Abstract
More than 70% of birch pollen-allergic patients develop allergic cross-reactions to the major allergen found in apple fruits (Malus domestica), the 17.5 kDa protein Mal d 1. Allergic reactions against this protein result from initial sensitization to the major allergen from birch pollen, Bet v 1. Immunologic cross-reactivity of Bet v 1-specific IgE antibodies with Mal d 1 after apple consumption can subsequently provoke severe oral allergic syndromes. This study presents the three-dimensional NMR solution structure of Mal d 1 (isoform Mal d 1.0101, initially cloned from ‘Granny Smith’ apples). This protein is composed of a seven-stranded antiparallel β-sheet and three α-helices that form a large internal cavity, similar to Bet v 1 and other cross-reactive food allergens. The Mal d 1 structure provides the basis for elucidating the details of allergic cross-reactivity between birch pollen and apple allergens on a molecular level.
Keywords: structure, allergen, Mal d 1, Malus domestica
Introduction
In central and northern Europe as well as in North America a significant proportion of patients who suffer from birch pollen allergy develop intolerance to certain kinds of fruits and vegetables.1 Such birch pollen-related food allergies are the result of initial sensitization to the major birch pollen allergen, Bet v 1, and subsequent immunologic cross-reactivity of the Bet v 1-specific IgE antibodies with structurally homologous food proteins. Among the most frequent triggers of birch pollen-related food allergies are apples, with >70% of all individuals that are sensitized to birch pollen developing allergic reactions when consuming apples.2 Symptoms typically occur locally at the site of food contact and within minutes after apple consumption, including itching and swelling of the lips, tongue, and throat (oral allergic syndromes, OAS).3 Frequently, allergic patients can also exhibit symptoms of food-induced rhinoconjunctivitis and dyspnea.2
In apples (Malus domestica), the major allergen that is responsible for birch pollen-related food allergies is the 17.5 kDa protein Mal d 1.4 Mal d 1 belongs to group 10 of pathogenesis-related (PR) proteins that are activated in plants in response to different kinds of stress.5 The concentration of Mal d 1 in apples is highly dependent on the cultivar and also influenced by various biotic and abiotic factors, storage conditions, and storage duration.6,7 Typically, 1–30 μg of Mal d 1 per gram of fresh apple (accounting for up to 7% of total soluble protein) is present directly after harvest.7,8 After storage, these values can rise to values exceeding 100 μg Mal d 1 per gram of apple.7,9 Although Mal d 1 has been found in both the pulp and peel of apples, higher concentrations are present in the peel.8,10 On the basis of this observation, and because Mal d 1 appears to be up-regulated upon biotic stress, it has been speculated that this protein may play a role in plant defense response to pathological situations.11
Mal d 1 is encoded by a multigene family, and a number of isoforms of Mal d 1 have been identified to date, which are clustered into four groups on the basis of their DNA sequence similarities, that is, Mal d 1.01, Mal d 1.02, Mal d 1.03, and Mal d 1.04.12 PCR screening and mass spectrometric studies showed that Mal d 1 isoforms are not cultivar specific and that mixtures of isoforms are present in apple fruits.13,14 Along with Mal d 1.02, and depending on the cultivar, isoforms from the Mal d 1.01 cluster are by far the most abundant isoforms found in apples.8 Within the Mal d 1.01 cluster, protein sequence identities between known isoforms are >97%.15 Of note, immunologic investigations of naturally occurring Mal d 1 isoforms revealed only small differences of their IgE binding capacities and it appears that divergent allergenicities of apple strains are predominantly determined by different Mal d 1 expression levels.13
Whereas the immunological properties of Mal d 1 suggest that this protein has a three-dimensional structure and IgE binding epitopes that are similar to those of Bet v 1 and other members of the PR-10 protein family, experimental structural data for Mal d 1 have not been available to date. As a first step toward structural characterization, we recently assigned the NMR backbone and side chain 1H, 13C, and 15N chemical shifts of the isoform Mal d 1.0101.16 Mal d 1.0101, initially cloned from ‘Granny Smith’ apples, and Mal d 1.0102, from ‘Golden Delicious’, were the first isoforms for which the DNA sequence was determined and are identical at the amino acid level.4,17 Here we report the NMR solution structure of this protein.
Materials and Methods
NMR Spectroscopy
The DNA of Mal d 1.0101 (GenBank nucleotide code X83672, protein code CAA58646) was cloned into the expression vector pET28b by using the restriction sites NcoI and XhoI.16 Construct integrity was ensured by DNA sequencing (Microsynth AG, Balgach, Switzerland), and the protein was expressed in Escherichia coli BL21 Star (DE3). Mal d 1.0101 was purified by anion exchange and size exclusion chromatography as described in detail elsewhere.16 The mass and the amino acid sequence of purified Mal d 1.0101 were confirmed by mass spectrometry using a 7 T Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer (Bruker Daltonics, Bremen, Germany) with an attached electrospray ionization (ESI) source.
Protein concentrations for NMR spectroscopic experiments for structure determination were 0.5 mM for 15N/13C-labeled and 0.8 mM for 15N-labeled samples in 91% H2O/9% D2O (v/v) at pH 6.9, 10 mM sodium phosphate, and 7 or 11.2 mM l-ascorbic acid, respectively. All NMR experiments were carried out at 298 K, using either a 500 MHz Agilent DirectDrive spectrometer (Agilent Technologies, Santa Clara, CA, USA) equipped with a room temperature probe or a 600 MHz Bruker Avance II+ spectrometer (Bruker BioSpin, Karlsruhe, Germany) equipped with a Prodigy CryoProbe. NMR resonance assignments of Mal d 1.0101 were made using standard triple-resonance methods16 and were deposited at the Biological Magnetic Resonance Data Bank (BMRB) under accession no. 25968. Three-dimensional 15N and 13C edited NOESY-HSQC experiments (mixing times of 150 ms) were recorded for derivation of distance restraints. NMR data were processed using NMRPipe18 and analyzed with CcpNmr.19
For measuring protein translational diffusion, we employed a stimulated echo pulsed field gradient NMR experiment.20 Experimental details were identical to those reported for Bet v 1.21 For the determination of the hydrodynamic radius of Mal d 1.0101, we used dioxane as a standard reference under identical buffer conditions, assuming a hydrodynamic radius of 2.12 Å.22
Structure Calculation
Structure calculations were performed with the program XPLOR-NIH 2.4223,24 using a simulated annealing protocol. An initial structural model was generated with CS-ROSETTA25 using the BMRB CS-Rosetta server.26 A total of 2079 distance restraints were obtained from 3D 15N and 13C edited NOESY-HSQC spectra. NOE values were converted on the basis of peak intensities into distances with upper limits of 3.0 Å (strong), 4.0 Å (medium), 5.0 Å (weak), and 6.0 Å (very weak). Dihedral angle restraints were predicted using TALOS+27 and CS-ROSETTA.25 In all regular secondary structure elements hydrogen bonds were included for backbone amide protons, if the 15N edited NOESY-HSQC spectra did not show a water exchange cross peak. Of 100 generated structures, the 20 lowest energy structures were picked and further refined in explicit solvent with the AMBER14 simulation package28 using pmemd.cuda29 and the AMBER force field 99SB-ILDN.30 Each structure was soaked into a truncated octahedral solvent box of TIP3P water molecules with a minimum wall distance of 10 Å. For the refinement, hydrogen atoms and water molecules were minimized with fixed heavy atoms. The temperature was increased from 0 to 300 K, where the structures were simulated using the NOE distance restraints, minimized again, and validated using the protein structure validation software (PSVS) suite (Table 1).31 The coordinates of the Mal d 1.0101 structures were deposited in the Protein Data Bank under the accession code number 5MMU. Graphics were prepared using the program MOE.32
Table 1. Summary of Restraints Used for NMR Structure Determination of Mal d 1.0101 and Structure Refinement Statistics.
| experimental restraintsa | |
| total no. of NOE-based distance restraints | 2079 |
| intraresidue [i = j] | 658 |
| sequential [|i–j| = 1] | 678 |
| medium range [1 < |i – j| < 5] | 307 |
| long-range [|i–j| ≥ 5] | 436 |
| dihedral angle restraints | 308 |
| hydrogen bond restraints | 131 |
| total no. of restraints | 2518 |
| total no. of restraints per residue | 15.9 |
| long-range restraints per residue | 3.2 |
| restraint violationsb | |
| distance violations/structure | |
| 0.1–0.2 Å | 14.3 |
| 0.2–0.5 Å | 2.75 |
| >0.5 Å | 0 |
| RMS of distance violation/restraint | 0.02 Å |
| max distance violationc | 0.50 Å |
| dihedral angle violations/structure | |
| 1–10° | 0.2 |
| >10° | 0 |
| RMS of dihedral angle violation/constraint | 0.06° |
| max dihedral angle violation | 2.60° |
| RMSD valuesd | |
| backbone atoms | 0.4 Å |
| heavy atoms | 1.0 Å |
| bond lengths | 0.010 Å |
| bond angles | 1.4° |
| Ramachandran plot statistics | |
| most favored regions | 92.7% |
| allowed regions | 6.6% |
| disallowed regions | 0.7% |
Numbers are given for all residues (1–158).
Calculated for all residues, using sum over r– 6.
Largest violation among all 20 reported structures.
Generated using the PSVS software suite.31
Results and Discussion
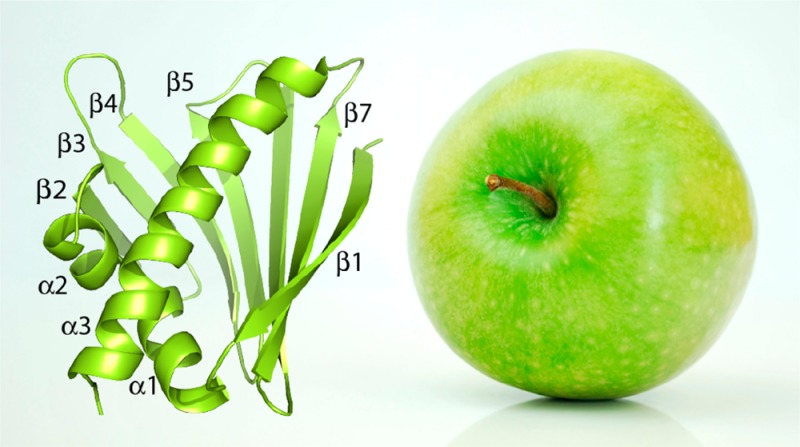
The three-dimensional structure of Mal d 1.0101 consists of a curved, seven-stranded antiparallel β-sheet (β1-β7) embracing a long helix at the C-terminus of the protein (α3) and two consecutive short helices (α1, α2) (Figure 1). The edges of the β-sheet are formed by strands β1 and β2, which are connected by helices α1 and α2 that form a V-shaped support for the C-terminal part of helix α3. In total, Mal d 1 contains ca. 35% β-sheet and ca. 25% helical structure, agreeing well with secondary structure estimates from infrared and circular dichroism.33−35 As in other proteins from the PR-10 family, strands β2 and β3 are connected by a glycine-rich loop motif (Gly46-Asn47-Gly48-Gly49-Pro50-Gly51). Together, these structural elements create the large internal cavity that is typical for the canonical PR-10 fold. From Figure 1B it is evident that in our NMR structural ensemble of Mal d 1, secondary structure elements are very well-defined and conformationally homogeneous in all 20 structural models. Only slightly elevated levels of conformational heterogeneity are observed for some of the solvent-exposed loops that connect secondary structure elements and the C-terminus of the protein.
Figure 1.
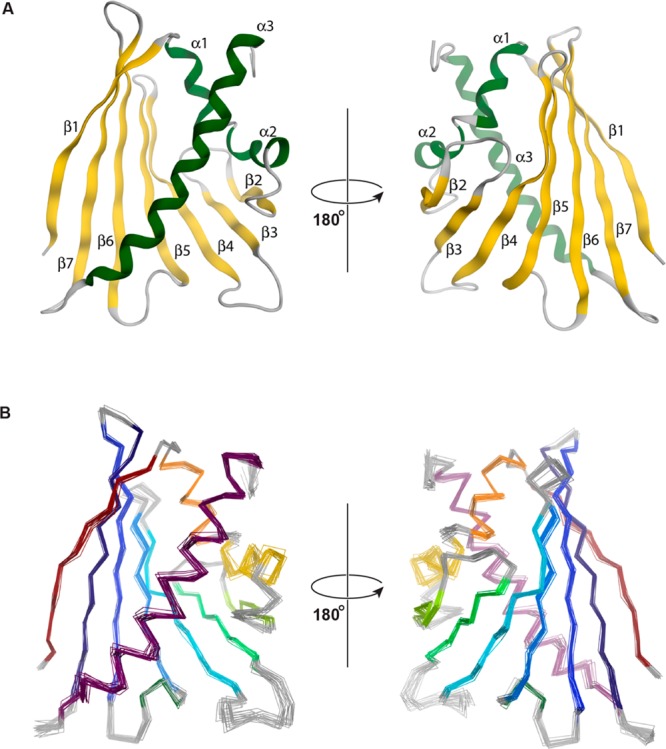
NMR solution structure of the major apple allergen Mal d 1.0101 (PDB accession code 5MMU). (A) Ribbon representation of the lowest energy structure. Secondary structure elements are labeled β1 (Val2–Ser11), β2 (Gln40–Glu45), β3 (Ile53–Thr57), β4 (Tyr66–Ile74), β5 (Ser80–Gly88), β6 (Glu96–Val105), β7 (Ser111–Thr121), α1 (Pro15–Val23), α2 (Ala26–Ile33), α3 (Lys128–Asp152). β-Strands and α-helices are colored in gold and green, respectively. (B) Backbone overlay of the ensemble of the 20 lowest energy structures of Mal d 1.0101. Secondary structure elements are colored from red (N-terminus) to purple (C-terminus).
A peculiar feature of the PR-10 fold is the large internal cavity. In Mal d 1.0101, the volume of this cavity36 is ca. 2230 Å3, which is comparable in size to those of other PR-10 proteins.5 As found in the birch pollen allergen Bet v 1 and other homologous food allergens, in Mal d 1 the majority of amino acids that form the surface of the cavity are hydrophobic (Figure 2). A large proportion of the inner cavity surface is formed by amino acid residues in the β-sheet whose hydrophobic side chains are located at the protein interior (Ile56 (β3), Val67 (β4), Ile71 (β4), Tyr81 (β5), Tyr83 (β5), Leu85 (β5), Ile98 (β6), Tyr100 (β6), Ile113 (β7)) along with inward-pointing residues in the long amphiphilic helix α3 (Val132, Val134, Ala139, Leu142, Phe143, Ile146), the two short helices α1 (Phe22, Val23) and α3 (Ala26, Ile30), and loop regions (Ile38, Phe58, Tyr64, Ala90). In addition, a few polar and charged side chains are located at the inside of the molecule and form part of the cavity surface, such as Asp27 (α2), His69 (β4), Ser115 (β7), and Lys138 (α3), so that the cavity itself is actually amphiphilic, as noted before for the major birch pollen allergen, Bet v 1.37 In crystal structures of other PR-10 proteins the cavity is occupied by water, amphiphilic ligand molecules, or components of the crystallization buffer.5 For Mal d 1, it is currently not known whether ligands bind specifically to the cavity or what the biological function of ligand binding could be.
Figure 2.
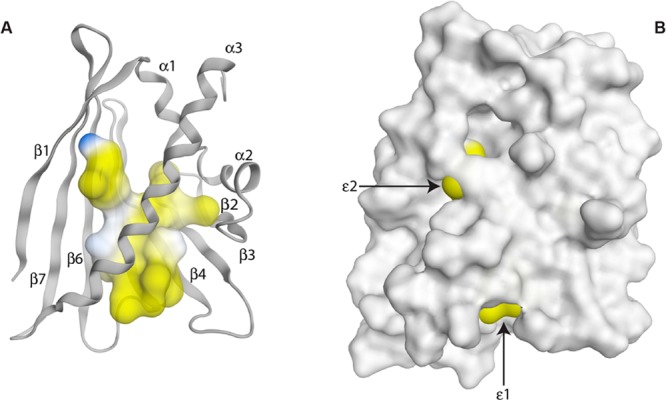
(A) Internal cavity of Mal d 1.0101, colored according to the lipophilic potential as implemented in MOE,32 where hydrophilic regions are colored in blue and lipophilic regions are colored in yellow. (B) Surface representation of the lowest energy solution structure of Mal d 1.0101. The two amphiphilic entrances to the internal cavity are indicated as ε1 (between the N-terminal end of helix α3 and the loops connecting strands β3−β4 and β5−β6) and ε2 (between the edge of the β-sheet and the C-terminal end of helix α3).
The internal cavity in Mal d 1 can be reached by two openings (Figure 2). One entrance to the protein interior, ε1, is shaped by residues in the N-terminal half of helix α3 (His131, Val134) along with the loops connecting strands β3−β4 (Gln63, Tyr64) and strands β5−β6 (Asp89). Together, these amino acids create an amphiphilic access route to the protein interior. A second amphiphilic entrance, ε2, is present at the edge of the β-sheet between helix α3 (Lys136, His140, Lys144, and Glu147) and strand β1 (Asn7, Phe9, and Ser 11). In the NMR solution structures of Mal d 1 this access route is partly obstructed by the side chain of His140. Of note, entries to the internal cavity at similar locations have also been described for other members of the PR-10 protein family.5
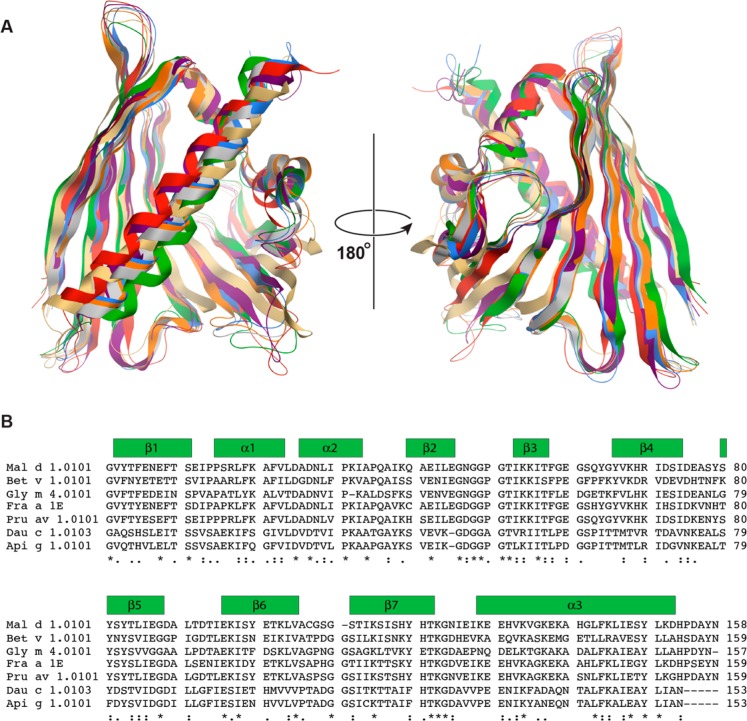
Figure 3 shows a comparison of Mal d 1 with Bet v 1 and birch pollen-related food allergens from the PR-10 family whose structures have been determined so far. Despite the fact that sequence identities between these proteins are only slightly higher than 50% in some cases, their three-dimensional structures are generally very similar, with backbone rmsd values for secondary structures typically below 2 Å.38 In light of the observed immunologic cross-reactivity between Mal d 1 and the major birch pollen allergen, Bet v 1, the structural comparison of these two proteins is of particular interest. The backbone rmsd between Mal d 1.0101 and the hyperallergenic isoform Bet v 1.0101 (61% sequence identity) of the birch pollen allergen is 2.13 Å (1.70 Å for secondary structure elements). Of note, Mal d 1 and Bet v 1 differ in length by one amino acid, and divergent presumptions have been made about the location of the gap in Mal d 1. On the basis of sequence alignments of PR-10 food allergens it has been proposed that either the loop right before39,40 or right after4,34,41,42 strand β7 is one residue shorter in Mal d 1. Our solution structure shows that the loop right before strand β7 is the one that is shorter in Mal d 1.0101. Strands β6 (Glu96–Val105 in both Mal d 1.0101 and Bet v 1.0101) and β7 (Ser111–Thr121 in Mal d 1.0101 and Ser112–Thr122 in Bet v 1.0101) occupy identical positions and have equal hydrogen bonding patterns in the antiparallel β-sheets of these proteins. They are connected via loops consisting of four residues (Cys-Gly-Ser-Gly in Mal d 1) and five residues (Thr-Pro-Asp-Gly-Gly in Bet v 1), respectively, which produces a small structural difference in these loop segments between the two proteins.
Figure 3.
Comparison of PR-10 food and plant allergens with known structures. (A) Overlay of the lowest energy structure of Mal d 1.0101 (green, PDB accession code 5MMU) with the structures of the major birch pollen allergen Bet v 1.0101 (blue, 4A88), the carrot allergen Dau c 1.0103 (orange, 2WQL), the celery allergen Api g 1.0101 (gray, 2BK0), the soybean allergen Gly m 4.0101 (yellow, 2K7H), the strawberry allergen Fra a 1E (red, 2LPX), and the cherry allergen Pru av 1.0101 (purple, 1E09). (B) Multiple sequence alignment of these allergens obtained with Clustal Omega.53 Amino acids are marked with asterisks (identical), colons (conserved), and dots (semiconserved). Secondary structure elements as present in Mal d 1.0101 are indicated.
Mal d 1 is known to have a tendency for cysteine-mediated dimerization, as shown for the isoform Mal d 1.0108 by nonreducing gel electrophoresis and size exclusion chromatography.35 Like Mal d 1.0108, the isoform Mal d 1.0101 contains a single cysteine residue, Cys107. In the three-dimensional solution structure of Mal d 1.0101 Cys107 is located at the C-terminal tip of strand β7, with its side chain oriented toward the protein surface. To probe the oligomerization state of Mal d 1.0101 under the conditions that we employed for NMR structure determination (pH 6.9, 10 mM sodium phosphate, 14 mol equiv of l-ascorbic acid, 298 K) we performed pulsed-field-gradient NMR diffusion experiments. We obtained a value of 21.6 ± 0.8 Å for the hydrodynamic radius of Mal d 1.0101, which is comparable to the hydrodynamic radius of monomeric Bet v 1.0101 (20.1 Å) under similar experimental conditions.21 This is consistent with our observation that, using the same buffer, Mal d 1.0101 elutes from a size exclusion column with a retention time that is virtually identical to that of Bet v 1.0101. These results were further verified by FT-ICR mass spectrometry, which shows that Mal d 1.0101 does not form dimers or higher order aggregates.
The NMR solution structure of Mal d 1 shows that this protein consists of a highly curved antiparallel β-sheet and three α-helices forming a large internal cavity, very similar in fashion to other PR-10 proteins.5 This is in agreement with the observed immunologic cross-reactivity between Mal d 1 and the major birch pollen allergen, Bet v 1, as well as other food allergens from the PR-10 protein family.4,43 In most patients Bet v 1 is the sensitizing agent, whereas Bet v 1-specific IgE antibodies subsequently cross-react with Mal d 1 and elicit an allergic response, as reflected by the clinical observation that apple allergy develops only after the onset of birch pollinosis.44
Along these lines, cross-inhibition experiments of Mal d 1 using sera from apple-allergic patients showed that Mal d 1 shares IgE epitopes with the major birch pollen allergen, Bet v 1.4,43 From a structural perspective, limited information about the exact nature of binding epitopes of Mal d 1 and Bet v 1 is available. Detailed structural information about a sequentially discontinuous (i.e., conformational) B-cell epitope in Bet v 1 was obtained by cocrystallizing the particular isoform Bet v 1.0112 with an antigen-binding fragment (Fab) derived from the murine monoclonal IgG antibody BV16.45 This epitope is formed by the segment between Glu42 and Thr52 (including the glycine-rich loop motif between strands β2 and β3), along with Arg70, Asp72, His76, Ile86, and Lys97 of Bet v 1, covering approximately 10% (≈900 Å2) of the entire protein surface. Binding of BV16 to this epitope measurably reduces serum IgE interactions, indicating that IgE and monoclonal IgG BV16 compete for overlapping binding surfaces on Bet v 1.46 Moreover, mutation of a central residue (Glu45→Ser) significantly reduced the IgE binding capacity of Bet v 1, confirming the significance of this particular epitope for interactions with IgE.46
Figure 4A shows the molecular interaction surface that corresponds with the BV16 epitope in the apple allergen. In Mal d 1 these residues form a contiguous surface patch along with a somewhat distal residue (Glu76), similar in shape and size to the BV16 epitope of Bet v 1. Moreover, the contributing amino acids are largely conserved between Mal d 1 and Bet v 1. Thirteen of the 16 amino acids in the BV16 epitope are identical, whereas only 3 residues are different in Mal d 1.0101 and Bet v 1.0101 (Figure 4B). These data thus provide a structural rationale for the observed allergic cross-reactivity between birch pollen and apple allergens. Interestingly, mutational studies indicate that the ability of Mal d 1 to bind serum IgE from patients with birch pollen allergies can be increased by increasing the similarity of the BV16 epitope in Mal d 1 to that of Bet v 1, indicating that these amino acids are indeed involved in binding of Bet v 1 specific to Mal d 1.39
Figure 4.
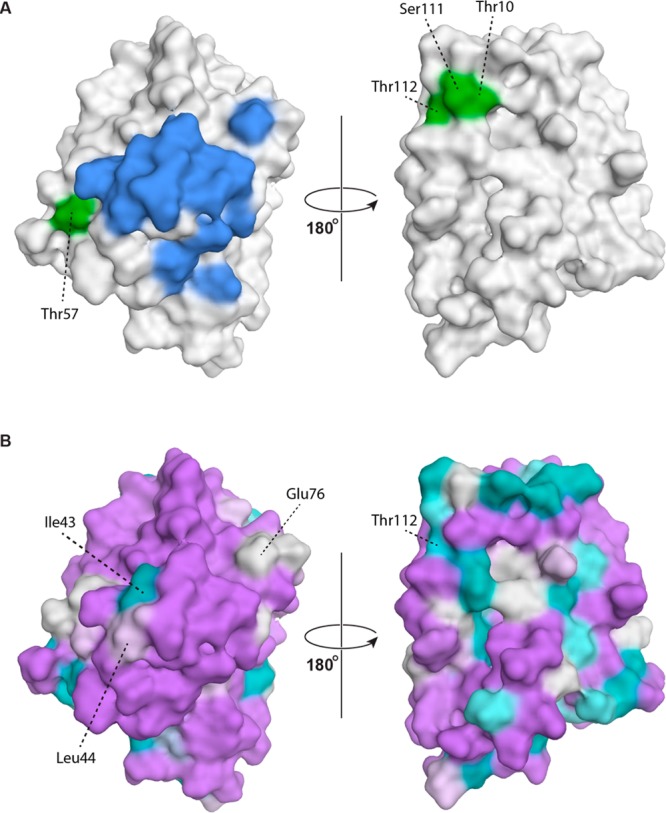
(A) Conformational epitopes of Mal d 1. Amino acid residues that correspond to the molecular interaction surface between monoclonal IgG BV16 and Bet v 1.0112 (residues Glu42–Thr52, Arg70, Asp72, Glu76, Ile86, and Lys97 in Mal d 1.0101) are colored in blue.45 Amino acid positions that were shown to be crucial for IgE recognition of Mal d 1 in mutational analyses (Thr10, Ile30, Thr57, Ser111, Thr112, and Ile113) are shown in green (Ile30 and Ile113 are located in the protein interior and do not contribute to the surface).13,34 (B) Amino acid similarities between Bet v 1.0101 and Mal d 1.0101 using a color gradient from lilac (highly similar) to teal (highly dissimilar). Epitope residues that are different between Bet v 1.0101 and Mal d 1.0101 are labeled. Similarities were calculated on the basis of substitution matrix scores (BLOSUM62) as implemented in MOE.32
It is likely that Mal d 1 contains more than a single conformational epitope.47 A number of amino acid positions that are relevant for IgE recognition have been identified by mutational analysis.13,34 For a five-point mutant of Mal d 1.0108 (Thr10→Pro, Ile30→Val, Thr57 →Asn, Thr112→Cys, and Ile113→Val) a markedly reduced capacity for binding Mal d 1-specific IgE was found in vitro.34 Skin prick tests in apple-allergic patients comparing wild-type Mal d 1 with the five-point mutant further showed a significantly lower ability of the mutant protein to induce skin reactions in vivo.48 Further experiments showed that the T-cell recognition level of wild-type Mal d 1 is conserved in the five-point mutant.34 Because these five amino acids are likely involved in IgE interactions not only in Mal d 1 but also in Bet v 1, they could well be part of common cross-reacting epitopes in these two allergens.49 This is corroborated by mutational studies, which showed that peptide stretches encompassing these residues are indeed involved in immunological cross-reactivity between Mal d 1 and Bet v 1.50 In addition, in an independent study, Ser111 was identified as being essential for IgE binding to Mal d 1, and a Ser111→Cys mutation resulted in significantly reduced affinity for IgE in immunoblotting experiments.13
Figure 4A shows that these six residues are fairly dispersed on the protein surface of Mal d 1 and that neither of these amino acids overlaps with the BV16 epitope. Amino acids Thr10, Ser111, and Thr112 form a common patch on the protein surface, whereas Thr57 is located approximately 37–39 Å away and close to the BV16 epitope. Considering that an epitope of typical size (∼600–900 Å2)39 and circular shape would have an arc length of 28–34 Å on the Mal d 1 surface, residues Thr10, Ser111, and Thr112 are probably too far away from Thr57 to be part of a common binding epitope. The remaining two residues, Ile30 and Ile113, do not reach the protein surface in Mal d 1. Whereas Ile113 is close in space to the Thr10-Ser111-Thr112 patch, its hydrophobic side-chain is pointing toward the interior of the protein, where it participates in a small hydrophobic core located at the inner end of the proteins’ cavity (between helices α1 and α3 and the β-sheet). Residue Ile30 is also located in the protein interior with its aliphatic side chain forming part of the internal cavity and does not contribute to the protein surface.
Of note, because the loop between strands β6 and β7 is shorter by one residue in Mal d 1 than in Bet v 1, Ser111 and Thr112 of β7 in Mal d 1 occupy the β7 positions of Ser112 and Ile113 in Bet v 1. The surface patch formed by Thr10, Ser111, and Thr112 in Mal d 1 thus appears to be less hydrophobic than the corresponding surface patch in Bet v 1 (Thr10, Ser112, and Ile113). As a matter of fact, also the protein surface surrounding these three residues displays a considerably lower level of similarity between Mal d 1.0101 and Bet v 1.0101 than other parts of the protein surface, as can be seen in Figure 4B. This might in part be responsible for the different IgE binding properties of these allergens. It has been noted, on the other hand, that epitope coincidence between Bet v 1 and Mal d 1 may be limited,47 as exemplified by a recent study describing the isolation of human IgE binding to Bet v 1 but not to Mal d 1.51 Moreover, different Mal d 1 isoforms contain amino acid substitutions within potential IgE interaction surfaces,39 suggesting that they may influence the immunologic reaction.
It is clear that high-resolution structural data provide the basis to determine and compare structural details of (cross-reactive) binding epitopes in allergenic proteins. In addition, grafting of conformational epitopes by transferring stretches of residues between homologous allergens has become a valuable experimental tool. Epitope grafting was used to characterize the role of the BV16 epitope in Mal d 1 by recreating this epitope on the Mal d 1 surface, confirming its importance for IgE binding and cross-reactivity with Bet v 1.39 In an orthogonal approach, several Mal d 1 stretches encompassing residues that are crucial for IgE binding were transferred to Bet v 1 to investigate the role of these structural segments for cross-reactivity,50 and chimeras of Bet v 1 and Mal d 1 were created to map the epitope of a human monoclonal IgE, which was isolated from a phage library, to the C-terminus of Bet v 1.51 In addition, epitope grafting provides access to chimeric allergens with fine-tuned antigenic properties, such as reduced IgE binding capacitites, for molecule-based allergy diagnosis and specific immunotherapy.52 Knowledge of the structural details of these allergens elements is required to generate correctly folded chimeras, because transfer of (partly) mismatching stretches of secondary structure between different allergens may well be the reason for a loss of protein fold and, consequently, reduction of IgE-binding capacities.41 The three-dimensional structure of Mal d 1.0101 presented here provides the biophysical basis for elucidating the molecular details of immunological cross-reactivity in great detail.
Acknowledgments
We thank Dr. Kathrin Breuker (University of Innsbruck) for mass spectrometric analyses.
Glossary
Abbreviations Used
- PR-10
pathogenesis-related proteins of class 10
- NOE
nuclear Overhauser effect
- PFG
pulsed field gradient
- FT-ICR
Fourier transform ion cyclotron resonance
- ESI
electrospray ionization
This work was supported by the Austrian Science Fund FWF (P26849) and the European Regional Development Fund (Interreg V-A Italien–Österreich, ITAT1013).
The authors declare no competing financial interest.
References
- Ballmer-Weber B. K. Food allergy in adolescence and adulthood. Chem. Immunol. Allergy 2015, 101, 51–58. 10.1159/000371669. [DOI] [PubMed] [Google Scholar]
- Geroldinger-Simic M.; Zelniker T.; Aberer W.; Ebner C.; Egger C.; Greiderer A.; Prem N.; Lidholm J.; Ballmer-Weber B. K.; Vieths S.; Bohle B. Birch pollen-related food allergy: clinical aspects and the role of allergen-specific IgE and IgG4 antibodies. J. Allergy Clin. Immunol. 2011, 127, 616–622. 10.1016/j.jaci.2010.10.027. [DOI] [PubMed] [Google Scholar]
- Mari A.; Ballmer-Weber B. K.; Vieths S. The oral allergy syndrome: improved diagnostic and treatment methods. Curr. Opin. Allergy Clin. Immunol. 2005, 5, 267–273. 10.1097/01.all.0000168793.27948.b0. [DOI] [PubMed] [Google Scholar]
- Vanek-Krebitz M.; Hoffmann-Sommergruber K.; Machado M. L. D.; Susani M.; Ebner C.; Kraft D.; Scheiner O.; Breiteneder H. Cloning and sequencing of Mal d 1, the major allergen from apple (Malus domestica), and its immunological relationship to Bet v 1, the major birch pollen allergen. Biochem. Biophys. Res. Commun. 1995, 214, 538–551. 10.1006/bbrc.1995.2320. [DOI] [PubMed] [Google Scholar]
- Fernandes H.; Michalska K.; Sikorski M.; Jaskolski M. Structural and functional aspects of PR-10 proteins. FEBS J. 2013, 280, 1169–1199. 10.1111/febs.12114. [DOI] [PubMed] [Google Scholar]
- Schmitz-Eiberger M.; Matthes A. Effect of harvest maturity, duration of storage and shelf life of apples on the allergen Mal d 1, polyphenoloxidase activity and polyphenol content. Food Chem. 2011, 127, 1459–1464. 10.1016/j.foodchem.2011.01.101. [DOI] [Google Scholar]
- Matthes A.; Schmitz-Eiberger M. Apple (Malus domestica L. Borkh.) allergen Mal d 1: effect of cultivar, cultivation system, and storage conditions. J. Agric. Food Chem. 2009, 57, 10548–10553. 10.1021/jf901938q. [DOI] [PubMed] [Google Scholar]
- Marzban G.; Puehringer H.; Dey R.; Brynda S.; Ma Y.; Martinelli A.; Zaccarini M.; van der Weg E.; Housley Z.; Kolarich D.; Altmann F.; Laimer M. Localisation and distribution of the major allergens in apple fruits. Plant Sci. 2005, 169, 387–394. 10.1016/j.plantsci.2005.03.027. [DOI] [Google Scholar]
- Sancho A. I.; Foxall R.; Browne T.; Dey R.; Zuidmeer L.; Marzban G.; Waldron K. W.; Van Ree R.; Hoffmann-Sommergruber K.; Laimer M.; Mills E. N. C. Effect of postharvest storage on the expression of the apple allergen Mal d 1. J. Agric. Food Chem. 2006, 54, 5917–5923. 10.1021/jf060880m. [DOI] [PubMed] [Google Scholar]
- Pagliarani G.; Paris R.; Arens P.; Tartarini S.; Ricci G.; Smulders M. M. J.; van de Weg W. E. A qRT-PCR assay for the expression of all Mal d 1 isoallergen genes. BMC Plant Biol. 2013, 13, 51. 10.1186/1471-2229-13-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puehringer H.; Moll D.; Hoffmann-Sommergruber K.; Watillon B.; Katinger H.; Machado M. L. D. The promoter of an apple Ypr10 gene, encoding the major allergen Mal d 1, is stress- and pathogen-inducible. Plant Sci. 2000, 152, 35–50. 10.1016/S0168-9452(99)00222-8. [DOI] [Google Scholar]
- Puehringer H. M.; Zinoecker I.; Marzban G.; Katinger H.; Laimer M. MdAP, a novel protein in apple, is associated with the major allergen Mal d 1. Gene 2003, 321, 173–183. 10.1016/S0378-1119(03)00822-9. [DOI] [PubMed] [Google Scholar]
- Son D. Y.; Scheurer S.; Hoffmann A.; Haustein D.; Vieths S. Pollen-related food allergy: cloning and immunological analysis of isoforms and mutants of Mal d 1, the major apple allergen, and Bet v 1, the major birch pollen allergen. Eur. J. Nutr. 1999, 38, 201–215. 10.1007/s003940050063. [DOI] [PubMed] [Google Scholar]
- Helsper J. P.; Gilissen L. J.; van Ree R.; America A. H.; Cordewener J. H.; Bosch D. Quadrupole time-of-flight mass spectrometry: a method to study the actual expression of allergen isoforms identified by PCR cloning. J. Allergy Clin. Immunol. 2002, 110, 131–138. 10.1067/mai.2002.125599. [DOI] [PubMed] [Google Scholar]
- Allergen Nomenclature. www.allergen.org (accessed Jan 30, 2017).
- Ahammer L.; Grutsch S.; Tollinger M. NMR resonance assignments of the major apple allergen Mal d 1. Biomol. NMR Assignments 2016, 10, 287–290. 10.1007/s12104-016-9685-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoening B.; Ziegler W. H.; Vieths S.; Baltes W. Apple allergy: The cDNA sequence of the major allergen of apple, determined by performing PCR with a primer based on the N-terminal amino acid sequence, is highly homologous to the sequence of the major birch pollen allergen. J. Sci. Food Agric. 1996, 71, 475–482. . [DOI] [Google Scholar]
- Delaglio F.; Grzesiek S.; Vuister G. W.; Zhu G.; Pfeifer J.; Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Vranken W. F.; Boucher W.; Stevens T. J.; Fogh R. H.; Pajon A.; Llinas M.; Ulrich E. L.; Markley J. L.; Ionides J.; Laue E. D. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins: Struct., Funct., Genet. 2005, 59, 687–696. 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Choy W. Y.; Mulder F. A.; Crowhurst K. A.; Muhandiram D. R.; Millett I. S.; Doniach S.; Forman-Kay J. D.; Kay L. E. Distribution of molecular size within an unfolded state ensemble using small-angle X-ray scattering and pulse field gradient NMR techniques. J. Mol. Biol. 2002, 316, 101–112. 10.1006/jmbi.2001.5328. [DOI] [PubMed] [Google Scholar]
- Grutsch S.; Fuchs J. E.; Freier R.; Kofler S.; Bibi M.; Asam C.; Wallner M.; Ferreira F.; Brandstetter H.; Liedl K. R.; Tollinger M. Ligand binding modulates the structural dynamics and compactness of the major birch pollen allergen. Biophys. J. 2014, 107, 2972–2981. 10.1016/j.bpj.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins D. K.; Grimshaw S. B.; Receveur V.; Dobson C. M.; Jones J. A.; Smith L. J. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry 1999, 38, 16424–16431. 10.1021/bi991765q. [DOI] [PubMed] [Google Scholar]
- Schwieters C. D.; Kuszewski J. J.; Tjandra N.; Clore G. M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003, 160, 65–73. 10.1016/S1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Schwieters C. D.; Kuszewski J. J.; Clore G. M. Using Xplor-NIH for NMR molecular structure determination. Prog. Nucl. Magn. Reson. Spectrosc. 2006, 48, 47–62. 10.1016/j.pnmrs.2005.10.001. [DOI] [Google Scholar]
- Lange O. F.; Rossi P.; Sgourakis N. G.; Song Y.; Lee H. W.; Aramini J. M.; Ertekin A.; Xiao R.; Acton T. B.; Montelione G. T.; Baker D. Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 10873–10878. 10.1073/pnas.1203013109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BMRB CS-Rosetta server. https://csrosetta.bmrb.wisc.edu.
- Shen Y.; Delaglio F.; Cornilescu G.; Bax A. TALOS plus: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 2009, 44, 213–223. 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case D.; Berryman J.; Betz R.; Cerutti D.; Cheatham T. I.; Darden T.; Duke R.; Giese T.; Gohlke H.; Goetz A.; Homeyer N.; Izadi S.; Janowsk i. P.; Kaus J.; Kovalenko A.; Lee T.; LeGrand S.; Li P.; Luchko T.; Luo R.; Madej B.; Merz K.; Monard G.; Needham P.; Nguyen H.; Nguyen H.; Omelyan I.; Onufriev A.; Roe D.; Roitberg A.; Salomon-Ferrer R.; Simmerling C.; Smith W.; Swails J.; Walker R.; Wang J.; Wolf R.; Wu X.; York D.; Kollman P.. AMBER 2015; University of California: San Francisco, CA, USA, 2015. [Google Scholar]
- Salomon-Ferrer R.; Gotz A. W.; Poole D.; Le Grand S.; Walker R. C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- Lindorff-Larsen K.; Piana S.; Palmo K.; Maragakis P.; Klepeis J. L.; Dror R. O.; Shaw D. E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins: Struct., Funct., Genet. 2010, 78, 1950–1958. 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A.; Tejero R.; Montelione G. T. Evaluating protein structures determined by structural genomics consortia. Proteins: Struct., Funct., Genet. 2007, 66, 778–795. 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- Molecular Operating Environment (MOE); Chemical Computing Group Inc.: Montreal, QC, Canada, 2014. [Google Scholar]
- Somkuti J.; Houska M.; Smeller L. Pressure and temperature stability of the main apple allergen Mal d1. Eur. Biophys. J. 2011, 40, 143–151. 10.1007/s00249-010-0633-8. [DOI] [PubMed] [Google Scholar]
- Ma Y.; Gadermaier G.; Bohle B.; Bolhaar S.; Knulst A.; Markovic-Housley Z.; Breiteneder H.; Briza P.; Hoffmann-Sommergruber K.; Ferreira F. Mutational analysis of amino acid positions crucial for IgE-binding epitopes of the major apple (Malus domestica) allergen, Mal d 1. Int. Arch. Allergy Immunol. 2006, 139, 53–62. 10.1159/000089756. [DOI] [PubMed] [Google Scholar]
- Roulias A.; Pichler U.; Hauser M.; Himly M.; Hofer H.; Lackner P.; Ebner C.; Briza P.; Bohle B.; Egger M.; Wallner M.; Ferreira F. Differences in the intrinsic immunogenicity and allergenicity of Bet v 1 and related food allergens revealed by site-directed mutagenesis. Allergy 2014, 69, 208–215. 10.1111/all.12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundas J.; Ouyang Z.; Tseng J.; Binkowski A.; Turpaz Y.; Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006, 34, W116–W118. 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler S.; Asam C.; Eckhard U.; Wallner M.; Ferreira F.; Brandstetter H. Crystallographically mapped ligand binding differs in high and low IgE binding isoforms of birch pollen allergen Bet v 1. J. Mol. Biol. 2012, 422, 109–123. 10.1016/j.jmb.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seutter von Loetzen C.; Hoffmann T.; Hartl M. J.; Schweimer K.; Schwab W.; Rosch P.; Hartl-Spiegelhauer O. Secret of the major birch pollen allergen Bet v 1: identification of the physiological ligand. Biochem. J. 2014, 457, 379–390. 10.1042/BJ20130413. [DOI] [PubMed] [Google Scholar]
- Holm J.; Ferreras M.; Ipsen H.; Wurtzen P. A.; Gajhede M.; Larsen J. N.; Lund K.; Spangfort M. D. Epitope grafting, re-creating a conformational Bet v 1 antibody epitope on the surface of the homologous apple allergen Mal d 1. J. Biol. Chem. 2011, 286, 17569–17578. 10.1074/jbc.M110.194878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiteneder H.; Ebner C. Molecular and biochemical classification of plant-derived food allergens. J. Allergy Clin. Immunol. 2000, 106, 27–36. 10.1067/mai.2000.106929. [DOI] [PubMed] [Google Scholar]
- Wallner M.; Hauser M.; Himly M.; Zaborsky N.; Mutschlechner S.; Harrer A.; Asam C.; Pichler U.; van Ree R.; Briza P.; Thalhamer J.; Bohle B.; Achatz G.; Ferreira F. Reshaping the Bet v 1 fold modulates T(H) polarization. J. Allergy Clin. Immunol. 2011, 127, 1571–1578. 10.1016/j.jaci.2011.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch R.; Bohle B.; Vollmann U.; Wiedermann U.; Jahn-Schmid B.; Krebitz M.; Breiteneder H.; Kraft D.; Ebner C. Bet v 1, the major birch pollen allergen, and Mal d 1, the major apple allergen, cross-react at the level of allergen-specific T helper cells. J. Allergy Clin. Immunol. 1998, 102, 679–686. 10.1016/S0091-6749(98)70287-8. [DOI] [PubMed] [Google Scholar]
- Ebner C.; Birkner T.; Valenta R.; Rumpold H.; Breitenbach M.; Scheiner O.; Kraft D. Common epitopes of birch pollen and apples-studies by western and northern blot. J. Allergy Clin. Immunol. 1991, 88, 588–594. 10.1016/0091-6749(91)90152-E. [DOI] [PubMed] [Google Scholar]
- Ebner C.; Hirschwehr R.; Bauer L.; Breiteneder H.; Valenta R.; Ebner H.; Kraft D.; Scheiner O. Identification of allergens in fruits and vegetables: IgE cross-reactivities with the important birch pollen allergens Bet v 1 and Bet v 2 (birch profilin). J. Allergy Clin. Immunol. 1995, 95, 962–969. 10.1016/S0091-6749(95)70096-X. [DOI] [PubMed] [Google Scholar]
- Mirza O.; Henriksen A.; Ipsen H.; Larsen J. N.; Wissenbach M.; Spangfort M. D.; Gajhede M. Dominant epitopes and allergic cross-reactivity: complex formation between a Fab fragment of a monoclonal murine IgG antibody and the major allergen from birch pollen Bet v 1. J. Immunol. 2000, 165, 331–338. 10.4049/jimmunol.165.1.331. [DOI] [PubMed] [Google Scholar]
- Spangfort M. D.; Mirza O.; Ipsen H.; Van Neerven R. J.; Gajhede M.; Larsen J. N. Dominating IgE-binding epitope of Bet v 1, the major allergen of birch pollen, characterized by X-ray crystallography and site-directed mutagenesis. J. Immunol. 2003, 171, 3084–3090. 10.4049/jimmunol.171.6.3084. [DOI] [PubMed] [Google Scholar]
- Holm J.; Baerentzen G.; Gajhede M.; Ipsen H.; Larsen J. N.; Lowenstein H.; Wissenbach M.; Spangfort M. D. Molecular basis of allergic cross-reactivity between group 1 major allergens from birch and apple. J. Chromatogr., Biomed. Appl. 2001, 756, 307–313. 10.1016/S0378-4347(01)00089-5. [DOI] [PubMed] [Google Scholar]
- Bolhaar S. T.; Zuidmeer L.; Ma Y.; Ferreira F.; Bruijnzeel-Koomen C. A.; Hoffmann-Sommergruber K.; van Ree R.; Knulst A. C. A mutant of the major apple allergen, Mal d 1, demonstrating hypo-allergenicity in the target organ by double-blind placebo-controlled food challenge. Clin. Exp. Allergy 2005, 35, 1638–1644. 10.1111/j.1365-2222.2005.02390.x. [DOI] [PubMed] [Google Scholar]
- Ferreira F.; Ebner C.; Kramer B.; Casari G.; Briza P.; Kungl A. J.; Grimm R.; Jahn-Schmid B.; Breiteneder H.; Kraft D.; Breitenbach M.; Rheinberger H. J.; Scheiner O. Modulation of IgE reactivity of allergens by site-directed mutagenesis: potential use of hypoallergenic variants for immunotherapy. FASEB J. 1998, 12, 231–242. [DOI] [PubMed] [Google Scholar]
- Klinglmayr E.; Hauser M.; Zimmermann F.; Dissertori O.; Lackner P.; Wopfner N.; Ferreira F.; Wallner M. Identification of B-cell epitopes of Bet v 1 involved in cross-reactivity with food allergens. Allergy 2009, 64, 647–651. 10.1111/j.1398-9995.2008.01844.x. [DOI] [PubMed] [Google Scholar]
- Hecker J.; Diethers A.; Schulz D.; Sabri A.; Plum M.; Michel Y.; Mempel M.; Ollert M.; Jakob T.; Blank S.; Braren I.; Spillner E. An IgE epitope of Bet v 1 and fagales PR10 proteins as defined by a human monoclonal IgE. Allergy 2012, 67, 1530–1537. 10.1111/all.12045. [DOI] [PubMed] [Google Scholar]
- Ferreira F.; Wolf M.; Wallner M. Molecular approach to allergy diagnosis and therapy. Yonsei Med. J. 2014, 55, 839–852. 10.3349/ymj.2014.55.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F.; Wilm A.; Dineen D.; Gibson T. J.; Karplus K.; Li W.; Lopez R.; McWilliam H.; Remmert M.; Soding J.; Thompson J. D.; Higgins D. G. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]