Abstract
Background
Advances in DNA sequencing technology have resulted in an abundance of personalized data with challenging clinical utility and meaning for clinicians. This wealth of data has potential to dramatically impact the quality of healthcare. Nurses are at the focal point in educating patients regarding relevant healthcare needs; therefore, an understanding of sequencing technology and utilizing these data are critical.
Aim
The objective of this paper is to explicate the role of nurses and nurse scientists as integral members of healthcare teams in improving understanding of DNA sequencing data and translational genomics for patients.
Approach
A history of the nurse role in newborn screening is used as an exemplar.
Discussion
This paper serves as an exemplar on how genome sequencing has been utilized in nursing science and incorporates linkages of other omics approaches used by nurses that are included in this special issue. This special issue showcased nurse scientists conducting multi-omic research from various methods, including targeted candidate genes, pharmacogenomics, proteomics, epigenomics and the microbiome. From this vantage point, we provide an overview of the roles of nurse scientists in genome sequencing research and provide recommendations for the best utilization of nurses and nurse scientists related to genome sequencing.
Keywords: genetic research, genetic testing, genome, nurses, precision medicine
Nurses are uniquely qualified to examine and understand patient reports based on DNA sequencing and to translate genomic information to patients and their families which can dramatically improve health outcomes. Recent advances have made it economically and technically feasible to employ high throughput DNA sequencing to identify potentially life-changing or life-altering therapies for individuals at high risk for illnesses with a strong underlying genetic basis. However, the path between raw sequence data and direct clinical utility presents a significant challenge. Content within this special issue includes examples of how nurses are working to bridge the gap from omics research results to clinical application.
DNA sequencing technologies are already in routine use in the clinic, and nurses play a crucial role in turning genetic information into concrete action plans for their patients. In the coming years the onslaught of information which will be reaching the bedside and the clinic will become a literal tidal wave. In order to give the best care possible to their patients, nurses need to understand this crucial information extremely well and be adequately prepared to support patients and families in using the new information to bring about the best possible patient care outcomes. Additionally, it is imperative that nurses consider how the omics data deluge will influence nursing research in order to develop new knowledge relevant to nursing practice. This paper will show how genome sequencing has been utilized in nursing science and will incorporate linkages other omics approaches used by nurses that are included in this special issue. This Nursing Research special issue on Omics in Nursing Science showcases nurse scientists conducting multi-omic research from various methods such as targeted candidate genes (Correa-Rodriguez, Rio-Valle & Blanca, 2017; Zahari et al,. 2017; Smoot et al., 2017; Han et al., 2017; Gillespie et al., 2017; Lin, Nunez, Johns & Shaio, 2017; Gonzales, Yu, & Shaio, 2017), pharmacogenomics (Aroke, Crawford, Dungan, 2017), proteomics (Voss, et al., 2017), epigenomics (Braid, Okrah, Shetty & Bravo, 2017), and the microbiome (Cong et al., 2017; Jordan et al., 2017; Ames, Ranucci, Moriyama & Wallen, 2017), and lastly, we provide an overview of the role of nurse scientists in genome sequencing here in this paper. We hope to inspire nurses to continue utilizing innovative omics approaches in their research, consider where the field of nursing research may be heading, and envision the use of multi-omic strategies as the basis for development of useful interventions and subsequent translation for nursing practice in the US and abroad.
Newborn Screening— A Framework for Genome Based Population Screening
Population-based genetic screening has great potential to improve health outcomes for individuals and health care costs for society. A striking example exemplifying this concept is mandated newborn screening, most of which involves testing the blood of newborns blood to identify disorders that are not readily observable upon physical exam shortly after birth (Association of Women’s Health, Obstetric and Neonatal Nurses [AWHONN], 2010; Berry, 2015). Newborn screening is known to bring great benefit to children and is considered without question to be cost effective in our current era of healthcare cost management (Centers for Disease Control and Prevention, 2008; Geelhoed, Hounsome & O’Leary, 2005). Newborn screening, which includes screening for various genetic disorders, is currently a standard of care and reimbursed in all 50 US states as well as many other countries around the world. The impact and value of newborn screening in the improvement of health care outcomes in populations around the world is without question due to the improved quantity and quality of lives of the infants identified with conditions that require early treatment for healthy development.
A brief review of the history of newborn screening and the roles nurses play in delivering life changing and life-saving care is instructive. Newborn screening first began in the 1950s based on a growing understanding of phenylketonuria (PKU), an inborn error in metabolism (Paul & Brosco, 2013). This autosomal recessive disorder that results in decreased levels of they enzyme that converts phenylalanine to tyrosine and affects approximately 1 in 15,000 children in the US. The enzyme deficiency present in individuals with PKU was first elucidated in the 1930s in Norway. By the 1950s research had led to the suggestion that a strict limitation on the amount of phenylalanine in the diet could completely reverse the effects of the genetic mutation. However, in order to be effective in preventing detrimental effects related to the enzyme deficiency this dietary based therapy must begin as soon after birth as possible. Furthermore, the dietary change could not simply eliminate phenylalanine from the diet since this amino acid is essential for life but rather provide the child with a diet very low in phenylalanine but adequate to allow normal amounts of protein synthesis. In the 1950s, a test for identifying infants with PKU using ferric chloride which reacts to the urine in a baby’s diaper was put into use. This test allowed infants with PKU to be identified shortly after birth in order to evaluate the effectiveness of early dietary intervention for PKU (Groves & Scholesser, 1964; Ragsdale & Koch, 1964).
Nurses are integral to the interdisciplinary teams that work with families for the implementation of this life changing intervention (Steele, 1989). The low phenylalanine diet was an intervention that did not fix everything in a single visit to the office but rather required the regular and engaged interaction between parents and their healthcare providers over the course of the entire growth to maturity of the child. The nurse was a pivotal player in the team supporting the family in efforts to save the child from serious brain damage. As dietary intervention proved to be so completely successful in PKU, it soon became a mainstay in nursing education for students to gain an intimate understanding of PKU and its therapy. As time moved on more efficient methods of screening for PKU were developed. For some period of time the innovation that changed newborn screening methods was a test based on bacterial growth that would allow the estimation of phenylalanine levels from a drop a blood from a heelstick on a filter paper disc. This “Guthrie card” method of screening newborns (named after the innovator of the bacterial growth test) soon became standard of care. Additional inborn errors in metabolism with corresponding effective interventions could be detected by the bacterial growth assay, but the next significant breakthrough came with the implementation of mass spectrometer based readouts from the Guthrie card. Now, a large number of metabolites could be quantitated in a single rapid and automated reading procedure. The number of tests that could now be read by a centralized laboratory and an automated report generated numbered in the tens or even hundreds of thousands per year. The impact of this discovery at point-of-care of this expansion were substantial. Nurses, are and have been critical in the counseling and teaching functions of providing parents with guidance and a resource in providing this form of positive health benefit were faced with an increasing challenge of diseases to become familiar with and therapeutic interventions to understand and develop. It is the multiplication of these challenges beyond the current form of newborn screening that we believe nurses will need to meet in the coming years. The reasons these new forms of information, intervention and therapy will without question enter the healthcare arena are based on the principles that currently guide newborn screening.
The principles which underlie newborn screening are that health providers bear a responsibility to identify and make available interventions which are life saving or may substantially improve the lives of infants with disorders detected through newborn screening. The first criterion that must be met for a condition to be included in a newborn screening panel is that the condition must be actionable (i.e., a definitive positive test result is directly associated with a clinical intervention that extends life and/or substantially improves the health of the infant). Beyond this basic principle there is considerable variability in the number of disorders tested for, how long samples are kept, and protocol for disseminating results. The protocols for each of these issues vary on a state-by-state basis in the US (see Table 1; Supplemental Digital Content 1) and a country-by-country basis worldwide.
TABLE 1.
Newborn Screening Protocol Variation in the 50 US States
| Measure | Range | n | (%) |
|---|---|---|---|
| Disorders screened (number) | 29−57a | ||
| Bloodspot storage (time) | ≥ 42 daysb | 50 | (100) |
| 0–6 monthsc | 16 | (32) | |
| 8–12 monthsd | 11 | (22) | |
| 1.5–5.0 years | 10 | (20) | |
| >5 yearse | 8 | (16) | |
| Not specified | 5 | (10) | |
| Research allowed on spots | |||
| No | 16 | (32) | |
| Yes | 23 | (43) | |
| Not specified/under review | 12 | (24) |
Note. Information based on data obtained from Baby’s First Test (2015) links to Public Health Departments of each State.
M = 43.4.
Includes indefinite storage.
Includes states that store “until testing complete.
Storage times may be longer for positive results.
Includes states that store bloodspots indefinitely.
There is now a burgeoning movement to incorporate whole-genome sequencing methods, which reads the entire genome of an individual, into the newborn screening process. Sequencing technologies that read small regions of an individual’s genome are already used for confirmatory and follow-up testing related to positive or ambiguous results from initial mass spectrometer based screening. Assessment of the analytic performance of using sequencing technologies as the initial screening test has determined results are comparable to mass spectrometry with fewer false positive (Baker et al., 2016; Bodian et al., 2016).
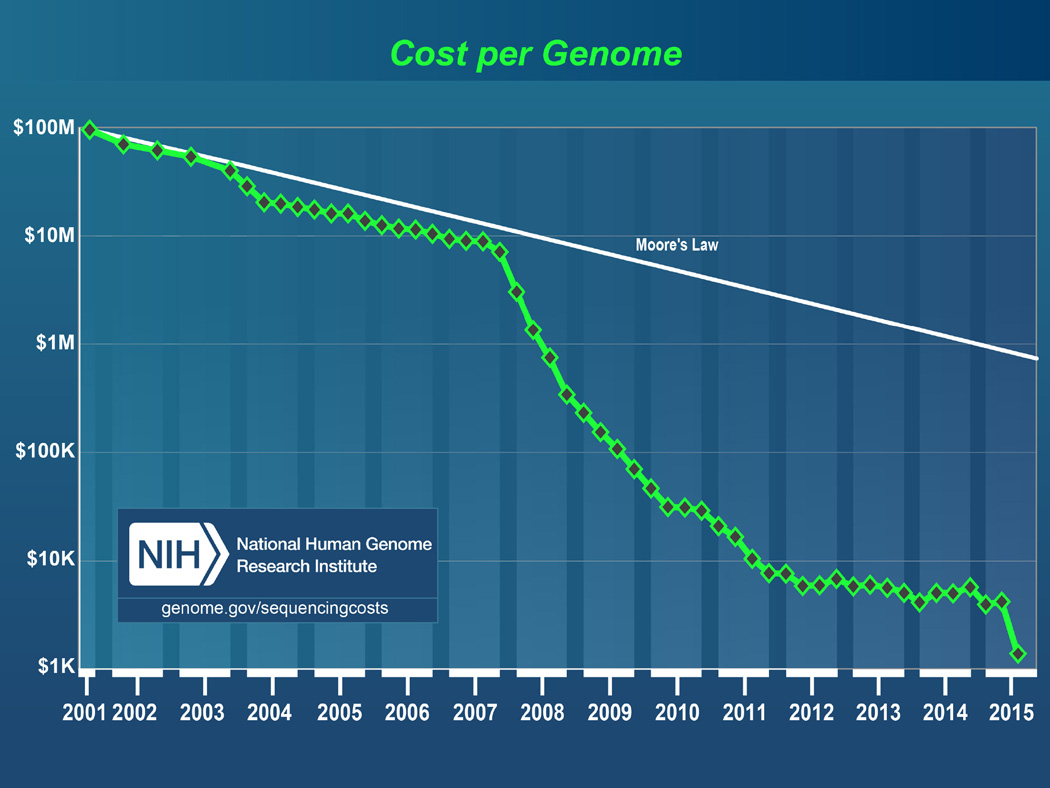
Testing for small molecules, such as proteins or other metabolites, created an economically feasible way to screen for many conditions at once because technology (mass spectrometry) detects molecules produced by targeted conditions (e. g., PKU, maple syrup disease). This testing format allows screening a group of conditions together, but would not be economically justified to screen only one at a time. Screening in newborns or at the population level in adults based on DNA sequencing has not been feasible until recently due to the relatively high cost. However, the cost of sequencing technologies has dramatically decreased (Figure 1) (Wetterstrand, 2015); several ongoing studies are focused on determining the best way to incorporate these approaches into standard care across the lifespan (www.genomes2people.org/g2p/). Since there is broad applicability across the lifespan, these technologies will be able to improve our understanding of etiology and care for both common and rare diseases. For example, cardiovascular disease occurring consequential to partial loss of function in the LDL receptor gene is estimated to have a frequency of 1 in 200 individuals, which is relatively common, and has a well-established therapeutic intervention (prescription of pharmacologic therapies such as use of statins or other medications based on an individual’s unique genomic profile) could be clearly identified and treated earlier. Sequencing technologies will also drive the identification of relatively rare but nevertheless life threatening conditions such as familial hypercholesterolemia (Wright, Housman & Taylor, 2016) and familial hemophagocytic lymphohistocytosis, for example.
FIGURE 1.
Cost for sequencing whole genomes over time. Source: National Human Genome Research Institute (genome.gov/sequencingcosts).
For these reasons, as the cost of sequencing continues to drop and the rationale for it continues to become more and more compelling, we believe that whole genome sequencing for screening and diagnostic purposes will become routine. Sequencing data will make a dramatic difference in the choice of therapeutic intervention appropriate to treat or forestall serious negative health outcomes. Nurses will often be implementing therapeutic interventions derived from sequence information from a genetic screening test. A strong understanding of the connection between the sequencing result and the individuals therapy based on that sequencing result will be essential for the most effective nurse-provided care and support for patients.
The International Society of Nurses in Genetics (ISONG) (International Society of Nurses in Genetics, 2012) issued a position statement on the role of nurses in the newborn screening process in 2012, which recommends the following for nursing:
Active participation in current knowledge of NBS as an advocate for patients and families,
Provide patient education prior to testing for NBS,
Offer counseling to families post testing,
Explain use of results, and if being utilized in research studies, explain their benefits, and
Include information about NBS in newborn care curriculum for patient education.
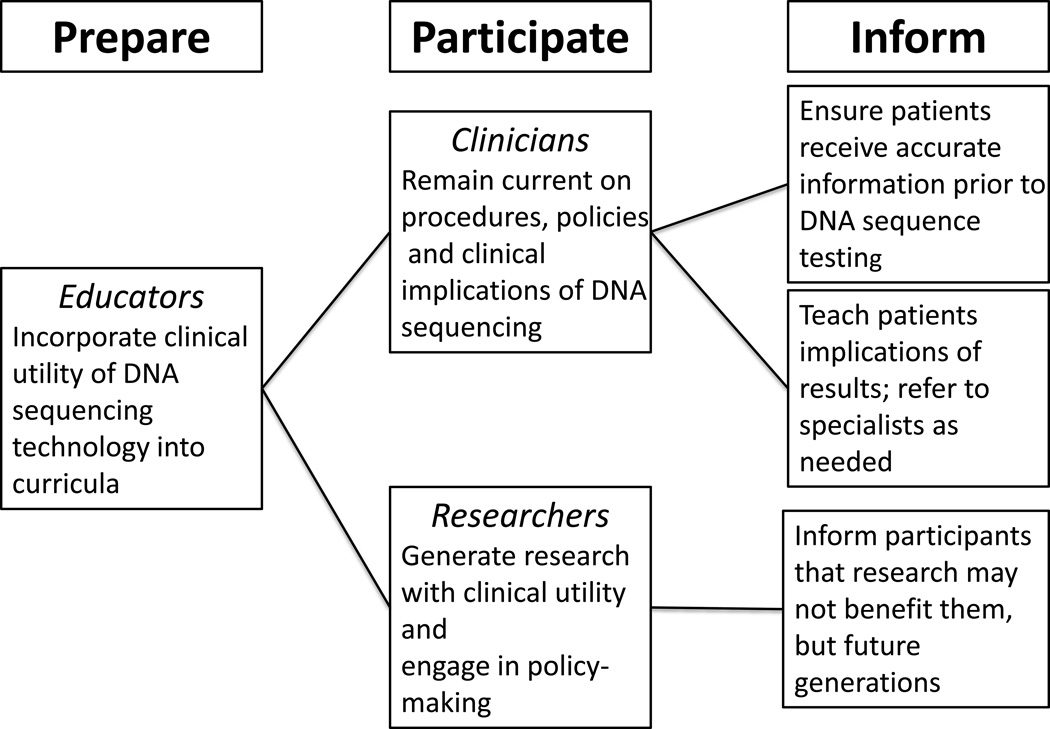
We have adapted key features of the ISONG newborn screening position statement to propose how nurses and nurse scientists can broadly utilize DNA sequencing technology, participate in policy development, translate findings obtained from these methods, and ensure adequate education and dissemination of information to patients and the public (Figure 2). Nurses have the knowledge and skills to be involved with clinical care that incorporates results from sequencing technologies, and must stay abreast of new developments to provide competent care for diverse patient populations. First, we must incorporate advances into nursing curricula so that clinical nurses and nurse scientists have the knowledge to develop and incorporate innovative interventions into practice to improve or maintain patient health. Ideally, there will be an exchange of information between nurse clinicians and nurse scientists so that new knowledge can be generated to serve patients most in need. For example, individuals of African Ancestry in the US suffer disproportionately from poor cardiovascular outcomes. Familial hypercholesterolemia is a genetic condition that can result in severe dysfunction at an early age. However, many of the genetic variants that have been identified are not present in patients of African ancestry and much of the sequencing research to date has been conducted in populations of European ancestry (Wright, Housman, & Taylor, 2016). Communication between clinicians and researchers will be vital in areas such as these to identify disease variants that are not consistent to realize the true potential of personalized medicine.
FIGURE 2.
Proposed roles of clinical nursing in DNA sequencing.
The Current Policy Landscape
Currently, there are two policy statements intended to establish guidelines for testing and reporting genome sequencing results to patients in the clinical setting. First, the American College of Medical Genetics and Genomics (ACMG Board of Directors, 2014; Green et al., 2013) recommended that testing be conducted by a trained healthcare professional. Additionally, when whole genome sequencing is conducted, patients must be clearly informed about all “actionable” variants that are tested, and the opportunity to opt out of this testing is provided (for both adults and children). (Actionable variants are mutations that are deleterious to health and have specific, evidence-based recommendations that can be implemented to improve health outcomes in the affected individual.) Currently, guidance on how and whether “incidental” or “secondary” findings should be reported remains ambiguous. A second statement on genome sequencing published by the Association of Genetic Nurses and Counsellors (AGNC) in the United Kingdom and Ireland (Middleton et al., 2014) is harmonious with the ACMG statement, but takes the issue of incidental findings a step further. Two striking differences between the statements are : (a) the AGNC refers to genome sequencing screening as “opportunistic” and cautions that screening should be done when benefits are judged to outweigh harms, and (b) the AGNC does not recommend screening children for ”adult onset” diseases.
As nurse scientists and genetic counselors consider the use of genome sequencing in clinical and community settings, it is important to consider the usefulness of this technology as a screening tool and how it has been used in the past to identify heritable disorders in programs such as prenatal screening. In 2012, the Genomic Nursing State of the Science Advisory Panel was held at the National Institutes of Health and made recommendations that research should focus on building capacity for using genomic information in a clinically meaningful way to reduce costs and improve health outcomes in diverse patient populations (Calzone et al., 2013).
Additionally, there is an increasing demand from the public for information regarding genetic predisposition for disease. However, the reliability, accuracy, and clinical manifestations associated with genetic variants identified by whole genome sequencing remain largely unknown because research studies focused on affected individuals do not evaluate population-based data or individuals with the variant who are unaffected (Williams, Cashion, & Veenstra, 2015). As public policy statements evolve, the practical implications of new developments in DNA sequencing for nurses will include the need to keep curricula in nursing education programs up-to-date (Calzone et al., 2013; Conley et al., 2015) as new sequencing and therapeutic modalities based on this screening are brought into practice (Wright, Housman & Taylor, 2016). For nurses already in practice, it is critical that in-service education programs be made available so that nurses will have useful resources to integrate the new information into their day-to-day nursing activities effectively.
Clear, effective and ethical communication of the relevance and impact of the results of these technologies is the most significant barrier to development of potentially life-saving therapeutic intervention strategies and dramatic overall improvements in health. Nurses should also be aware of the classification categories put out by the AMCG on how to interpret sequence variant data and the decision tree for use of these data in clinical care (Richards et al., 2008; Richards et al., 2015) (Table 2). Once DNA sequence variants are classified in the laboratory, the clinical report is created and submitted to healthcare personal for review and further follow-up and/or discussion with individuals and families for clinical use. By developing, evaluating and improving the use of sequencing technology to achieve this clinical utility, nurses are well-situated to change the landscape of precision healthcare by using sequencing to develop genomic screening tools for widespread implementation of population based use. Because nurses are at the forefront of patient interactions it seems only fitting that nurse scientists lead the charge in genomic research that can be translated from the bench-to-bedside and be the agent for change in health policy related to genomic screening recommendations worldwide.
TABLE 2.
Classification Categories for Sequence Dataa
| Category | Definition |
|---|---|
| Pathogenic | Previously reported and recognized as cause of the disorder. |
| Likely pathogenic | Previously unreported but expected to cause the disorder. |
| Variant: unknown significance | Previously unreported and may or may not cause the disorder. |
| Likely benign | Previously unreported and probably not causative of disease |
| Benign | Previously reported and is recognized as a neutral variant. |
| Variant of unknown significance—suspicious |
Not known or expected to be causative of disease, but is found to be associated with a clinical presentation. |
From the American College of Medical Genetics and Genomics (ACMG); see Richards et al. (2008).
Discussion
The development of increasingly powerful technology such as genome sequencing to understand and decode the human genome sequence is now having a dramatic and expanding impact on the ability to deliver effective and precisely targeted healthcare. Nurses are uniquely poised to carry out a critical role in this healthcare revolution. It is crucial that nurses are able to provide patients with a clear and understandable rationale for the therapeutic interventions dictated by genomic analysis and to counsel wisely as questions arise from patients and families in the course of treatment.
Based on our review of current policy, we would like to comment on two issues presented by the AGNC statement on opportunistic genome sequencing screening. Providing a label for a screening tool utilizing genome-screening technology as ‘opportunistic’ provides a negative connotation to testing and may be off-putting to patients when considering testing. Furthermore, genetic screening using sequencing may soon become routine practice and in fact adopted as standard of care. We recommend that the term opportunistic be restricted to its narrow sense use—the adventitious discovery of clinically significant information about one condition when sequencing is performed motivated by an effort to assign a genetic basis to another condition. If patients are to be educated and informed about the usefulness or lack thereof for genome sequencing, it should be made on an individual basis based on the premise of precision medicine initiatives and not on fear or negativity. The AGNC recommendation to not screen children for “adult-onset” diseases is challenging to understand or implement in its current form. There are many genetically-based conditions that are best addressed by early therapeutic intervention or clinical screening well prior to the age of 21, yet “onset” may be defined as occurring in early adulthood. These artificial distinctions related to “adult onset” diseases must be changed in our view, to relate to the point in life at which early intervention is clinically important. Clinical practice and scientific research in pediatrics has shown that disorders once thought to be ‘adult onset’ are increasingly being diagnosed in childhood (e.g., diabetes, hypertension, dyslipidemia, obesity), and genetic risk variants have been identified in children as young as three (Taylor, Sampson, Anderson, Caldwell, & Taylor, 2012; Taylor, Sun, Hunt, & Kardia, 2010). In fact, the prenatal screening program in the United States has been widely successful in screening and treating newborns for disorders that would otherwise be undiagnosed and ultimately saves lives of these children. To encourage clinical scientists to ignore the possibility of ignoring the heritable risks for development of disorders in children simply because it has yet to phenotypically express would be a missed opportunity to promote health and provide early interventions based on the child’s individual genetic profile. Although next generation sequencing is currently being used in clinical trials such as in the BabySeq (www.genomes2people.org/babyseqproject/) and the Clinical Sequencing Exploratory Research (CSER) consortium (https://cser-consortium.org/) and may be useful as a prediagnostic tool for use in clinical care, ethical issues remain (Lantos, 2016) and the challenge of data analytics on approximately 6 billion base pairs using whole genome sequencing (Wang, Lu, & Zhao, 2015) on each individual can be daunting.
Implications for Nurses and Nurse Scientists
Current clinical usage of genome sequencing technologies is confined to circumstances in which there is an index of suspicion that a genetic basis exists for the clinical phenotype exhibited by a particular patient. A potential impact of such technologies in healthcare is the identification of genetically vulnerable individuals through screening programs for whom intervention prior to the onset of pathology can delay or prevent a pathological condition. Although we recognize that screening itself is imperfect, it is the first necessary step in confirmatory testing and diagnostics. While the cost of screening programs based on whole genome sequencing prohibits their current use, as the cost of genome sequencing continues to decrease and the number of detectable and preventable conditions increases, the crossover point to economic viability of this methodology is inevitable. Because nurses are educated in genetics/genomics it seems as if they could play a more pivotal role with hands-on involvement in conducting genetic screening, analyzing results, and conducting the needed interventions and counseling for the families. Underutilization of nurses in genetic testing and patient education is a missed opportunity for health professionals and their patients. Nurses are especially astute at performing health histories, conducting testing, and providing needed health promotion/risk reduction education to patients. When nurses are utilized in this health promotion capacity, it will expand multi-disciplinary teams to identify areas where we can provide education to prevent exposures to specific risks based on an individual’s genomic profile. Translational clinical research must capitalize on what makes some people with a variant phenotypically express the disease and while others do not. More than 70–90% of chronic diseases are multifactorial and have strong environmental components (Yoon, Bastian, Anderson, Collins, & Jaffe, 2014). Nurses excel in patient teaching and are experts in creating plans of care that focus on health promotion. Nurses need to be more involved in every aspect of genetic testing such as sequencing and delivery of results in order to provide patients with the best possible information and strategies to improve their health.
Current recommendations and statements by the ACMG highlight some of the pros and cons of using genome sequencing as a screening tool. However, nurse scientists with a background in genomics and/or genetic epidemiology may be expertly positioned to lead in the area of genome sequencing and patient care in two very important areas. Advances in analytical protocols and processes for DNA sequence missense mutation variances (for example, MacArthur et al., 2014) and laboratory informatics linked with decision support (for example, Jones, Johnson, & Batstone, 2014) are moving genome sequencing to clinical care. These protocols will serve as the basis for screening individuals with genome sequencing and guide the laboratory processes and confirmatory analyses required for reporting results to patients in an ethical, legal and socially responsible manner via genetic nurse scientists and counselors. Currently, there are programs available that train nurse scientists in complex genomic data analysis and big data computation, including the National Institute of Nursing Research Big Data Bootcamp (www.ninr.nih.gov/training/trainingopportunitiesintramural/bootcamp) and the National Heart, Lung, and Blood Institute Programs to Increase Diversity Among Individuals Engaged in Health-Related Research (PRIDE) (www.nhlbi.nih.gov/research/training/PRIDE-research-programs). These programs may aid in positioning attendees for work in these areas of computation of next generation sequencing data for clinical use. By supporting and being an integral part of genome sequencing screening in clinical, community and resource-poor areas, genomic nurse scientists and counselors will make a great contribution to reducing morbidity and mortality among many vulnerable and underserved populations around the world in the long-term.
Conclusion
In addition to the education, advocacy and counseling roles, nurses’ frequent and direct interactions with patients provides ample opportunity to effectively translate powerful genomic technologies into positive health outcomes for the American population, and other populations worldwide. Currently, there are training programs available that educate nurse scientists in complex genomic data analysis and big data computation (NINR Big Data Bootcamp; NINR “Precision Health: From Omics to Data Science Bootcamp; NHLBI PRIDE programs). These programs are important resources for nurse scientists who aspire to respond to the challenge of analyzing large amounts of genomic data that multi-omic methods bring and then using those data for development of clinical interventions. Nurses also play a key role in bringing the benefits of genome-sequence based screening to the clinic by supporting the patient in understanding how DNA sequencing based findings can lead to therapeutic intervention which will lead to improved health over a lifetime and ultimately save lives.
Supplementary Material
Acknowledgments
Funding: Massachusetts Institute of Technology, Martin Luther King, Jr. Visiting Scholars Program (JYT & DEH). This work was also partially supported by the National Institutes of Health, National Institute for Nursing Research R01NR013520 (JYT).
Footnotes
Author Contributions: JYT conceptualized and drafted the manuscript; MLW, KTH and DEH added critical content and revisions. All authors approved the final manuscript.
The authors have no conflicts of interest to report.
Supplemental Digital Content 1. Table illustrating protocols on a state-by-state basis in the US. doc
Contributor Information
Jacquelyn Y. Taylor, Diversity and Inclusion, School of Nursing, Yale University, Orange, CT.
Michelle L. Wright, School of Nursing, Yale University, Orange, CT.
Kathleen T. Hickey, School of Nursing, Columbia University, New York, NY.
David Housman, D. K. Ludwig Scholar for Cancer Research, Department of Biology, Massachusetts Institute of Technology, Cambridge, MA.
References
- ACMG Board of Directors. ACMG policy statement: Updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genetics in Medicine. 2014;17:68–69. doi: 10.1038/gim.2014.151. [DOI] [PubMed] [Google Scholar]
- Ames NJ, Ranucci A, Moriyama B, Wallen GR. The human microbiome and understanding the 16SrRNA fene in translational nursing science [Review] Nursing Research. 2017;66:xx–xx. doi: 10.1097/NNR.0000000000000212. doi: xxx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroke EN, Crawford SL, Dungan JR. Pharmacogenetics of ketamine-induced emergence phenomena: A pilot study. Nursing Research. 2017;66:xx–xx. doi: 10.1097/NNR.0000000000000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Association of Women’s Health, Obstetric and Neonatal Nurses (AWHONN) Newborn screening [Position Statement] Journal of Obstetric, Gynecologic & Neonatal Nursing. 2011;40:136–137. [Google Scholar]
- Baby’s First Test. About newborn screening. What your state offers. 2015 Retrieved from http://www.babysfirsttest.org/newborn-screening/about-newborn-screening. [Google Scholar]
- Baker MW, Atkins AE, Cordovado SK, Hendrix M, Earley MC, Farrell PM. Improving newborn screening for cystic fibrosis using next-generation sequencing technology: A technical feasibility study. Genetics in Medicine. 2016;18:231–238. doi: 10.1038/gim.2014.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry SA. Newborn screening. Clinics in Perinatology. 2015;42:441–453. doi: 10.1016/j.clp.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Bodian DL, Klein E, Iyer RK, Wong WSW, Kothiyal P, Stauffer D, Solomon BD. Utility of whole-genome sequencing for detection of newborn screening disorders in a population cohort of 1,696 neonates. Genetics in Medicine. 2016;18:221–230. doi: 10.1038/gim.2015.111. [DOI] [PubMed] [Google Scholar]
- Braid SM, Okrah K, Shetty A, Bravo HC. DNA methylation patterns in cord blood across gestation age: Association with cell-type proportions. Nursing Research. 2017;66:xx–xx. doi: 10.1097/NNR.0000000000000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzone KA, Jenkins J, Bakos AD, Cashion AK, Donaldson N, Feero WG, Webb JA. A blueprint for genomic nursing science. Journal of Nursing Scholarship. 2013;45:96–104. doi: 10.1111/jnu.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Impact of expanded newborn screening—United Status, 2006. Morbidity and Mortality Weekly Report. 2008;57:1012–1015. Retrieved from www.cdc.gov/mmwr/preview/mmwrhtml/mm5737a2.htm. [PubMed] [Google Scholar]
- Cong X, Judge M, Xu W, Diallo A, Janton S, Brownell EA, Maas K, Graf J. Influence of feeding type on gut microbiome development in hospitalized preterm infants. Nursing Research. 2017;66:xx–xx. doi: 10.1097/NNR.0000000000000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley YP, Heitkemper M, McCarthy D, Anderson CM, Corwin EJ, Daack-Hirsch S, Voss J. Educating future nurse scientists: Recommendations for integrating omics content in PhD programs. Nursing Outlook. 2015;63:417–427. doi: 10.1016/j.outlook.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa-Rodríguez M, Rio-Valle JS, Blanca BR-M. RANKL/RANK/OPG polymorphisms and heel quantitative ultrasound in young adults. Nursing Research. 2017;66:xx–xx. doi: 10.1097/NNR.0000000000000202. [DOI] [PubMed] [Google Scholar]
- Geelhoed EA, Lewis B, Hounsome D, O’Leary PO. Economic evaluation of neonatal screening for phenylketonuria and congenital hypothyroidism. Journal of Paediatrics and Child Health. 2005;41:575–579. doi: 10.1111/j.1440-1754.2005.00725.x. [DOI] [PubMed] [Google Scholar]
- Gillespie SL, Neal JL, Christian LM, Szalacha LA, McCarthy DO, Salsberry PJ. Interleukin-1 receptor antagonist polymorphism and birth timing: Pathway analysis among African American women. Nursing Research. 2017;66:xx–xx. doi: 10.1097/NNR.0000000000000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales MC, Yu P, Shiao SPK. MTHFR gene polymorphism-mutations and air pollution as risk factors for breast cancer: A meta-prediction study. Nursing Research. 2017;66:xx–xx. doi: 10.1097/NNR.0000000000000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, Biesecker LG. ACMG Recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genetics in Medicine. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves R, Schloesser PT. A state program to control phenylketonuria. American Journal of Nursing. 1964;64(8):74–77. [PubMed] [Google Scholar]
- Han CJ, Kohen R, Jun S, Jarrett ME, Cain KC, Burr R, Heitkemper MH. COMT Val158Met polymorphism and symptom improvement following a cognitively-focused intervention for irritable bowel syndrome. Nursing Research. 2017;66:xx–xx. doi: 10.1097/NNR.0000000000000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Society of Nurses in Genetics. Newborn screening: The role of the nurse [Position Statement] 2012 Retrieved from http://www.isong.org/pdfs2013/PS_Newborn_Screenings.pdf. [Google Scholar]
- Jones RG, Johnson OA, Batstone G. Informatics and the clinical laboratory. Clinical Biochemist. Reviews. 2014;35:177–192. Retrieved from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4204239/ [PMC free article] [PubMed] [Google Scholar]
- Jordan S, Baker B, Dunn A, Edwards S, Ferranti E, Mutic AD, Rodriguez J. Maternal-child specimen microbiome collection, storage, and implications for research and practice [Review] Nursing Research. 2017;66:xx–xx. doi: 10.1097/NNR.0000000000000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantos JD. Introduction to Bioethics Special Supplement V: Ethical Issues in Genomic Testing of Children. Pediatrics. 2016;137(Suppl. 1):S1–S2. doi: 10.1542/peds.2015-3731B. [DOI] [PubMed] [Google Scholar]
- Lin Y-C, Nunez V, Johns R, Shiao SPK. APOA5 gene polymorphisms and cardiovascular diseases: Meta-prediction in global populations. Nursing Research. 2017;66:xx–xx. doi: 10.1097/NNR.0000000000000207. [DOI] [PubMed] [Google Scholar]
- MacAurthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Gunter C. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508(7497):469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton A, Patch C, Wiggins J, Barnes K, Crawford G, Benjamin C, Bruce A. Position statement on opportunistic genomic screening from the Association of Genetic Nurses and Counsellors (UK and Ireland) European Journal of Human Genetics. 2014;22:955–956. doi: 10.1038/ejhg.2013.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul DB, Brosco JP. The PKU paradox: A short history of a genetic disease. Baltimore, MD: Johns Hopkins University Press; 2013. [Google Scholar]
- Ragsdale N, Koch R. Phenylketonuria: Detection and therapy. American Journal of Nursing. 1964;64(1):90–96. [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Rehm HL. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Ward BE. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genetics in Medicine. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- Smoot B, Kober KM, Paul SM, Levine JD, Abrams G, Mastick J, Miaskowski C. Potassium channel candidate genes predict the development of secondary lymphedema following breast cancer surgery. Nursing Research. 2017;66:xx–xx. doi: 10.1097/NNR.0000000000000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele S. Phenylketonuria: Counseling and teaching functions of the nurse on an interdisciplinary team. Issues in Comprehensive Pediatric Nursing. 1989;12:395–409. doi: 10.3109/01460868909038047. [DOI] [PubMed] [Google Scholar]
- Taylor JY, Sampson DA, Anderson CM, Caldwell D, Taylor AD. Effects of parity on blood pressure among West African Dogon women. Ethnicity & Disease. 2012;22:360–366. [PMC free article] [PubMed] [Google Scholar]
- Taylor JY, Sun YV, Hunt SC, Kardia SLR. Gene-environment interaction for hypertension among African American women across generations. Biological Research for Nursing. 2010;12:149–155. doi: 10.1177/1099800410371225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss JG, Shagal AG, Tsuji JM, MacDonald JW, Bammler TK, Farin FM, St. Pierre Schneider B. Time course of inflammatory gene expression following crush injury in murine skeletal muscle. Nursing Research. 2017;66:xx–xx. doi: 10.1097/NNR.0000000000000209. [DOI] [PubMed] [Google Scholar]
- Wang Q, Lu Q, Zhao H. A review of study designs and statistical methods for genomic epidemiology studies using next generation sequencing. Frontiers in Genetics. 2015;6 doi: 10.3389/fgene.2015.00149. (Article 149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetterstrand KA. DNA sequencing costs: Data from the NHGRI Genome Sequencing Program (GSP) 2015 Retrieved from www.genome.gov/sequencingcosts.
- Williams JK, Cashion AK, Veenstra DL. Challenges in evaluating next-generation sequence data for clinical decisions. Nursing Outlook. 2015;63:48–50. doi: 10.1016/j.outlook.2014.08.007. [DOI] [PubMed] [Google Scholar]
- Wright ML, Housman DE, Taylor JY. A perspective for sequencing familial hypercholesterolaemia in African Americans. npj Genomic Medicine. 2016;1 doi: 10.1038/npjgenmed.2016.12. (Article 16012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon PW, Bastian B, Anderson RN, Collins JL, Jaffe HW. Potentially preventable deaths from the five leading causes of death —United States, 2008–2010. Morbidity and Mortality Weekly Report. 2014;63(17):369–374. [PMC free article] [PubMed] [Google Scholar]
- Zahari Z, Lee CS, Ibrahim MA, Musa N, Yasin MAM, Lee YY, Ismail R. Opioid-dependent men on methadone maintenance therapy: Cold pressor pain responses and ABCB1 gene polymorphisms. Nursing Research. 2017;66:xx–xx. doi: 10.1097/NNR.0000000000000204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.