acetonitrile/NH4HCO3 fractions. The resulting HLB fractions were lyophilized and desalted with C18 ZipTips prior to LC-MS/MS analysis. ZipTips were not used if working with trap column.
Enrichment of Phosphopeptides for Mass Spectrometric Analysis
To prepare the samples for phosphoproteomic analysis, the IMAC-HLB protocol was used as previously described.(25) Phos-Select iron affinity gel (IMAC beads) were purchased from Sigma-Aldrich (St Louis, MO). The beads were washed three times with a 10× volume of IMAC binding buffer composed of 40% acetonitrile and 25 mM formic acid in water. The beads were then resuspended in the binding buffer to generate a 50% gel slurry. The 3 mg peptide mixture was incubated with vigorous shaking for 60 min at room temperature in 120 µL of the IMAC binding buffer and 10 µL of the IMAC slurry in a 100:1 peptide-to-IMAC ratio. Following the incubation, the IMAC beads were washed three times in IMAC binding buffer to eliminate any non-specific binding. To elute the phosphopeptides, the IMAC beads were rinsed three times at room temperature with 40 µL of elution buffer (50 mM K2HPO4/NH4OH, pH10) while shaking. A solution of formic acid (40 µL of a 10% solution) was added to acidify the solution immediately after the elution. The second round of IMAC enrichment was performed with the IMAC flow-through from the first round of IMAC enrichment and the third round was performed with the flow-through from the second round. After three rounds of IMAC, the peptide solution was separated with the high-pH reverse phase (HLB) separation as previously described in the proteomic sample preparation. The HLB fractions were lyophilized and desalted with C18 ZipTips prior to LC-MS/MS analysis.
Mass spectrometric analysis
Prior to tandem mass spectrometric analysis (MS/MS), all peptide samples were lyophilized and resuspended in a mass spectrometry loading buffer (1% HPLC grade acetonitrile, 0.1% formic acid in HPLC grade water), and analyzed with a Q-Exactive mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) coupled to a nanoACQUITY Ultra Performance LC (UPLC) system from Waters Corporation (Milford, MA). The peptide samples were injected onto a C18 reverse phase column (100 µm × 100 mm, 1.7 µm particle size, BEH130) (Waters Corporation, Milford, MA) with 97% buffer A (0.1% formic acid in water) and 3% buffer B (0.1% formic acid in acetonitrile). A 73-min linear gradient was used to separate the peptides, with the gradient changing from 3% to 40% buffer B for non-enriched samples, and 3% to 30% for phosphopeptide enriched samples, at a flow rate of 1000 nL/min. The mobile phase composition was then maintained at 85% buffer B for 5 min before being re-equilibrated with 3% buffer B for 10 min before the next run.
A nanoelectrospray source was used on the Q-Exactive and was operated at a source voltage of 1.8 kV. The ion transfer tube temperature was maintained at 280 °C. Full MS scans were acquired with an m/z range of 350–1800 in the Orbitrap mass analyzer with a mass resolution of 70000 at m/z=200. The automatic gain control (AGC) value was set at 1×106, with maximum fill times of 250 ms. For MS/MS scans, the top 12 most intense parent ions were selected with an 2.0 m/z isolation window and fragmented with a normalized collision energy (NCE) of 30%. The AGC value for MS/MS was set to a target value of 1×106, with a maximum fill time of 120 ms. Parent ions with a charge state of z=1 or with unassigned charge states were excluded from fragmentation and the intensity threshold for selection was set to 8.3×104. Fragmentation was performed with an HCD collision cell (mass resolution 35000 at m/z=200). A dynamic exclusion period of 20s was used after 1 repeat count. All samples were run in technical duplicate.
Data Analysis
The .raw files acquired using the Q-Exactive were analyzed with Proteome Discoverer 1.3 software (Thermo Scientific, Bremen, Germany) with MASCOT 2.2.4 as the search algorithm. The searches were against the SwissProt human database modified with the addition of common contamination sequences (updated on 05/2012, 86758 sequences). The peptide false discovery rate (FDR) was calculated with searches against the corresponding reverse database. Searches were carried out with a precursor peptide mass tolerance of 10 ppm and a fragment ion mass tolerance of 0.02Da. Two missed trypsin cleavages were allowed in the searches. Cysteine carbamidomethylation was set as a fixed modification. Oxidation of methionine and phosphorylation of serine, threonine, and tyrosine were all set as variable modifications. Any peptides that were the same mass but with different oxidation states were treated as the same peptide in all data analysis in case the oxidation was a result of sample manipulation. A value of 0.01 for the FDR was used in all searches. Confident phosphosite identifications had PhosphoRS scores higher than 0.99. The resulting peptide lists from the searches were exported to Microsoft Excel and Venn diagrams were generated with the online tool Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html). (26) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and Gene Ontology (GO) analysis was performed with R package Cluster Profiler. (27) Network analysis was performed with Search Tool for the Retrieval of Interacting Genes/Proteins (STRING). (28) Only those proteins/phosphoproteins that were quantified with more than 2-fold change in both biological replicates were used for GO, STRING, and KEGG analysis. To maintain as much information of STRING network but avoid making the network too complicated, the following interaction score cutoff were applied for each network: 0.9 for up regulated proteome; 0.7 for down regulated proteome; 0.9 for uncorrected phosphoproteome; and 0.5 for corrected phosphoproteome. For GO analysis, the p-value cutoff of 0.01 and q-value cutoff of 0.01 were applied for enriched GO terms.
RESULTS AND DISCUSSION
2D and 3D Cultured Cells Show Different Protein Expression and Phosphorylation
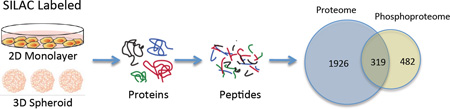
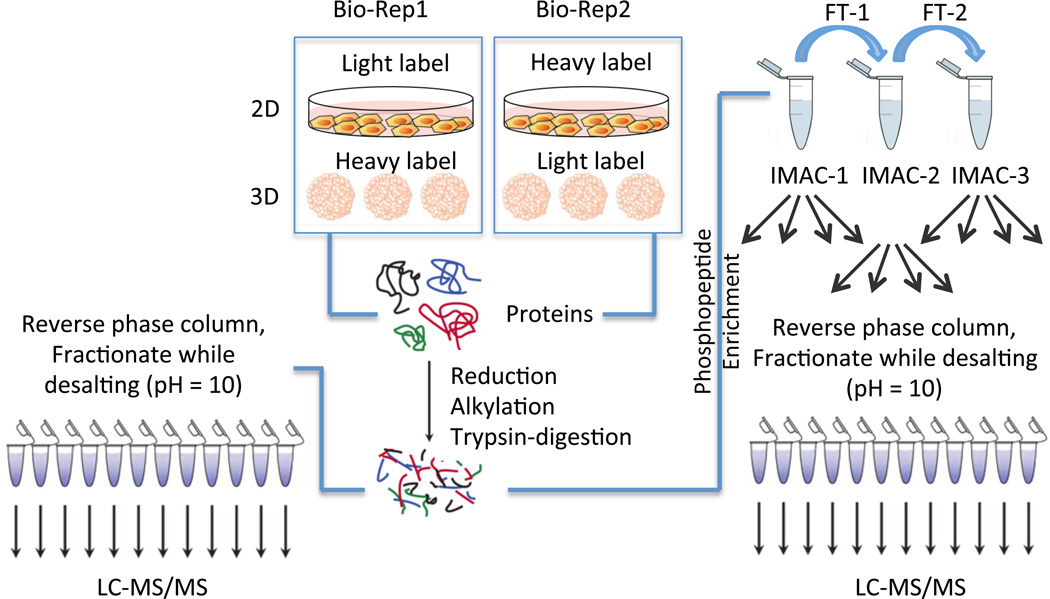
The differences in protein expression and phosphorylation levels between 2D and 3D cultures were analyzed with a SILAC labeling strategy. As illustrated in Figure 1, 2D and 3D cultured cells were differently labeled with “light” or “heavy” amino acids and lysed separately to extract labeled proteins. In order to minimize the side effects from labeling, we used a reverse label strategy and included two biological replicates. The cell lysates were then mixed with a 1:1 ratio. After alkylation and trypsin digestion, the pooled SILAC peptides were divided into two samples. One sample was fractionated with high pH reverse phase columns for proteomic analysis. The second portion of the pooled sample was treated with IMAC to enrich for phosphopeptides, and the enriched phosphopeptides were further fractionated with high pH reverse phase column for mass spectrometric analysis of the phosphoproteome (Figure 1).
Figure 1.
Workflow for the experiments. HT29 cells were grown either as a 2D monolayer or in 3D spheroids in two biological replicates. In one biological replicate, the 3D spheroids were labeled with the heavy SILAC media while the monolayers were prepared in light SILAC media. Reverse labeling was used for the second biological replicate. Proteins were harvested from the 2D and 3D samples in each biological replicate and combined in a 1:1 ratio. The combined protein fractions were then aliquoted for proteomic and phosphoproteomic enrichment, with 500 µg used for the proteomic analysis and 3 mg of protein allocated for the phosphoproteomic enrichment. The proteins were then reduced and alkylated prior to tryptic digestion. For the proteomic analysis, peptides were fractionated on HLB reverse-phase columns and then analyzed by LC-MS/MS. For the phosphoproteomic analysis, peptides were enriched first by three rounds of IMAC, followed by high pH reverse phase fractionation prior to LC-MS/MS analysis, as described previously. (25)
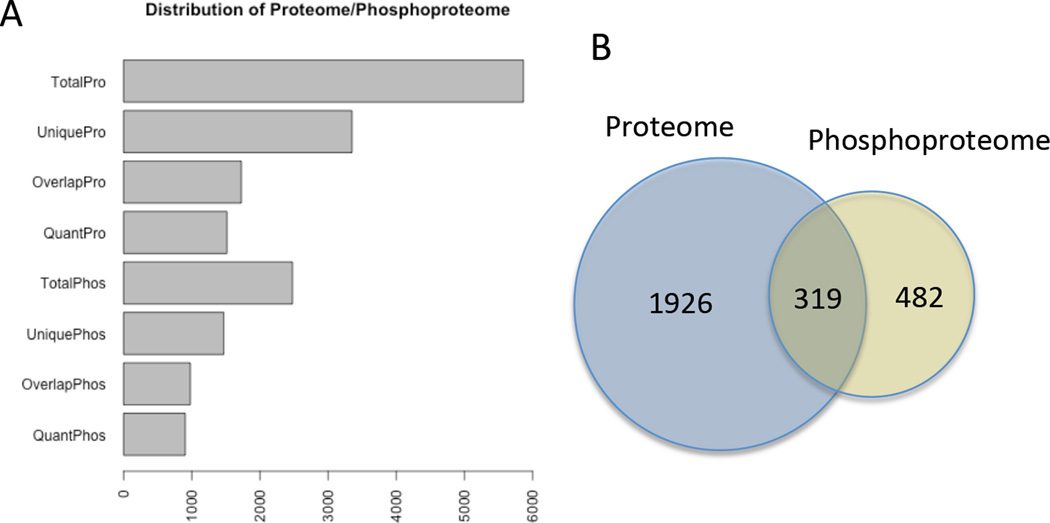
We identified and quantified notable numbers of proteins and phosphoproteins from both the 2D and 3D samples. The identification and quantification results are summarized in Figure 2 and Table 1. We identified a total of 5867 protein groups, including 2523 phosphoprotein groups and 8733 phosphopeptides. Among these identified results, 3348 were unique protein groups, 1536 were unique phosphoprotein groups, and 4468 were unique phosphopeptides. The overlap between biological replicates indicated good reproducibility of the experiment, as 2492 out of 3348 unique protein groups (74.4%) and 987 out of 1536 unique phosphoprotein groups (64.3%) were identified in both biological replicates (Table 1). Of all these identifications, 319 protein groups were identified in both non-phosphorylated and phosphorylated states (Figure 2B). In order to compare the protein expression and phosphorylation levels in the 2D and 3D culture systems, we screened for those that are quantifiable in both biological replicates. This analysis revealed 2245 quantifiable protein groups, 801 quantifiable phosphoproteins, and 1534 quantifiable phosphopeptides that were quantifiable in both biological replicates. Among these, in the 3D culture system, we quantified 225 protein groups and 73 phosphoproteins, corresponding to 120 phosphopeptides were up regulated. By contrast, 116 protein groups and 236 phosphoproteins corresponding to 374 phosphopeptides were down regulated at least two-fold (Table 1). For the phosphoproteins that were identified with their non-phosphorylated counterparts, a correction with the non-phosphorylated protein expression was performed to determine the corrected phosphorylation levels, which showed that 29 phosphoproteins and 59 phosphopeptides were up regulated in 3D cultures, while 80 phosphoproteins and 130 phosphopeptides that were down regulated in 3D cultures with the corrected phosphorylation levels (Table 1). The complete lists of up and down regulated proteins are detailed in the Supporting Information.
Figure 2. Distribution of Proteins and Phosphoproteins Detected.
A) Numbers of proteins and phosphoproteins identified. TotalPro is the total number of proteins detected in the proteome; UniquePro is the number of unique proteins that were only detected in one biological replicate; OverlapPro is the number of proteins that were identified in both biological replicates; QuantPro is the number of proteins, identified in both biological replicates with at least a Log2 fold change difference in expression. The same terminology is used for the phosphoproteome numbers. B) Venn diagram showing the number of unique proteins identified in the proteome (blue circle) with the number of unique phosphoproteins (yellow). The intersection shows proteins identified in both the proteome and phosphoproteome.
Table 1.
Number of protein groups, phosphoproteins groups and phosphopeptides identified. All up /down regulated proteins/phosphoproteins/phosphopeptides displayed at least a Log2-fold change in both biological replicates.
| Description | Number of Protein Groups |
Number of Phosphoprotein Groups |
Number of Phosphopeptides |
|---|---|---|---|
| Total | 5867 | 2523 | 8733 |
| Unique | 3348 | 1536 | 4468 |
| Identified in both bio- replicates |
2492 | 987 | 1667 |
| Quantifiable in both bio-replicates |
2245 | 801 | 1534 |
| Up-regulated in 3D (2-fold cutoff)* |
225 | 73 (29**) | 120 (59**) |
| Down-regulated in 3D (2-fold cutoff)* |
116 | 236 (80**) | 374 (130**) |
Up-/down-regulated proteome/phosphopeptides are decided with 2-fold changes in both biological replications.
Indicate up-/down-regulated phosphoprotein groups or phosphopeptides corrected with their correspondent protein expression levels.
A previous report by Poland et al examined the differential proteome by combining 2-dimensional gel electrophoresis with MALDI to compare proteomic differences between 2D and 3D conditions, using the same cell line as we used in this research. (29) The authors reported that 14-3-3 β, 14-3-3 η, α-Tubulin, β-Tubulin, and Rho-GDI were down regulated in 3D cultures, while calreticulin precursor and peroxiredoxin were up regulated in 3D cultures. Our data agrees well with all these findings, indicating good reproducibility for comparing 2D and 3D cultures. In addition, our dataset identified more protein targets as well as provided the phosphorylation information that was absent from Poland et al.’s work.
Observed Differences Between 2D and 3D Cultured Cells are Reproducible in Biological Replicates
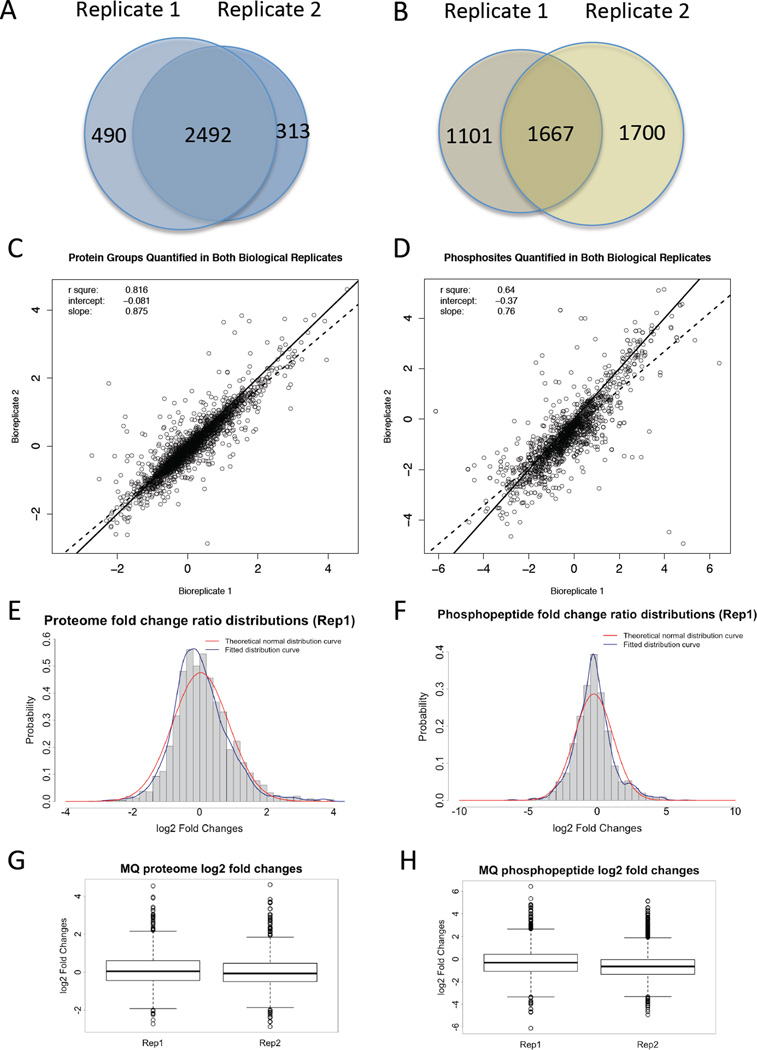
The data quality of the mass spectrometric proteomic analysis and the reliability of the identified and quantified protein groups and phosphopeptides were examined by comparing the two biological replicates for each analysis (Figure 3). The overlap between biological replicate 1 and biological replicate 2 shows 2492 identifiable protein groups, and 2245 quantified protein groups (Figure 3A & Table 1). The correlation of biological replicate 1 and biological replicate 2 showed a linear trend and the R squared value is 0.816, indicating a high correlation between the two biological replicates (Figure 3C). Interestingly, most of the quantified protein groups did not show much change between the 2D and 3D cultured cells (Figure 3E & G). Two-fold change was selected as a threshold value to determine whether a protein has differential expression levels or not in the 2D and 3D cultured cells.
Figure 3. Data Quality Control.
A) and B) Venn diagram showing protein groups or phosphoprotein groups detected in the proteomic analyses of two biological replicates. C) and D) Scatter plot of the fold changes for all protein groups or phosphoprotein groups quantified in both biological replicates in the proteomic analyses. The solid line represents a 1:1 correlation between the replicates. The dashed line is the calculated correlation in the dataset. E) and F) Theoretical normal distribution of fold changes versus calculated distribution of fold changes for the proteome or phosphoproteome dataset. G) and H) Box plots of the MaxQuant Log2 fold change results for biological replicate 1 and 2 for the proteome or phosphoproteome dataset. Panels A, C, E, G represent the results of proteome datasets, while panels B, D, F, H represent the result of phosphoproteome datasets.
The data quality of phosphoproteomic analysis is shown in Figure 3B, D, F, H. Similar to proteomic analysis, the correlation of biological replicate 1 and biological replicate 2 showed a linear trend and the R squared value is 0.655 for phosphopeptides, indicating some correlation between the 2 biological replicates (Figure 3D). The overlap between biological replicate 1 and biological replicate 2 shows 1667 identified phosphopeptides, and 1534 quantifiable phosphopeptides (Figure 3B & Table 1), and most of the quantifiable phosphopeptides did not show notable abundance changes between the 2D or 3D cultured cells (Figure 3F & H). We chose two-fold change as the threshold to determine whether a phosphopeptide has different expression level or not in 2D and 3D cultured cells.
One interesting observation is that the overall protein expression levels and phosphorylation level show the contrary trend, with 225 up regulated protein groups and 116 down regulated protein groups in 3D cultures, in contrast to 120 up regulated phosphopeptides and 374 down regulated phosphopeptides in 3D cultures. These data correspond to 59 up regulated phosphopeptides and 130 down regulated phosphopeptides in the dataset with phosphorylation levels corrected by corresponding non-phosphorylated counterparts (Table 1). Although the distribution of fold changes for both protein groups and phosphopeptides shows a fitted Gaussian distribution for each biological replicate and did not show shifting to one side (Figure 3E & F), the overlapping segment with protein groups and phosphopeptides quantified in both biological replicates shows a decreased overall phosphorylation level in 3D cultures.
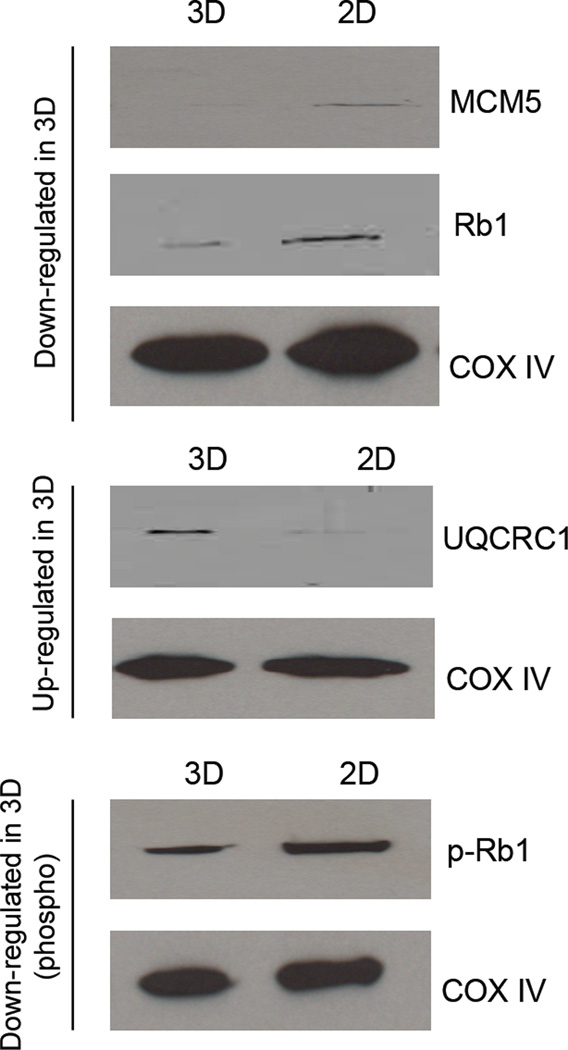
Western blot analysis was performed in lysates harvested from an additional 3D and 2D biological replicate to verify the abundance of selected proteins. The proteins MCM5, Rb1, and p-Rb1 were down regulated in the 3D cultures as compared to the 2D cultures. In comparison, UQCRC1 was shown to be up regulated in 3D cell culture when compared to 2D. These results are shown in Figure 4 and match the SILAC results obtained.
Figure 4. Western blot verifies relative protein quantitation found in SILAC proteomic results.
MCM5, Rb1, and p-Rb1 were down regulated while UQCRC1 was up regulated in 3D cell culture when compared to 2D. COX IV was used as a loading control throughout the experiment. Protein amounts of COX IV was confirmed to be constant using Image J.
Gene Ontology and Network Analysis for Protein Groups
Gene Ontology (GO) analysis was performed to examine the gene functions in the up and down regulated proteins with more than two-fold expression changes in the 3D cultures (Table 2). The enriched GO terms indicate that the proteins with differential expression in 2D and 3D cultures are primarily correlated with RNA binding, nucleic acid binding, enzyme binding, cytoskeletal protein binding, and histone binding for their molecular functions (MF). While for the biological process (BP), the differences mainly exist in the process of cell cycle, cytoskeleton organization, and DNA metabolic process. On the other hand, for cellular components (CC), the enriched GO terms between 2D and 3D cultures that differ include nuclear part, organelles, nucleus, as well as cytosol, cytoplasm, and cytoskeleton components.
Table 2. Enriched GO terms.
Gene Ontology terms that are enriched between the 3D and 2D phosphoproteins datasets. All protein groups that were changed more than Log2 either up or down regulated in the 3D compared to the 2D dataset were included. GO terms for molecular functions, biological processes, and cellular components with p values < 0.01 and Q values < 0.01 are included.
| Molecular Functions (MF) | Biological Process (BP) | Cellular Components (CC) |
|---|---|---|
| protein binding | cellular component organization or biogenesis |
nuclear part |
| poly(A) RNA binding | cell cycle | nuclear lumen |
| RNA binding | cytoskeleton organization | non-membrane-bounded organelle |
| nucleic acid binding | multi-organism cellular process | organelle lumen |
| heterocyclic compound binding | cellular response to heat | membrane-enclosed lumen |
| organic cyclic compound binding | macromolecular complex assembly | nucleus |
| enzyme binding | cellular component assembly | intracellular part |
| cytoskeletal protein binding | regulation of catalytic activity | nucleoplasm |
| macromolecular complex binding |
positive regulation of metabolic process | intracellular |
| actin binding | cellular response to stress | organelle part |
| histone binding | ribonucleoprotein complex biogenesis | macromolecular complex |
| response to heat | intracellular membrane- bounded organelle |
|
| DNA metabolic process | cytosol | |
| endomembrane system organization | cytoplasm | |
| nucleic acid metabolic process | cytoskeleton | |
| protein complex subunit organization | adherens junction | |
| metabolic process | ||
| chromatin assembly or disassembly | ||
| actin cytoskeleton organization |
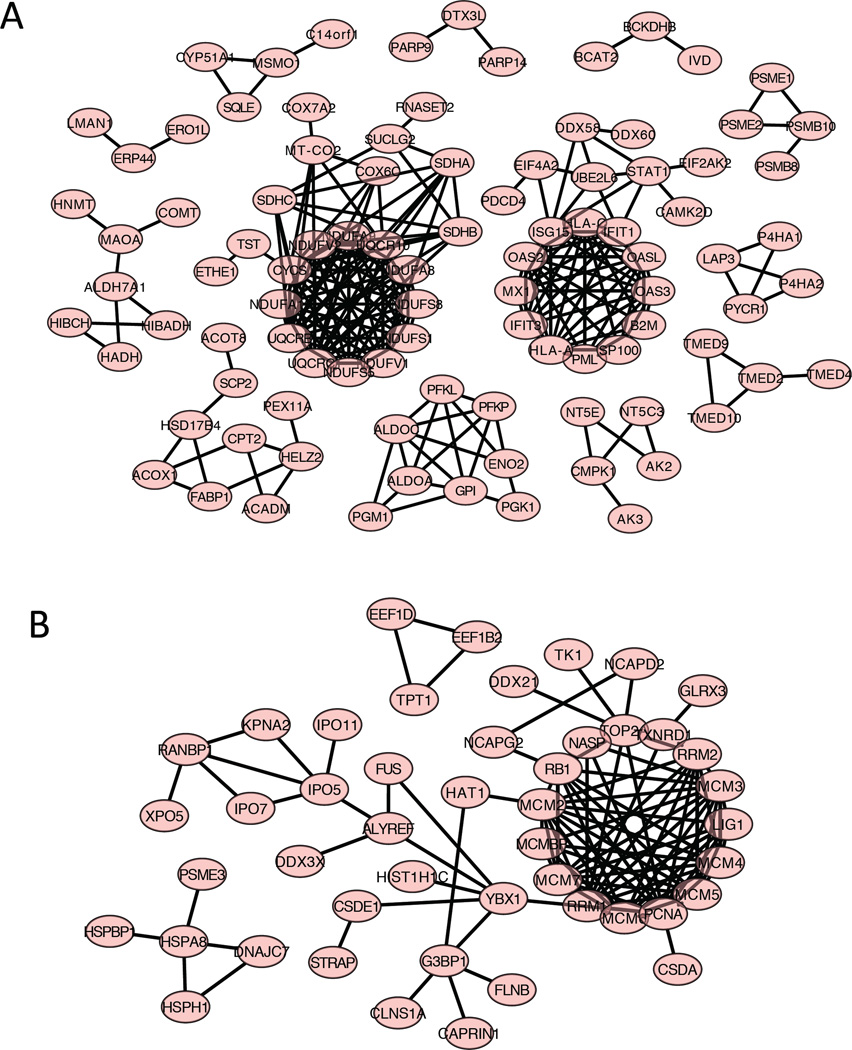
To determine the possible protein-protein interactions, those proteins that have more than two-fold expression changes in 3D versus 2D cultures were analyzed by STRING (Figure 5). The proteins that are up regulated in 3D cultures show several clusters. One large cluster is composed of protein components in the mitochondrial respiratory chain complex, including NDUFA1 (NADH: ubiquinone oxidoreductase subunit A1), NDUFS5, NDUFS8, SDHA (succinate dehydrogenase complex iron sulfur subunit A), SDHB, SDHC, etc. This cluster contains components that mainly function in the mitochondrial respiratory chain, and are involved in energy conversion in mitochondria. Another large cluster includes interferon related proteins, including ISG15 (interferon-stimulated protein, 15 KDa), IFIT1 and IFIT3 (interferon-induced protein with tetracopeptide repeats 1, and repeats 3), etc. These proteins are functional in pathways to regulate the energy supply for cells. The up regulation of these protein networks in 3D cultures indicate that in 3D spheroids, cells are undergoing more oxidative phosphorylation and are providing more energy to support cell functions. By contrast, the proteins down regulated in 3D cultures contains the MCM (minichromosome maintenance complex) family proteins, including MCM 2, 3, 4, 5, 6, 7, and MCMBP (Figure 5B). The MCM proteins are essential for initiation of eukaryotic genome replication, (30) and the down regulation of MCM proteins in 3D cultures indicates that cell DNA replication and cell proliferation is down regulated in 3D systems. This result is logical since in 3D cultures the cell proliferation is restrained in the necrotic core, and cells are mostly quiescent in the middle layer. (16, 17, 31–39)
Figure 5. Networks of up or down regulated proteins in 3D versus 2D in the proteomic dataset.
A) Proteins that were up regulated with a fold change of at least Log2 in both biological replicates in the 3D samples as compared to the 2D samples. The protein data was uploaded to STRING and the functional protein interactions were predicted as a several protein networks. B) Proteins that were down regulated with a fold change of at least Log2 in both biological replicates in the 3D samples as compared to the 2D samples. The protein data was uploaded to STRING and the functional protein interactions were predicted as a several protein networks.
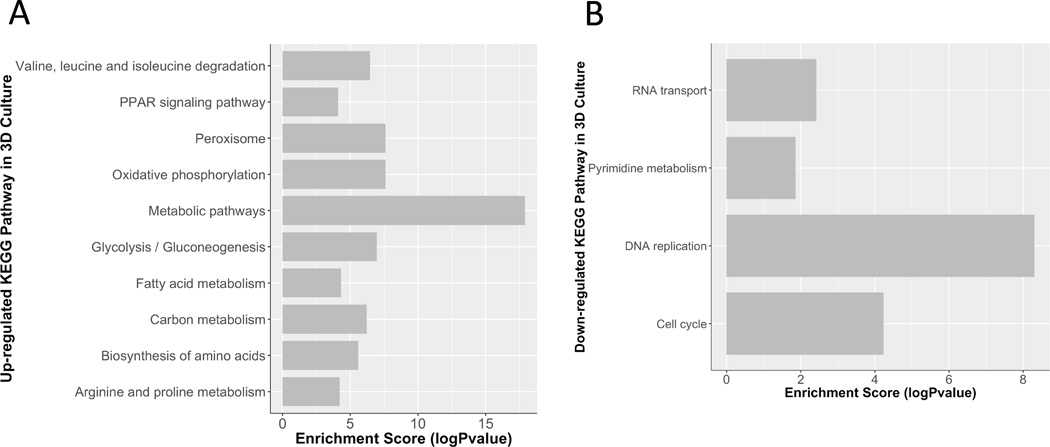
The KEGG pathway analysis of proteins showed similar trends (Figure 6). The up-regulated pathways in 3D cultures include the oxidative phosphorylation pathway, metabolic pathways, peroxisome pathways, and biosynthesis of amino acids (Figure 6A). The peroxisome pathways are crucial for biosynthesis of ether lipids, (40) which is an important composition of cell membranes. Up regulation of these pathways will provide more energy for cells to maintain their functions. On the other hand, the down-regulated pathways in 3D cultures include DNA replication, cell cycle, and RNA transport (Figure 6B). Down regulation of these pathways in 3D cultures would result in decrease of cell proliferation, which is in agreement of the STRING network analysis. Tables of identified pathways are detailed in the Supporting Information.
Figure 6. KEGG Pathway Analysis.
The up and down regulated proteins in the proteomic dataset were analyzed by KEGG Pathway analysis to examine the expression changes in well-annotated pathways. A) The table shows pathways enriched in the up regulated proteins groups in the 3D sample compared to the 2D sample are shown, including their enrichment score. B) The table shows pathways enriched in the down regulated proteins groups in the 3D sample compared to the 2D sample are shown, including their enrichment score. Many of the down regulated pathways in the 3D versus the 2D sample are related to cell growth.
Network Analysis for Phosphoproteins
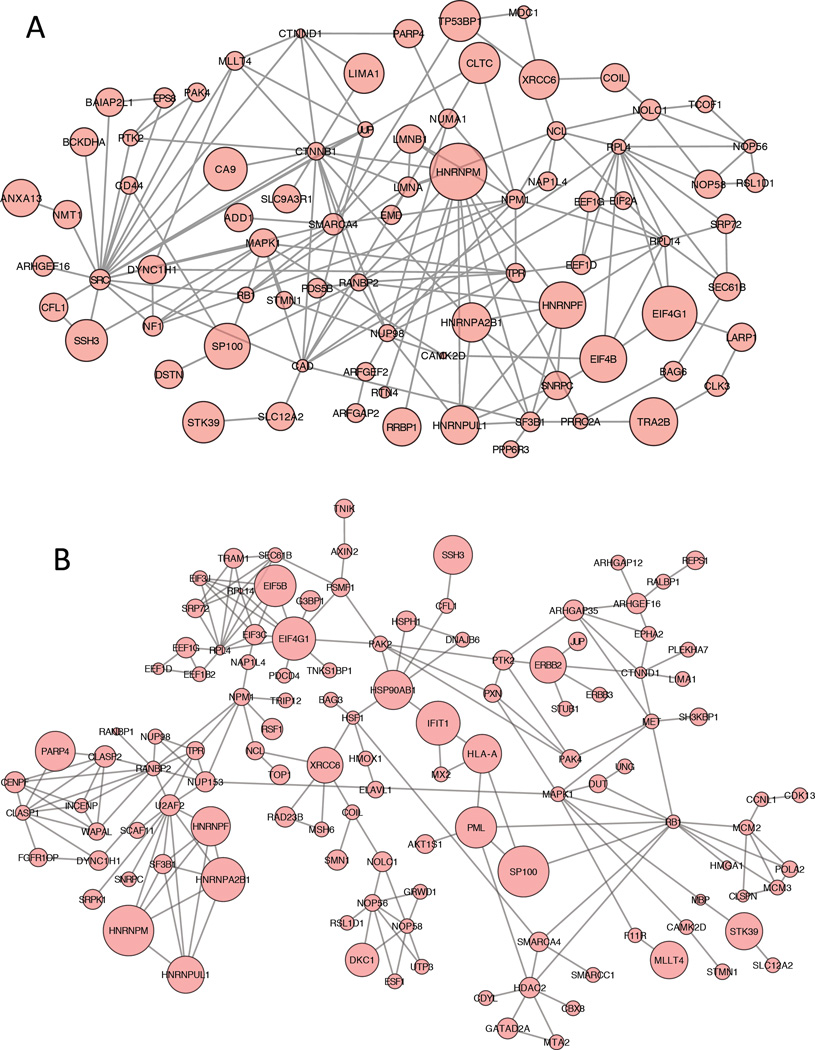
For the phosphoprotein dataset, only the phosphoproteins that have corresponding protein information were used for network analysis, and phosphorylation levels were corrected with the corresponding protein levels. The STRING algorithm was applied to phosphoproteins that were either up or down regulated by more than 2-fold with their corrected phosphorylation levels. For phosphoproteins that have multiple phosphopeptides quantified, only those that had same trends of up or down regulation for all quantified phosphopeptides were used, and the phosphorylation levels were represented with the average value of all quantified phosphopeptides from the same phosphoprotein. The STRING networks with corrected phosphorylation levels are shown in Figure 7A, while the STRING networks with uncorrected phosphorylation levels are shown in Figure 7B.
Figure 7. Network of up and down regulated proteins in 3D versus 2D in the phosphoproteomic dataset.
A) Corrected phosphoproteins that were altered with a fold change of at least Log2 in both biological replicates in the 3D samples as compared to the 2D samples. The protein data was uploaded to STRING and the functional protein interactions were predicted a protein network. Up regulated corrected phosphoproteins in the 3D versus the 2D samples are shown as large circles and down regulated corrected phosphoproteins are shown as small circles. B) STRING network of uncorrected phosphoproteins generated with similar strategies as in A).
Some of the interesting phosphoproteins in the corrected network include MAPK1 (mitogen-activated protein kinase 1), PTK2 (protein tyrosine kinase 2), and PAK4 (p21-activated kinase 4), which were all down regulated in 3D cultures. The phosphorylation of MAPK1 regulates cell growth, adhesion, survival and differentiation. (41) PAK phosphorylation has multiple roles including cytoskeleton regulation, cell motility, cell cycle progression, and also apoptosis and proliferation. (42) PTK2, also known as FAK1 (focal adhesion kinase 1), is essential for regulation of cell migration, adhesion, formation of focal adhesions, and cell cycle progression. (43) The down regulation of phosphorylation levels on these proteins in 3D cultures as compared to 2D cultures correlate with the slower doubling time, and thus slower growth rate, for the 3D cultured cells. They also indicate that the 3D cultured cells have lower cell-to extracellular matrix interactions.
One interesting phosphoprotein, MSH6 (mutS homolog 6), was found only in the phosphoproteome dataset but not in the proteome dataset. MSH6 is a DNA mismatch repairing protein and is essential for repairing DNA. (44) Decreased phosphorylation levels of MSH6 imply that in 3D culture, less MSH6 was required for DNA repair, which might be a result of the slower growth rates for cells in 3D cultures, or because better resistance to DNA damage for cells in 3D cultures. To analyze whether cells in 3D cultures are more resistant to DNA damage, further research is required.
CONCLUSION
This is the first report of a large-scale comparison of 2D and 3D cell cultured systems examining both the protein expression and phosphorylation levels. In this paper, we identified 3348 unique protein groups and 4468 unique phosphopeptides from SILAC-labeled 2D and 3D cultures. We quantified 225 up regulated and 116 down regulated protein groups, and 120 upregulated and 374 down regulated phosphopeptides in 3D cultures in comparison to 2D cultures. These protein groups and phosphoproteins interact with each other and form complicated networks. These networks regulate cell functions such as proliferation, cell cycle, DNA damage repair, and DNA replication, and metabolic. In summary, our work suggests that in 3D cultures, cells have slower growth rates, require more energy to maintain cellular functions, have less DNA replication and fewer cells are progressing in cell cycle. Given that many biomedical studies have been performed in 2D instead of 3D cultures over the years, our study provides an assessment of the proteins and pathways that should be reevaluated from studies conducted from cells grown in 2D monolayers. While 2D cultures are a valuable model system, this study clearly indicates that there are substantial proteomic and phosphoproteomic differences when cells are cultured in 3D instead of 2D. Given the fact that in vivo, cells do not proliferate and expand as rapidly as 2D cultures, as evidenced by the fact that tissues remain constant in size, the 3D cultures are better representations of in vivo conditions. The results presented in this study might be applied to many areas of in vitro study such as drug screening and aging.
Supplementary Material
Acknowledgments
The work was funded by Department of Defense Visionary Postdoctoral Fellowship Award to XY (USAMRAA W81XWH-12-1-0412). Salary support for ABH was provided by the Walther Cancer Foundation and an NSF Early CAREER award (CHE-1351595). We thank the staff from the Mass Spectrometry and Proteomics Facility at the University of Notre Dame, especially Dr. Bill Boggess, for their helpful discussions and advice, and for their help maintaining the mass spectrometry equipment.
Footnotes
ASSOCIATED CONTENT:
SUPPORTING INFORMATION: The following files are available free of charge at ACS website http://pubs.acs.org: Supporting Information. The file contains tables detailing the Max Quant data and pathways analysis.
REFERENCES
- 1.Katt ME, Placone AL, Wong AD, Xu ZS, Searson PC. In Vitro Tumor Models: Advantages, Disadvantages, Variables, and Selecting the Right Platform. Front Bioeng Biotechnol. 2016;4:12. doi: 10.3389/fbioe.2016.00012. PubMed PMID: 26904541; PMCID: PMC4751256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lengyel E, Burdette JE, Kenny HA, Matei D, Pilrose J, Haluska P, Nephew KP, Hales DB, Stack MS. Epithelial ovarian cancer experimental models. Oncogene. 2014;33(28):3619–3333. doi: 10.1038/onc.2013.321. PubMed PMID: 23934194; PMCID: 3990646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batalov I, Feinberg AW. Differentiation of Cardiomyocytes from Human Pluripotent Stem Cells Using Monolayer Culture. Biomark Insights. 2015;10(Suppl 1):71–76. doi: 10.4137/BMI.S20050. PubMed PMID: 26052225; PMCID: PMC4447149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ham SL, Joshi R, Thakuri PS, Tavana H. Liquid-based three-dimensional tumor models for cancer research and drug discovery. Exp Biol Med (Maywood) 2016 doi: 10.1177/1535370216643772. PubMed PMID: 27072562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilead A, Neeman M. Dynamic remodeling of the vascular bed precedes tumor growth: MLS ovarian carcinoma spheroids implanted in nude mice. Neoplasia. 1999;1:226–230. doi: 10.1038/sj.neo.7900032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zietarska M, Maugard CM, Filali-Mouhim A, Alam-Fahmy M, Tonin PN, Provencher DM, Mes-Masson AM. Molecular description of a 3D in vitro model for the study of epithelial ovarian cancer (EOC) Mol Carcinog. 2007;46(10):872–885. doi: 10.1002/mc.20315. PubMed PMID: 17455221. [DOI] [PubMed] [Google Scholar]
- 7.Raghavan S, Ward MR, Rowley KR, Wold RM, Takayama S, Buckanovich RJ, Mehta G. Formation of stable small cell number three-dimensional ovarian cancer spheroids using hanging drop arrays for preclinical drug sensitivity assays. Gynecol Oncol. 2015;138(1):181–189. doi: 10.1016/j.ygyno.2015.04.014. PubMed PMID: 25913133; PMCID: 4480341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin RZ, Chang HY. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol J. 2008;3(9–10):1172–1184. doi: 10.1002/biot.200700228. PubMed PMID: 18566957. [DOI] [PubMed] [Google Scholar]
- 9.Grun B, Benjamin E, Sinclair J, Timms JF, Jacobs IJ, Gayther SA, Dafou D. Three-dimensional in vitro cell biology models of ovarian and endometrial cancer. Cell Prolif. 2009;42(2):219–228. doi: 10.1111/j.1365-2184.2008.00579.x. PubMed PMID: 19222485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein OJ, Bhayana B, Park YJ, Evans CL. In vitro optimization of EtNBS-PDT against hypoxic tumor environments with a tiered, high-content, 3D model optical screening platform. Mol Pharm. 2012;9(11):3171–3182. doi: 10.1021/mp300262x. PubMed PMID: 22946843; PMCID: PMC3538815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu YM, Tang J, Zhao P, Chen ZN, Jiang JL. Morphological changes and molecular expressions of hepatocellular carcinoma cells in three-dimensional culture model. Exp Mol Pathol. 2009;87(2):133–140. doi: 10.1016/j.yexmp.2009.07.003. PubMed PMID: 19615356. [DOI] [PubMed] [Google Scholar]
- 12.Sutherland RM, Sordat B, Bamat J, Gabbert H, Bourrat B, Mueller-Klieser W. Oxygenation and differentiation in multicellular spheroids of human colon carcinoma. Cancer Res. 1986;46(10):5320–5329. PubMed PMID: 3756881. [PubMed] [Google Scholar]
- 13.Nath S, Devi GR. Three-dimensional culture systems in cancer research: Focus on tumor spheroid model. Pharmacol Ther. 2016 doi: 10.1016/j.pharmthera.2016.03.013. PubMed PMID: 27063403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai Y, Cheng K, Kisaalita W. Three dimensional neuronal cell cultures more accurately model voltage gated calcium channel functionality in freshly dissected nerve tissue. PLoS One. 2012;7(9):e45074. doi: 10.1371/journal.pone.0045074. PubMed PMID: 23049767; PMCID: 3458113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bryce NS, Zhang JZ, Whan RM, Yamamoto N, Hambley TW. Accumulation of an anthraquinone and its platinum complexes in cancer cell spheroids: the effect of charge on drug distribution in solid tumour models. Chem Commun (Camb) 2009;(19):2673–2675. doi: 10.1039/b902415h. PubMed PMID: 19532917. [DOI] [PubMed] [Google Scholar]
- 16.Liu X, Hummon AB. Quantitative determination of irinotecan and the metabolite SN-38 by nanoflow liquid chromatography-tandem mass spectrometry in different regions of multicellular tumor spheroids. J Am Soc Mass Spectrom. 2015;26(4):577–586. doi: 10.1007/s13361-014-1071-0. PubMed PMID: 25604392; PMCID: PMC4361235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Weaver EM, Hummon AB. Evaluation of therapeutics in three-dimensional cell culture systems by MALDI imaging mass spectrometry. Anal Chem. 2013;85(13):6295–6302. doi: 10.1021/ac400519c. PubMed PMID: 23724927; PMCID: PMC4118837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eetezadi S, De Souza R, Vythilingam M, Lessa Cataldi R, Allen C. Effects of Doxorubicin Delivery Systems and Mild Hyperthermia on Tissue Penetration in 3D Cell Culture Models of Ovarian Cancer Residual Disease. Mol Pharm. 2015;12(11):3973–3985. doi: 10.1021/acs.molpharmaceut.5b00426. PubMed PMID: 26394060. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Hu MG, Pan K, Li CH, Liu R. 3D Spheroid Culture Enhances the Expression of Antifibrotic Factors in Human Adipose-Derived MSCs and Improves Their Therapeutic Effects on Hepatic Fibrosis. Stem Cells Int. 2016;2016:4626073. doi: 10.1155/2016/4626073. PubMed PMID: 27022400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Y, Shi T, Xu A, Zhang L. 3D spheroid culture enhances survival and therapeutic capacities of MSCs injected into ischemic kidney. J Cell Mol Med. 2016 doi: 10.1111/jcmm.12651. PubMed PMID: 26914637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaedtke L, Thoenes L, Culmsee C, Mayer B, Wagner E. Proteomic analysis reveals differences in protein expression in spheroid versus monolayer cultures of low-passage colon carcinoma cells. J Proteome Res. 2007;6(11):4111–4118. doi: 10.1021/pr0700596. PubMed PMID: 17918984. [DOI] [PubMed] [Google Scholar]
- 22.He W, Kuang Y, Xing X, Simpson RJ, Huang H, Yang T, Chen J, Yang L, Liu E, He W, Gu J. Proteomic comparison of 3D and 2D glioma models reveals increased HLA-E expression in 3D models is associated with resistance to NK cell-mediated cytotoxicity. J Proteome Res. 2014;13(5):2272–2281. doi: 10.1021/pr500064m. PubMed PMID: 24742303. [DOI] [PubMed] [Google Scholar]
- 23.Chan YH, Huang TW, Young TH, Lou PJ. Human salivary gland acinar cells spontaneously form three-dimensional structures and change the protein expression patterns. J Cell Physiol. 2011;226(11):3076–3085. doi: 10.1002/jcp.22664. PubMed PMID: 21302307. [DOI] [PubMed] [Google Scholar]
- 24.Levin VA, Panchabhai S, Shen L, Baggerly KA. Protein and phosphoprotein levels in glioma and adenocarcinoma cell lines grown in normoxia and hypoxia in monolayer and three-dimensional cultures. Proteome Sci. 2012;10(1):5. doi: 10.1186/1477-5956-10-5. PubMed PMID: 22276931; PMCID: 3317865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yue XS, Hummon AB. Combination of multistep IMAC enrichment with high-pH reverse phase separation for in-depth phosphoproteomic profiling. J Proteome Res. 2013;12(9):4176–4186. doi: 10.1021/pr4005234. PubMed PMID: 23927012. [DOI] [PubMed] [Google Scholar]
- 26.Oliveros JC. VENNY. An interactive tool for comparing lists with Venn Diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html. [Google Scholar]
- 27.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. PubMed PMID: 22455463; PMCID: PMC3339379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–D815. doi: 10.1093/nar/gks1094. PubMed PMID: 23203871; PMCID: PMC3531103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poland J, Sinha P, Siegert A, Schnolzer M, Korf U, Hauptmann S. Comparison of protein expression profiles between monolayer and spheroid cell culture of HT-29 cells revealed fragmentation of CK18 in three-dimensional cell culture. Electrophoresis. 2002;23(7–8):1174–1184. doi: 10.1002/1522-2683(200204)23:7/8<1174::AID-ELPS1174>3.0.CO;2-O. PubMed PMID: 11981867. [DOI] [PubMed] [Google Scholar]
- 30.Kang S, Warner MD, Bell SP. Multiple functions for Mcm2–7 ATPase motifs during replication initiation. Mol Cell. 2014;55(5):655–665. doi: 10.1016/j.molcel.2014.06.033. PubMed PMID: 25087876; PMCID: PMC4156824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahlf DR, Masyuko RN, Hummon AB, Bohn PW. Correlated mass spectrometry imaging and confocal Raman microscopy for studies of three-dimensional cell culture sections. Analyst. 2014;139(18):4578–4585. doi: 10.1039/c4an00826j. PubMed PMID: 25030970. [DOI] [PubMed] [Google Scholar]
- 32.Ahlf Wheatcraft DR, Liu X, Hummon AB. Sample preparation strategies for mass spectrometry imaging of 3D cell culture models. J Vis Exp. 2014;(94) doi: 10.3791/52313. PubMed PMID: 25549242; PMCID: PMC4396945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feist PE, Sun L, Liu X, Dovichi NJ, Hummon AB. Bottom-up proteomic analysis of single HCT 116 colon carcinoma multicellular spheroids. Rapid Commun Mass Spectrom. 2015;29(7):654–658. doi: 10.1002/rcm.7150. PubMed PMID: 26212283; PMCID: PMC4763982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keithley RB, Weaver EM, Rosado AM, Metzinger MP, Hummon AB, Dovichi NJ. Single cell metabolic profiling of tumor mimics. Anal Chem. 2013;85(19):8910–8918. doi: 10.1021/ac402262e. PubMed PMID: 24011091; PMCID: PMC3816011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LaBonia GJ, Lockwood SY, Heller AA, Spence DM, Hummon AB. Drug penetration and metabolism in 3D cell cultures treated in a 3D printed fluidic device: assessment of irinotecan via MALDI imaging mass spectrometry. Proteomics. 2016;16(11–12):1814–1821. doi: 10.1002/pmic.201500524. PubMed PMID: 27198560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Hummon AB. Imaging mass spectrometry of three-dimensional cell culture systems. Anal Chem. 2011;83(22):8794–8801. doi: 10.1021/ac202356g. PubMed PMID: 21992577. [DOI] [PubMed] [Google Scholar]
- 37.Liu X, Hummon AB. Mass spectrometry imaging of therapeutics from animal models to three-dimensional cell cultures. Anal Chem. 2015;87(19):9508–9519. doi: 10.1021/acs.analchem.5b00419. PubMed PMID: 26084404; PMCID: PMC4766864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weaver EM, Hummon AB. Imaging mass spectrometry: from tissue sections to cell cultures. Adv Drug Deliv Rev. 2013;65(8):1039–1055. doi: 10.1016/j.addr.2013.03.006. PubMed PMID: 23571020. [DOI] [PubMed] [Google Scholar]
- 39.Weaver EM, Hummon AB, Keithley RB. Chemometric analysis of MALDI mass spectrometric images of three-dimensional cell culture systems. Anal Methods. 2015;7(17):7208–7219. doi: 10.1039/C5AY00293A. PubMed PMID: 26604989; PMCID: PMC4654961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braverman NE, Moser AB. Functions of plasmalogen lipids in health and disease. Biochim Biophys Acta. 2012;1822(9):1442–1452. doi: 10.1016/j.bbadis.2012.05.008. PubMed PMID: 22627108. [DOI] [PubMed] [Google Scholar]
- 41.Hashimoto M, Nasser H, Chihara T, Suzu S. Macropinocytosis and TAK1 mediate anti-inflammatory to pro-inflammatory macrophage differentiation by HIV-1 Nef. Cell Death Dis. 2014;5:e1267. doi: 10.1038/cddis.2014.233. PubMed PMID: 24874739; PMCID: PMC4047869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kouwenhoven A, Minassian VD, Marsh JW. HIV-1 Nef mediates Pak phosphorylation of Mek1 serine298 and elicits an active phospho-state of Pak2. Curr HIV Res. 2013;11(3):198–209. doi: 10.2174/1570162x113119990039. PubMed PMID: 23746211. [DOI] [PubMed] [Google Scholar]
- 43.Dong JM, Lau LS, Ng YW, Lim L, Manser E. Paxillin nuclear-cytoplasmic localization is regulated by phosphorylation of the LD4 motif: evidence that nuclear paxillin promotes cell proliferation. Biochem J. 2009;418(1):173–184. doi: 10.1042/BJ20080170. PubMed PMID: 18986306. [DOI] [PubMed] [Google Scholar]
- 44.Roa S, Li Z, Peled JU, Zhao C, Edelmann W, Scharff MD. MSH2/MSH6 complex promotes error-free repair of AID-induced dU: G mispairs as well as error-prone hypermutation of A:T sites. PLoS One. 2010;5(6):e11182. doi: 10.1371/journal.pone.0011182. PubMed PMID: 20567595; PMCID: PMC2887398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.