Abstract
Chromatin remodeling complexes couple the energy released from ATP hydrolysis to facilitate transcription, recombination and repair mechanisms essential for a wide variety of biologic responses. While recombinant expression of the regulatory subunits of these enzymes is possible, measuring catalytic (ATPase) activity of the intact complexes recovered from normal or mutant cells is critical for understanding their mechanisms. SWI/SNF-like remodeling complexes can be megadaltons in size and include many regulatory subunits, making reconstitution of purified subunits challenging for recapitulating in vivo function. The protocol in this article defines the first highly quantitative ATPase assay for intact remodeling complexes that does not require radiation or reconstitution of recombinantly expressed subunits. This protocol is specifically useful for defining the catalytic role of active-site mutations in the context of other regulatory subunits and quantitatively rank-ordering inactivating catalytic-site mutations.
Keywords: Chromatin remodeling, epigenetic regulators, ATPase activity, luciferase, catalysis
INTRODUCTION
The need for a quantitative highly reproducible ATPase assay for intact chromatin remodeling complexes
A radioactive method for measuring ATPase activity of chromatin remodeling complexes has been previously reported by Bultman and co-workers (Bultman et al., 2005). The authors characterized a Brg hypomorph allele from a mutagenesis screen, and co-immunoprecipitated the BAF complex to run an on-bead assay measuring the conversion of gamma-labeled 32P-ATP to inorganic 32Pi and ADP. This method was modified from previous methodologies for catalysis assays for epigenetic regulatory enzymes (Cote et al., 1994; Laurent et al., 1993; Zhao et al., 1998). It has been used widely (Dykhuizen et al., 2013) since, and is an effective method for determining relative hydrolytic capacities of remodelers. Unfortunately, this method is prone to certain limitations for running many different conditions in parallel which is difficult for sensitive radiometric experiments, day-to-day variability related to the batch of gamma-labeled 32P-ATP, and limited by the sensitivity of quantification related to determining saturation. While we acknowledge the utility of previously reported methodologies in measuring hydrolytic rate of chromatin remodelers, we have developed a non-radioactive luciferase-coupled ATPase assay for remodeling enzymes that addresses certain limitations.
Initial efforts were inspired by readily available luminescence-based assays for purified kinase enzymes. Many assays are available for monitoring activities of kinases and specifically provide platforms for asking questions about the initial step in kinase activation, liberation of inorganic phosphate from ATP. Chromatin-remodeling enzymes can be studied in vitro by capturing the same information about relative rates of production of inorganic phosphate, but because of experimental limitations on studying purified intact chromatin remodeling enzymes (see above), it has been difficult. We therefore developed and optimized a general method for coupling purification of intact chromatin-remodeling complexes with luciferase-based measurements of production of ADP from ATP.
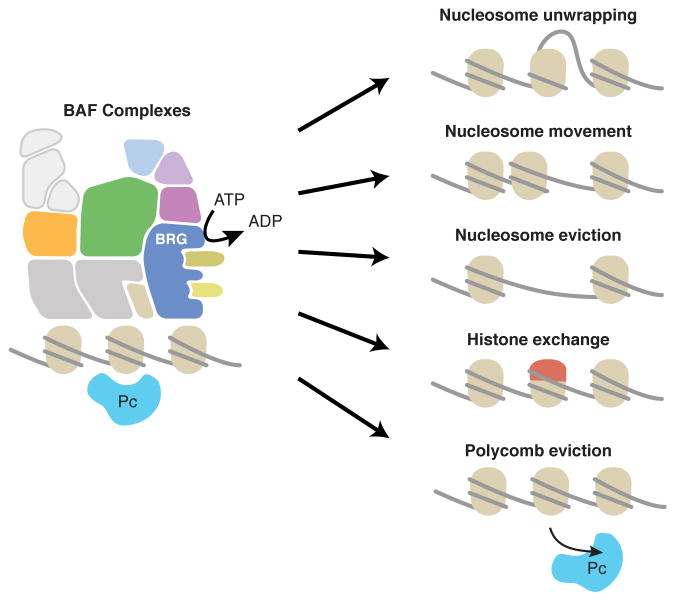
This method is useful for SWI/SNF-like remodelers that convert energy from ATP-hydrolysis into various cellular activities including altering the chromatin-DNA interface through several potential pathways (Figure 1). The enzymes intended for use in our assay are assembled into large complexes that cannot be studied in vitro through recombinant expression of certain subunits, but rather must be measured in the context of intact complexes. For example, the mammalian SWI/SNF-like BAF remodeling complex is approximately ~2 MDa, and it is difficult to study the cloned and bacterially expressed fragments of the ATPase domain alone. Therefore, remodeling complexes can be purified by co-immunoprecipitation and analyzed for hydrolytic activity on beads. Our method couples the purification of intact remodeling complexes with immunoprecipitations with a luciferase-based quantitative assay for making many measurements in parallel. This method is well suited for study of complexes containing mutant subunits recovered from human tissues or cell lines and can also be used to measure the effects of protein or small molecule modulators.
Figure 1. The BAF complex hydrolyzes ATP and alters chromatin structure.
Recent studies have revealed that SWI/SNF complexes are capable of altering chromatin accessibility, nucleosome positioning, and Polycomb activity. It is presently unknown whether these chromatin-remodeling activities rely strongly on the ATPase activity of the complexes or whether catalytic mutations may alter the ability of SWI/SNF complexes to regulate chromatin structure with unique downstream effects.
STRATEGIC PLANNING
Adapting our ATPase assay method should not require extensive implementation for most labs equipped for standard biochemistry. A cold room for overnight incubation of immunoprecipitations is necessary, as are magnetic racks for washing Dynabeads and purifying chromatin remodeling complexes. A luminescence plate reader should be available, but our protocol has luminescence levels that are stable for over an hour at room temperature, so a plate reader is not necessarily required in the same lab space as long as it is accessible nearby. Most investigators with a basic knowledge of biochemistry should be able to perform these experiments. The main limitations of our protocol are the cost of reagents, availability of specific antibodies for immunoprecipitations and background levels of luminescence.
Basic Protocol 1. MEASURING ENZYMATIC ACTIVITY OF INTACT ATP-DEPENDENT CHROMATIN REMODELING COMPLEXES
This method is especially useful for making standardized measurements for related mutations of remodeling enzymes that can be efficiently immunoprecipitated with intact complexes and tested side-by-side. Because of limitations in making large numbers of side-by-side measurements with radioactivity-based ATPase assays, this method is preferable. Traditionally, TLC-plate based measurements of radioactive 32Pi require overnight incubation with phosphoimager screen. While x-ray film based measurements of 32Pi can also be used for traditional ATPase assays of chromatin remodelers, the data are frequently out of the linear range of detection, and it is difficult to determine appropriate linear range of detection, making precise quantification problematic. Our method involves same-day immunopreciptiation and detection, allowing for more streamlined experimental design.
Materials
REAGENTS
293T cells
DNA encoding desired chromatin remodeler (e.g., Brg [ Mut ] V5)
PEI (Polysciences Cat # 24765-2)
Dynabeads Protein A (Life Technologies, Cat#10001D)
Brg (G7) Antibody (Santa Cruz Biotechnology, Cat.# sc-17796)
Opti-MEM (ThermoFisher, Cat# 51985091)
DMEM 1X (Life Technologies, Cat# 11960-077)
Fetal Bovine Serum (Omega Scientific, Cat# FB-01)
ADP-Glo MAX Assay (Promega Cat# V7001)
Ultra-Pure Glycerol (Thermo Fisher Cat# 15514-011)
Opaque-bottom non-treated round bottom 96-well plates (Corning Cat# CLS3789-100EA)
Chymostatin, protease inhibitor (EMD Millipore, Cat# 230790)
Leupeptin, protease inhibitor (EMD Millipore, Cat# 108975)
Pepstatin, protease inhibitor (EMD Millipore, Cat# 516481)
ssDNA (for use in 1X ATPase Reaction Buffer): CAAGCAGAAGACGGCATACGAGATGTAGAGAGGTCTCGTGGGCTCGGAGATGT (IDT, www.idtdna.com)
10N Sodium Hydroxide solution (VWR, Cat# VW3247-1)
EQUIPMENT
EnVision Plate Reader (Perkin Elmer Cat# 2104-0010)
Eppendorf Thermomixer (Fisher Scientific Cat# 05-400-201)
STEPS AND ANNOTATIONS
Plate 293T cells in a 10 cm tissue-culture dish (6 million cells, the evening before PEI transfection).
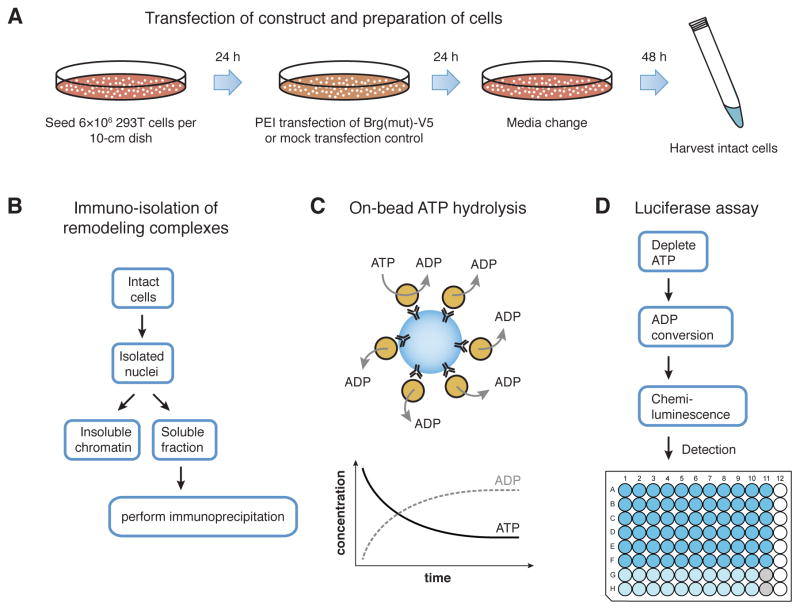
Prepare 12 μg DNA encoding desired chromatin remodeler (Brg [ Mut ] V5) and mix with 2 mL Opti-MEM. Mix well and incubate at room temperature for 2 minutes (Figure 2A).
Add 45 μL 1mg/mL buffered PEI (pH 7.0) to the DNA/Opti-MEM mixture and mix well. Incubate the DNA/PEI/Opti-MEM mixture at room temperature for 10 minutes.
Change the media on the 293T cells during the 10 minute incubation of DNA/PEI/Opti-MEM. Add the DNA/PEI/Opti-MEM mixture dropwise to the 293Ts after changing the media, swirling the dish to equilibrate the transfection solution.
Change media on the transfected 293T cells 24h after transfection (Figure 2A).
Wait 48h after media change, then trypsin digest and dissociate and prepare nuclear extracts by incubation with 10 mL Buffer A on ice 7 min.
After centrifugation at 500 × g for 5 minutes at 4° C, repeat Buffer A incubation on ice for 3 min and centrifuge again.
For each 50 million cell equivalent, incubate with 200–300 μL Lysis Buffer and incubate at 4° C with end-over-end rotation for 30 min.
Spin microfuge tubes at 16,000 × g for 10 minutes at 4° C. Collect supernatant and use for subsequent immunoprecipitations.
Perform immunoprecipitations of Brg with 18 μL G7 antibody per 50 million cell equivalent. Incubate at 4° C with end-over-end rotation overnight.
Immediately before use, resuspend a slurry of protein A Dynabeads. Prepare 33 μL for each immunoprecipitation, and wash twice with Lysis Buffer (including 1 mM DTT and 10 μg/mL PI). For each immunoprecipitation add 30 μL protein A Dynabeads suspended in Lysis Buffer to immobilize complexes (Figure 2B) by incubating at 4° C for 4 hours with end-over-end rotation.
Wash beads twice with 1 mL Wash Buffer and buffer exchange into 25 μL reactions in 1X ATPase Assay Reaction Buffer. Incubate at 37°C for 1 hour.
Immobilize beads by magnet and use the 25 μL supernatant directly in the ATP-coupled luciferase reaction by adding 25 μL ADP-Glo reagent. Incubate at room temperature for 1 h with gentle agitation.
Add 50 μL ADP-Glo Detection Substrate and incubate at room temperature for 1 hour. Measure luciferase activity with EnVision Plate Reader using 1 second integration time (Figure 2C). 1X ATPase Assay Reaction Buffer should be used as a negative control for subsequent normalization of luciferase activity.
Figure 2. Experimental design and workflow for measuring ATPase activity.
(A) Cells for use in the ATPase assay protocol are plated and prepared for expression of unique alterations of remodeling enzymes, which will be incorporated into intact ATP-dependent chromatin remodeling complexes for subsequent isolation. (B) The soluble fraction is isolated from harvested nuclei for immunoprecipitation reactions. (C) The isolated chromatin remodeling complexes are found to retain catalytic activity while bound to the beads used in the immunoprecipitations. This allows catalytic activities to be measured for native, intact remodeling complexes that remain bound to other subunits. (D) The luciferase assay is performed in small volumes in 96-well opaque-bottom plates for sensitive quantitation of catalytic activity.
REAGENTS AND SOLUTIONS
-
PEI, 1 mg/ml
Resuspend in distilled water at a concentration of 2 mg/mL and adjust with 10N Sodium Hydroxide solution to pH 7.0. As the pH approaches 7.0, a diluted 1N Sodium Hydroxide solution may be optimal for accuracy.
Dilute the pH 7.0 solution with distilled water to 1 mg/mL PEI.
Filter the PEI solution using 0.2 μm pore size. The PEI can be stored at −20 °C for up to two years, and stored at 4 °C for two weeks.
-
Protease Inhibitors (PI)
Using a balance, measure 100 mg Leupeptin, 100 mg Chymostatin, and 100 mg Pepstatin.
Mix the Leupeptin, Chymostatin, and Pepstatin together and dissolve in 10 mL DMSO to make a 10 mg/mL (1000X) stock.
Aliquot and freeze at −20 °C for subsequent use. The protease inhibitor aliquots can be stored for up to two years at −20 °C.
-
Buffer A for harvest of nuclei
Prepare Buffer A for nuclear isolation (25 mM HEPES pH 8.0, 5 mM MgCl2, 25 mM KCl, 0.05 mM EDTA pH 8.0, 10% Glycerol, 0.1% NP-40).
Sterile Filter Buffer A through 0.2 μm pore filter apparatus.
Store Buffer A at 4 °C, add 1 mM DTT and 10 μg/mL PI immediately before use. Buffer A can be stored without DTT or PI at 4 °C for up to one year.
-
Lysis Buffer for immunoprecipitation
Prepare Low-Salt Lysis Buffer (50 mM Tris pH 8.0, 150 mM NaCl, 0.1% NP-40).
Sterile Filter the Lysis Buffer through 0.2 μm pore filter apparatus.
Store Lysis Buffer at 4 °C, add 1 mM DTT and 10 μg/mL PI immediately before use.
-
ATPase Assay Wash Buffer
Prepare Wash Buffer (10 mM Tris pH 7.5, 50 mM NaCl, 5 mM MgCl2).
Sterile Filter the Lysis Buffer through 0.2 μm pore filter apparatus.
Store Lysis Buffer at 4 °C, add 1 mM DTT and 10 μg/mL PI immediately before use. Lysis Buffer can be stored without DTT or PI at 4 °C for up to one year.
-
2X ATPase Assay Reaction Buffer
Prepare 2X Reaction buffer (20 mM Tris pH 7.5, 100 mM NaCl, 10 mM MgCl2, 40% Glycerol).
Do not sterile filter this solution as it will not easily pass through a 0.2 μm membrane.
Store 2X ATPase Reaction Buffer at 4 °C for up to one year.
-
Dynabeads Protein A
It is important to never let Dynabeads dry during wash steps.
Avoid centrifugation of Dynabeads.
-
ADP-Glo MAX Assay (Promega Cat# V7001)
Thaw ADP-Glo Reagent at room temperature and aliquot (store 500 μL aliquots at −20 °C for up to two years).
-
Reconstitute ADP-Glo Detection Substrate and aliquot (store 250 μL aliquots at −20 °C for up to two years).
Just before use, equilibrate ADP-Glo reagent (25 μL x desired number of ATPase reactions) and Detection Substrate (50 μL x desired number of ATPase reactions) to room temperature.
COMMENTARY
Background Information
One limitation of the measurement of ATPase activity of intact chromatin remodeling complexes is that information regarding the productivity of the catalysis is not captured with these methods. Certain catalytic site mutations in conserved residues (Figure 3A–D) may appear to be catalytically unperturbed, but this is not a measure of the coupling of ATPase activity to chromatin regulatory capacity. It is important to note the possibility that in some cases catalytic site mutations may uncouple hydrolytic capacity from chromatin regulation. Further studies of chromatin accessibility or long-range chromatin interactions may yield further insight into the coupling of hydrolysis to chromatin regulation in this context. Other methods exist for measuring ATPase activity of including fluorometric-based approaches for cloned and purified ATPase enzymes. However, none of the existing methods has been useful for measuring ATPase activity of intact chromatin remodeling complexes.
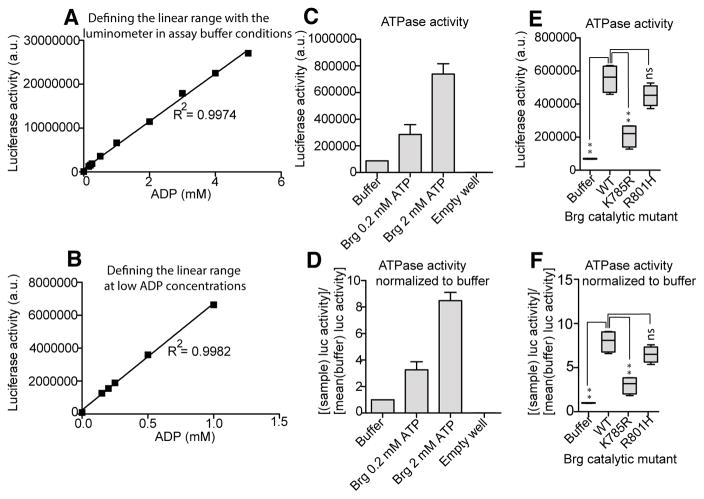
Figure 3. Quantification of ATPase assay results and normalization.
(A) The linear range was determined for a range of ADP concentrations, designed to simulate complete efficiency of enzymatic hydrolysis of ATP in order to define limiting conditions for the linear range at high and low (B) substrate concentrations. (C) BrgWT-expressing cells were harvested, and nuclei were extracted in order to immunopurify Brg-containing SWI/SNF complexes. It was found that concentrations 200 μM ATP and 2 mM ATP produced on-bead hydrolysis levels that could be readily differentiated by the assay conditions. Raw luciferase values (obtained from 1-second exposures per well) directly from the luminometer and (D) buffer-normalized values are shown. Raw (E) and buffer-normalized (F) ATPase assay data were obtained for Brg catalytic-site mutations. The highly quantitative ATPase assay allows for direct comparison and differentiation of unique point mutations based on either raw or normalized luciferase-coupled ATPase activity.
Critical Parameters
PEI Transfection
Attaining consistent results in the PEI-mediated transfection of 293T cells is critical for reproducibility. The timing and order of addition of the components during the transfection step should be performed such that the DNA/Opti-MEM is mixed by pipetting half the mixture volume (1 mL) at least five times before the introduction of PEI . After PEI is added to the DNA/Opti-MEM mixture, mix again by pipetting 1 mL five times. It is critical that the DNA/Opti-MEM/PEI mixture is incubated at room temperature for 10 minutes before addition to the 293T cells. Toxicity may be observed after transfection if the DNA/Opti-MEM/PEI mixture is allowed to incubate for longer periods of time. If the transfection mixture is preincubated for different periods of time across conditions, variability may result from inconsistent expression levels. Additionally, the 293T media should be changed within 24 hours after transfection or toxicity may result. PEI should not be left at room temperature after use in transfection experiments and should be stored at 4 °C for up to two weeks or at −20 °C for long-term storage.
Immunoprecipitations with Dynabeads
To ensure consistent results from independent immunoprecipitation experiments it is critical that Dynabeads are not allowed to dry. Thus, it is important that if independent immunoprecipitation reactions are washed in parallel, that the immobilizations on the magnetic rack are carried out in small batches of no more than six samples. If more than six immunoprecipitations are washed in parallel, drying may result during the aspiration steps while the beads are immobilized. If drying results during wash steps, this may reduce signal in the luciferase assay and cause variability or false negatives. When measuring catalytic activity it is critical that the association between Dynabeads and immunopurified chromatin-remodeling complexes is not perturbed by centrifugation or drying of the beads during any step of the protocol.
Assay Buffer Preparation & Reagent Preparation
Immediately before starting the ATPase reactions, it is critical to adjust the 2X Reaction Buffer composition to include BSA, ATP, DTT, PI, and ssDNA for a final 1X Reaction Buffer composition (10 mM Tris pH 7.5, 50 mM NaCl, 5 mM MgCl2, 20 % Glycerol, 1 mg/mL BSA, 4 mM ATP, 0.5 μg/μL ssDNA, 1 mM DTT, and PI). For long-term storage, the 2X Reaction Buffer should not include BSA, ATP, DTT, PI or ssDNA but rather should be adjusted to the 1X composition immediately before use. Other buffers used in immunoprecipitation reactions should be sterile-filtered for long-term storage at 4 °C. There may be some precipitate in the ATPase assay Detection Substrate even after equilibration to room temperature before use. It is critical to mix the Detection Substrate very well before use in the ATPase assay. After addition of the Detection Substrate to the individual assay reactions in the 96-well plate, it is critical to cover and spin the plate for 30 seconds at 300 × g to remove buffer on well edges.
Troubleshooting
STEP4
Problem: Observation of significant cell death
Possible Reason: PEI incubation was over 10 minutes
Solution: Repeat transfections with shorter PEI:DNA:OptiMEM incubation (can optimize with GFP-positive control plasmid)
STEP 14
Problem: Low signal after Detection Substrate added
Possible Reason: insufficient mixing of ADP-Glo substrate in Step 13
Solution: Cover, and briefly vortex assay plate after completing Step 13, then spin 30 seconds at 300 × g.
STEP 14
Problem: Low signal after Detection Substrate added
Possible Reason: did not thaw Detection Substrate sufficiently
Solution: Thaw ADP-Glo and Detection Substrate at room temperature for at least 30 min before Step 13.
STEP 14
Problem: High variability between replicates
Possible Reason: Batch-variability in antibody used in Step 10.
Solution: Always use the same batch number and lot number for co-immunoprecipitations so luminescence values in in Step 13 can be directly compared across experiments.
STEP 14
Problem: High variability between replicates
Possible Reason: Different DNA used in 1X ATPase Assay Reaction Buffer
Solution: Use a defined ssDNA PCR ~50-mer primer for each set of experiments. ATP is a substrate of chromatin remodelers, but DNA is also a substrate and can affect enzyme activity.
STEP 14
Problem: High variability between replicates
Possible Reason: Different promoters used to drive expression of chromatin remodeler in Step 2.
Solution: When comparisons in catalytic activity are desired, across a certain class of mutations in a defined chromatin remodeler, always compare experimental conditions when remodeler ATPase are expressed from the same vector backbone.
Anticipated Results
Results from the remodeler ATPase assay will be quantitative when in the linear range (Figure 3A,B). In our testing, statistical tests are generally independent of whether data are expressed in raw luminescence activity (Figure 3C,E) or instead normalized to buffer (Figure 3D,F). The sensitivity in measurements afforded by this assay allowed us to make the observation that many mutations tiled across the Brg ATPase domain were associated with catalytic defects that clustered with highly conserved motifs. The on-bead ATPase assay provides enough sensitivity to measure the hydrolytic capacities for weak enzymes, or enzymes that may have catalytic dysfunction as a result of mutations, and also provides a platform for comparison of point mutants in chromatin-remodeling ATPases. The assay provides a rigorous quantitative method to compare active-site mutants of the Brg1 remodeler ATPase enzyme, which often appear in human cancers. Because the transfected cells will express the protein complexes at high levels, background ATPase activity will be negligible, as can be evidenced by comparing ATPase activity of many of the Brg catalytic mutants to wild type and buffer control.
Time Considerations
Step 1, 24h
Step 2, 1h;
Step 3, 24h;
Step 4+5, 10 min;
Step 6, 48h;
Step 7, 3 min;
Step 8, 40 min;
Step 10, 16h;
Step 11, 60min;
Step 12+13, 2h.
Total: 4.5 days from plating cells for transfection to quantification of luciferase signal.
Acknowledgments
We thank Martha J. Zaslow, Ary Shalizi, Vincent D’Andrea, for assistance, and Warren J. Leonard (NHLBI) for shared equipment. We wish to honor the memory of Joseph P. Calarco with lasting gratitude for his encouragement and assistance. The work was supported by Division of Intramural Research of NHLBI, NIH (K.Z.), NIH grants R37NS046789 (G.R.C.) and R01CA163915 (G.R.C.), and HHMI (G.R.C.). An NCI career transition award K99CA187565 provided support for C.H.
Footnotes
Conflicts of Interest
The authors declare they have no competing financial interests relating to the research herein.
Literature Cited
- Bultman SJ, Gebuhr TC, Magnuson T. A Brg1 mutation that uncouples ATPase activity from chromatin remodeling reveals an essential role for SWI/SNF-related complexes in beta-globin expression and erythroid development. Genes & development. 2005;19:2849–2861. doi: 10.1101/gad.1364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- Dykhuizen EC, Hargreaves DC, Miller EL, Cui K, Korshunov A, Kool M, Pfister S, Cho YJ, Zhao K, Crabtree GR. BAF complexes facilitate decatenation of DNA by topoisomerase IIalpha. Nature. 2013;497:624–627. doi: 10.1038/nature12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nature genetics. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent BC, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes & development. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- Son EY, Crabtree GR. The role of BAF (mSWI/SNF) complexes in mammalian neural development. Am J Med Genet C Semin Med Genet. 2014;166C:333–349. doi: 10.1002/ajmg.c.31416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]