Abstract
CRISPR-based genetic screens are accelerating biological discovery, but current methods have inherent limitations. Widely used pooled screens are restricted to simple readouts including cell proliferation and sortable marker proteins. Arrayed screens allow for comprehensive molecular readouts such as transcriptome profiling, but at much lower throughput. Here we combine pooled CRISPR screening with single-cell RNA sequencing into a broadly applicable workflow, directly linking guide-RNA expression to transcriptome responses in thousands of individual cells. Our method for CRISPR droplet sequencing (CROP-seq) enables pooled CRISPR screens with single-cell transcriptome resolution, which will facilitate high-throughput functional dissection of complex regulatory mechanisms and heterogeneous cell populations.
Keywords: CRISPR genome editing, single-cell RNA-seq, Drop-seq, CROP-seq, genetic screening, pooled screens, arrayed screens, single-cell CRISPR screens, transcriptome signatures, bioinformatic cell state inference
Pooled CRISPR screening is a powerful and widely used method for identifying genes involved in biological mechanisms such as cell proliferation1, 2, drug resistance3, and viral infection4. Cells are transduced in bulk with a library of guide-RNA (gRNA)-encoding vectors, and the distribution of gRNAs is measured before and after applying a selective challenge (Fig. 1a). Pooled CRISPR screens work well for mechanisms that affect cell survival and proliferation, and they can be extended to measure the activity of individual genes (e.g., by using engineered reporter cell lines). However, they do not support complex molecular readouts such as transcriptome profiling, which is one of the most comprehensive and informative measures of cellular response5. Arrayed CRISPR screens, in which only one gene is targeted at a time, make it possible to use RNA-seq as the readout6. But this comes at the cost of a much lower throughput because cells transduced with individual gRNAs need to be physically separated (Fig. 1b).
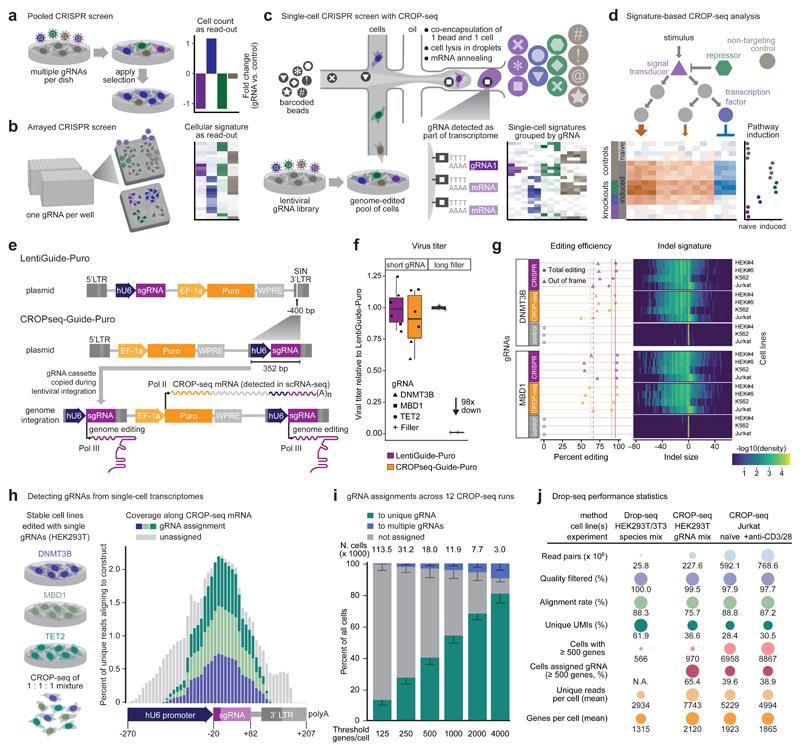
Figure 1. CROP-seq enables pooled CRISPR screening with single-cell transcriptome readout.
a) Pooled screens detect changes in gRNA abundance among bulk populations of cells, which limits them to simple readouts based on cell frequencies. b) Arrayed screens support complex readouts such as transcriptome profiling, but cells transduced with different gRNAs have to be physically separated. c) CROP-seq uses droplet-based single-cell RNA-seq to profile each cell’s transcriptome together with the expressed gRNA, and knockout signatures are derived by averaging across cells that express gRNAs for the same target gene. d) Data analysis identifies pathway signature genes and quantifies the effect of specific gRNAs on these signatures. e) The CROP-seq lentiviral construct includes a gRNA cassette within the 3’ long terminal repeat (LTR), which is duplicated during viral integration. It expresses an RNA polymerase III transcript for genome editing and a polyadenylated RNA polymerase II transcript detected by single-cell RNA-seq. f) Cloning the hU6-gRNA cassette into the 3’ LTR to generate CROPseq-Guide-Puro does not compromise lentiviral function for gRNAs. In contrast, 1,885 bp of filler DNA result in a 98-fold reduction of the viral titer. g) Genome editing efficiencies and indel signatures are highly similar between LentiGuide-Puro and CROPseq-Guide-Puro. h) CROP-seq can detect gRNAs from single-cell transcriptomes. i) The rate of successful gRNA assignments is associated with single-cell transcriptome quality, expressed as the number of detected genes per cell. Most cells were assigned to one gRNA, except for a small fraction of cell doublets. Error bars, 95% CI. j) Performance statistics across all CROP-seq experiments.
Here we propose a third and complementary screening paradigm, single-cell CRISPR screens, based on the idea that gRNAs and their induced cellular responses are already compartmentalized within single cells. Detecting each cell’s gRNA along with its single-cell transcriptome would thus provide gene expression signatures for individual gene knockouts in a complex pool of cells (Fig. 1c). Implementing this concept, our CROP-seq method combines four key components: a gRNA vector that makes individual gRNAs detectable in single-cell RNA-seq experiments, a high-throughput assay for single-cell RNA-seq7, a computational pipeline for assigning single-cell transcriptomes to gRNAs, and a bioinformatic method for analyzing and interpreting gRNA-induced transcriptional profiles. CROP-seq thereby enables pooled screens with single-cell transcriptome readout, providing a scalable method for dissecting complex signaling pathways and other biological mechanisms that are not easily reduced to a single selectable marker (Fig. 1d).
Results
Direct detection of gRNAs from single-cell transcriptome data
CRISPR gRNAs are typically transcribed by RNA polymerase III from a human U6 promoter (hU6). They lack a polyadenylated (poly-A) tail and are not detectable with current single-cell RNA-seq assays. We thus re-engineered a popular construct for pooled CRISPR screening (LentiGuide-Puro)8 to include the gRNA in a polyadenylated mRNA transcript (Fig. 1e, Supplementary Data, and Supplementary Fig. 1). Because LentiGuide-Puro was originally derived from a lentiviral vector rendered self-inactivating by a 400-bp deletion of key promoter elements, we hypothesized that this position within the 3’ long terminal repeat (LTR) would tolerate the insertion of the similarly sized hU6-gRNA cassette. At this position, the gRNA becomes part of the puromycin-resistance mRNA transcribed by RNA polymerase II and is detectable by RNA-seq protocols that use poly-A enrichment. Functional gRNAs continue to be expressed from the hU6 promoter. In addition, the entire hU6-gRNA cassette is copied to the 5’ LTR during reverse transcription and integration of the virus (Supplementary Fig. 1). This results in a second copy upstream of the possibly interfering EF-1a promoter – a design reminiscent of an early shRNA expression vector9. CROP-seq thereby solves the challenge of detecting gRNAs in single-cell transcriptomes at the vector level, which makes it compatible with various single-cell RNA-seq assays and with widely used cloning protocols for pooled screening.
Having assembled our CROPseq-Guide-Puro plasmid from four PCR products using the ligase cycling reaction10 (Supplementary Fig. 1 and Supplementary Table 1), we extensively validated its performance and suitability for CRISPR screening. The hU6-gRNA insertion into the 3’ LTR had no adverse effects on the formation of lentiviral particles nor on puromycin resistance of transduced cells. We produced virus from LentiGuide-Puro and CROPseq-Guide-Puro containing one of three validated gRNAs (targeting DNMT3B, MBD1, or TET2) or a longer filler construct that was expected to interfere with viral activity, and we observed comparable functional titers for all tested gRNA sequences (Fig. 1f). Genome editing efficiencies were benchmarked in three Cas9-expressing cell lines (K562, Jurkat, and two HEK293T clones), using two gRNAs and two complementary technologies – the T7 endonuclease assay11 (Supplementary Fig. 2) and next-generation sequencing of PCR amplicons (Fig. 1g). Genome editing was highly efficient for both constructs, averaging at 95.5% (LentiGuide-Puro) and 90.5% (CROPseq-Guide-Puro), with the expected two thirds (66.2% and 62.4%) of molecules carrying out-of-frame insertions and deletions. For each locus and cell line, we also observed characteristic editing signatures, which were highly similar between both constructs (Fig. 1g). CROPseq-Guide-Puro thus provides excellent results both in terms of the quantity and quality of induced genome editing events.
Next, we tested whether single-cell RNA-seq could detect expressed gRNAs in cells transduced with CROPseq-Guide-Puro (Fig. 1h and Supplementary Fig. 3). We generated three knockout cell lines from HEK293T cells, targeting DNMT3B, MBD1, and TET2 with validated gRNAs. By performing the transductions independently, we ensured that each cell received a unique gRNA – even in case of multiple infection events. When we applied Drop-seq7 on a 1:1:1 mixture of the three cell lines, the resulting single-cell transcriptomes provided high sequencing coverage for the gRNAs, and we were able to assign single cells to gRNAs at the expected ratios (Fig. 1h).
Combining all CROP-seq data from this study, we assessed the confidence of our gRNA assignments (Fig. 1i), which depended on the number of detected genes per cell. For example, 38.7% of cells with at least 500 and 78.9% of cells with at least 4,000 detected genes were uniquely assigned. Few cells matched more than one gRNA (e.g., 2.7% for a threshold of 500 detected genes), although this rate increased with the detected number of genes per cell (e.g., 9.8% for a threshold of 4,000). This rate increase was most likely due to rare cell doublets (Supplementary Fig. 3d) that release twice the amount of RNA, resulting in more detected genes and multiple gRNA assignments. Furthermore, we excluded any cells that were assigned to multiple gRNAs from the downstream analysis, and CROP-seq is robust toward potentially undetected doublets because it combines data across all single cells assigned to the same gRNA.
Single-cell CRISPR screening for T-cell-receptor induction
Having established and validated CROP-seq as a method for single-cell CRISPR screens (Fig. 1j provides detailed performance statistics across all 12 CROP-seq experiments), we tested our method in a proof-of-concept screen of T-cell receptor (TCR) activation in Jurkat cells (Fig. 2a). We designed a gRNA library for six high-level regulators of TCR signaling and 23 transcription factors, targeting each gene with three distinct gRNAs (Supplementary Table 2). We also included 20 nontargeting gRNAs as negative controls and 9 gRNAs for essential genes2 as positive controls. Jurkat cells that stably express Cas9 were transduced with lentivirus produced from this CROPseq-Guide-Puro gRNA library, and genome-edited cells were selected with puromycin. At day 10 post-transduction, the surviving pool of cells was serum starved, split, and subjected to either TCR stimulation via anti-CD3 and anti-CD28 antibodies or to continued starvation; and both cell populations were analyzed with CROP-seq.
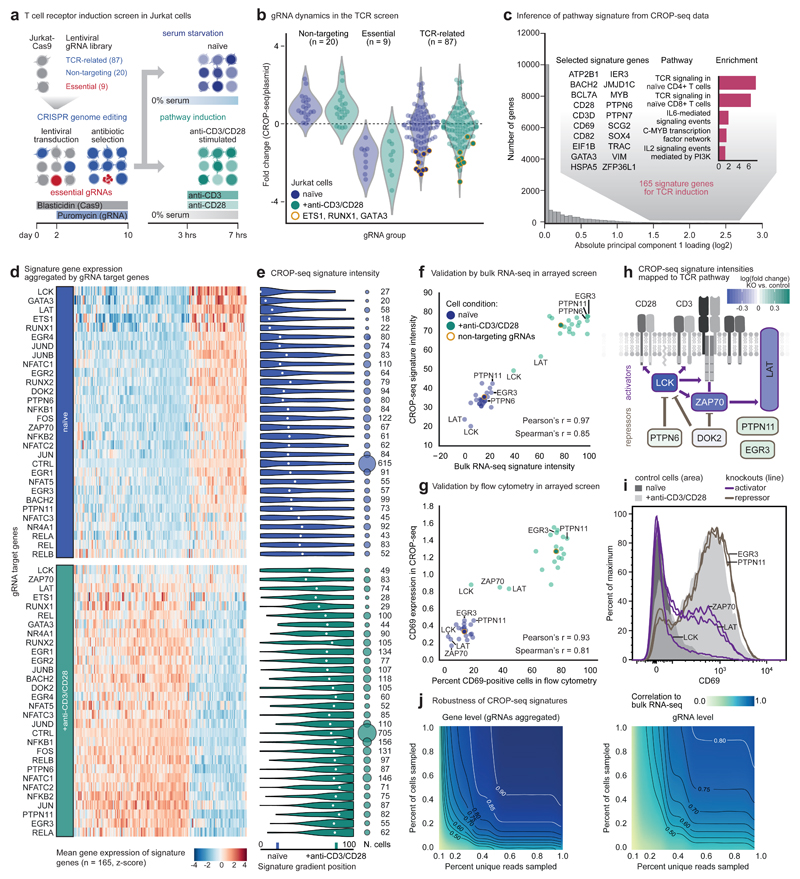
Figure 2. CROP-seq analysis of T cell receptor signaling.
a) Experimental design of a single-cell CRISPR screen for T cell receptor (TCR) pathway induction. b) Fold change of gRNA abundance between cell assignments from CROP-seq and gRNA counts from plasmid library sequencing. Values were normalized to the total of assigned cells or reads, respectively. c) Inference of pathway signature from CROP-seq data. Single-cell transcriptomes were aggregated by gRNA target genes, and principal component analysis separated naive and anti-CD3/CD28-stimulated cells. Genes with absolute loading values for principal component 1 that exceeded the 99th percentile were included in the TCR induction signature (n = 165). The signature was enriched for genes with a known role in TCR signaling (inset). d) Median relative expression (column z-score) across the 165 pathway signature genes (columns), aggregating cells that express gRNAs targeting the same gene (rows). e) Distribution of signature intensity across single cells (left) and number of cells (right) for each gRNA target gene. The median is indicated with a white dot. f) Gene signature concordance between CROP-seq and bulk RNA-seq in an arrayed validation screen. Known positive and negative regulators of the TCR pathway are highlighted. g) Concordance of the CD69 marker of TCR induction between CROP-seq and an arrayed validation screen with flow cytometry readout. h) Changes in TCR pathway induction detected by CROP-seq mapped onto a schematic of the T-cell receptor with key downstream regulators. i) CD69 marker levels in control cells and knockouts for important TCR activators or repressors. j) Robustness of CROP-seq signatures in a downsampling analysis at the gene and gRNA levels, evaluated against bulk RNA-seq data.
Comparing gRNA assignments based on CROP-seq with gRNA counts obtained by sequencing the plasmid library, we observed a consistent depletion of positive controls, confirming efficient editing (Fig. 2b and Supplementary Fig. 4). The gRNAs in our TCR library showed more diverse patterns; gRNAs targeting ETS1, RUNX1, and GATA3 were depleted, suggesting that the encoded transcription factors are essential in Jurkat cells. In contrast, most gRNAs behaved similarly to the negative controls, indicating that they did not have strong antiproliferative effects. At the transcriptome level, pairwise distances between gRNAs targeting the same gene were smaller than those between gRNAs targeting different genes (Supplementary Fig. 5a,b), and transcriptome profiles assigned to specific gRNAs and target genes were distinguishable from those of cells expressing nontargeting gRNAs (Supplementary Fig. 5c,d).
We established a transcriptome signature of TCR induction directly from the CROP-seq data by applying dimensionality reduction to single-cell RNA-seq profiles (Supplementary Fig. 6a-c). Principal component analysis for cells grouped by gRNA target genes separated naive and TCR-induced Jurkat cells along the first principal component, which defined a TCR induction signature of 165 genes (Fig. 2c and Supplementary Fig. 6d,e). This signature was enriched for genes known to be relevant in TCR signaling (Fig. 2c and Supplementary Fig. 6f).
Aggregating single-cell RNA-seq profiles by their assigned target genes and gRNAs (Fig. 2d and Supplementary Fig. 7a,b), we found that induced cells expressing gRNAs for LCK and ZAP70 were similar to the naive group, indicating that knockout of these key mediators of TCR signaling interfered with TCR induction. For a more quantitative picture, we positioned each cell on a gradient defined by naive and induced cells that expressed nontargeting control gRNAs (Fig. 2e and Supplementary Fig. 7c-e). This analysis was based on 5,798 high-quality single-cell transcriptome profiles with unique gRNA assignments and a median of 80 cells per targeted gene, while 1,320 control cells with nontargeting gRNAs were used as reference.
Validation by bulk RNA-seq and flow cytometry
For validation, we performed an arrayed screen using 48 independent lentiviral constructs, targeting 20 genes with two gRNAs and including eight nontargeting controls (Supplementary Table 3). We measured TCR induction by bulk RNA-seq (87 RNA-seq libraries; Fig. 2f and Supplementary Fig. 8a,b) and flow cytometry (96 flow cytometry profiles; Fig. 2g). High correlations were observed between bulk RNA-seq and CROP-seq data, both for inferred TCR signature intensities and for global expression profiles (Fig. 2f and Supplementary Fig. 8c,d). Flow cytometry for surface proteins that were correlated with our TCR signature (CD69 and CD82) or previously reported as markers of T-cell activation (CD25, CD38, CD154, and PD-1) further validated the CROP-seq results (Fig. 2g and Supplementary Fig. 9).
The arrayed screen also confirmed the results for individual genes that CROP-seq identified as important for TCR signaling in Jurkat cells. Depletion of the kinases LCK and ZAP70 and the adaptor protein LAT had a strong negative effect on the TCR activation signature, consistent with their established role in TCR signal propagation and amplification12 (Fig. 2h,i and Supplementary Fig. 8e). Moreover, we observed that targeting known negative regulators of TCR signaling (such as PTPN6, PTPN11, and EGR3) slightly increased the TCR activation signature. While the focus of this screen was to validate CROP-seq as a method, we also identified a few transcription factors that may warrant further investigation of their potential functional roles during TCR induction (Fig. 2d,e and Supplementary Fig. 7).
To assess how well CROP-seq might scale to larger (and eventually genome-wide) gRNA libraries, we performed downsampling analysis and identified the minimum number of cells that resulted in concordant TCR signatures regarding the TCR signature intensities of the targeted genes (Fig. 2j). We found that 870 single cells (15% of the dataset) yielded results similar to those of the entire screen. However, the exact number of cells needed for future screens will also depend on the strength of the investigated biological signal.
Discussion
We have presented proof of concept for single-cell CRISPR screening as an approach that combines key strengths of pooled and arrayed screens, and we validated CROP-seq as a flexible and straightforward method for performing such screens. We showed that gene signatures can be derived directly from single-cell data using the transcriptomes of cells with nontargeting gRNAs, which makes the assay self-calibrating and independent of any prior knowledge about relevant marker genes.
While this paper was in review13, two alternative methods – Perturb-seq and CRISP-seq – were published that use transcribed barcodes for single-cell CRISPR screening14–16. Compared to these methods, CROP-seq has the key advantage that it reads the gRNA directly, which dramatically simplifies single-cell CRISPR screening with large gRNA libraries and which makes CROP-seq fully compatible with standard cloning protocols for pooled CRISPR screens.
We expect CROP-seq to be broadly relevant for studying biological mechanisms that are difficult to reduce to a simple readout needed for classical pooled screens. Applied to heterogeneous cell populations and in vivo tissue (e.g., in combination with Cas9-expressing mice), CROP-seq can localize cell-type-specific changes in complex organs and cellular differentiation hierarchies. Given the increasing throughput of single-cell transcriptomics17 and the advent of single-cell multi-omics technology18, CROP-seq has the potential to provide comprehensive characterization of large CRISPR libraries and constitutes a powerful method for dissecting cellular regulation at scale.
Online Methods
Step-by-step protocol
A step-by-step experimental protocol for CROP-seq is included as Supplementary Protocol. Future updates of the protocol will be shared via http://crop-seq.computational-epigenetics.org.
Cloning and validation of the CROPseq-Guide-Puro plasmid
To clone the CROPseq-Guide-Puro plasmid, we amplified four PCR products from LentiGuide-Puro (Addgene 52963) and assembled them in different order using the ligase cycling reaction (LCR)10. Bridge oligonucleotides for LCR were designed such that each side had a melting temperature of 70 °C. The sequences of all primer pairs, the respective annealing temperatures, and the overlapping bridge oligonucleotides are provided in Supplementary Table 1. Plasmid fragments were amplified by mixing 12.5 μl Q5 Hot Start High-Fidelity 2x Master Mix (NEB cat. no. M0494L), 1.25 μl of 10 μM FWD primer, 1.25 μl of 10 μM REV primer, 10 ng of plasmid template, and water to 25 μl, and incubating as follows: 98 °C for 30 s, 25x (98 °C for 10 s, Ta or 30 s, 72 °C for 1 min 30 s), 72 °C for 2 min, hold at 10 °C. PCR products were cleaned by a 2.0x AMPure XP bead cleanup (Beckman Coulter cat. no. A63880), measured in a Qubit dsDNA HS assay (Invitrogen cat. no. Q32854), and diluted to 20 nM. Next, we mixed 3.125 μl of the four diluted PCR products, added 1.5 μl of 10x Ampligase buffer and 1 μl of DpnI (20 U/μl, NEB cat. no. R0176S), and digested at 37 °C for 1 hour without heat-inactivation. LCR was started by mixing 15 μl of DpnI-digested PCR preparations, 1 μl of 10x Ampligase buffer, 1.5 μl of Ampligase enzyme (5 U/μl, Epicentre cat. no. A32250), 2.25 μl 5 M betaine (Sigma Aldrich cat. no. B0300-1VL), 2 μl DMSO (Sigma Aldrich cat. no. D9170-1VL), 0.5 μl each of the four bridge oligonucleotides (1.5 μM in water), and 1.25 μl of water. The reaction was incubated for 2 min at 94 °C, 50x (94 °C for 10 s, 55 °C for 30 s, 66 °C for 1 min), storage at 4 °C. 2.5 μl of the LCR assembly were transformed into 25 μl of NEB Stable E. coli (NEB cat. no. C3040H) according to the manufacturer’s high-efficiency protocol. We screened for correctly assembled clones by colony PCR and further validated the assembly by restriction digestion with either SphI or SapI as well as Sanger sequencing. Moreover, to validate the duplication of the hU6-gRNA cassette, genomic DNA from cells infected with LentiGuide-Puro lentivirus, CROPseq-Guide-Puro lentivirus, or the CROPseq-Guide-Puro plasmid pool were PCR amplified with the primers hU6_FWD1 and hU6_REV2 (Supplementary Table 1), which are oriented in opposite directions. In this PCR, productive amplification occurs only when the gRNA cassette is duplicated during lentiviral reverse transcription and integration or when amplifying from a circular plasmid.
Cloning of individual gRNAs into the CROPseq-Guide-Puro plasmid
gRNA cassettes were annealed from two oligonucleotides (top: 5’-CACCG(N)20-3’, bottom: 5’-AAAC(N)20C-3’) by combining 1 μl of each 100 μM oligonucleotide with 1 μl of 10x T4 ligation buffer (NEB cat. no. B0202S), 6.5 μl of water, and 0.5 μl of T4 polynucleotide kinase (NEB cat. no. M0201S), incubating as follows: 37 °C for 30 min (oligonucleotide phosphorylation), 95 °C for 5 min, then ramping from 90 °C to 25 °C at 5 °C/min. Plasmid backbone was prepared by digesting 1 μg of CROPseq-Guide-Puro with 10 units of BsmBI (NEB cat. no. R0580L) in a volume of 30 μl 1x NEB buffer 3.1, incubating for 1 hour at 55 °C. To dephosphorylate the digested plasmid, we added 2 μl of shrimp alkaline phosphatase (rSAP, NEB cat. no. M0371L), incubating for 1 hour at 37 °C followed by heat inactivation (both BsmBI and rSAP) for 20 min at 80 °C. Ligation reactions were set up as follows: 1.6 μl of rSAP reaction, 1 μl gRNA cassette (diluted 1:200 in water), 5 μl 2x Quick Ligase buffer, 2.4 μl water, and 1 μl Quick Ligase (NEB cat. no. M2200S), incubated at 25 °C for 15 min. The ligation reaction was chemically transformed into NEB Stable competent E. coli (NEB cat. no. C3040H) following the manufacturer’s high-efficiency protocol.
Amplification-free cloning of pooled gRNA libraries into the CROPseq-Guide-Puro plasmid
Vector backbone was prepared by digesting 5 μg of CROPseq-Guide-Puro with 20 units of BsmBI (NEB cat. no. R0580L) in a total volume of 25 μl 1x NEB buffer 3.1, incubating for 1 h at 55 °C and inactivating the restriction enzyme for 20 min at 80 °C. The 8,333 bp fragment was purified using the SNAP UV-free gel purification kit (Invitrogen cat. no. K200025). gRNAs were synthesized by Sigma Aldrich as 74 base oligonucleotides with 18 and 35 bases of homology to the hU6 promoter and guide RNA backbone, respectively. Oligonucleotides were diluted to 100 μM and pooled in equal amounts. The oligonucleotide pool was further diluted 1:1,000 for a final molarity of 100 nM for assembly into the vector backbone. gRNA libraries were cloned by Gibson’s isothermal assembly: 11 fmoles (56.7 ng) of CROPseq-Guide-Puro backbone and 200 fmoles of pooled ssDNA oligonucleotides (2 μl of the 100 nM solution) were combined with 10 μl of NEBuilder HiFi DNA assembly master mix (NEB cat. no. E2621S) and water to 20 μl. After 1 h of incubation at 50 °C, reactions were desalted by filter dialysis (Merck cat. no. VMWP04700), and 10 μl of the reaction was electroporated into 25 μl of Lucigen Endura E. coli cells (Lucigen cat. no. 60242-2) using prechilled 1 mm electroporation cuvettes (BioRad cat. no. 1652089) in a BioRad GenePulser I machine set to 25 μF, 200 Ω and 1.5 kV. Within seconds after the pulse, 1 ml of 37 °C Recovery Medium (Lucigen) was added and bacteria were grown in round-bottom tubes for 1 h at 37 °C while shaking at 180 r.p.m. Then, 1 ml of the bacterial culture was plated on a 25 x 25 cm bioassay plate containing LB medium (Miller) with 100 µg/ml carbenicillin. Plates were incubated at 32 °C for 22 h, then LB medium was added and cells were scraped off the plate. Bacterial cells were pelleted by 15 min of centrifugation at 5,000 RCF at 4 °C, and plasmid DNA was extracted with Qiagen’s EndoFree Plasmid Mega kit (Qiagen cat. no. 12381). Library coverage was estimated by counting the number of bacterial colonies on a 1:1,000 dilution plate. The T-cell receptor library was cloned at 853x coverage.
Lentivirus production for single gRNAs and pooled CROP-seq screens
HEK293T cells were plated onto six-well plates at 1.2 million cells per well in 2 ml of lentivirus packaging medium (Opti-MEM I (Gibco cat. no. 51985-034), 5% FCS (Sigma), 200 μM sodium pyruvate (Gibco cat. no. 11360-070), no antibiotics). The next morning, cells were transfected with lipofectamine 3000 (Invitrogen cat. no. L3000015) using 1.7 μg of CROPseq-Guide-Puro (containing single gRNAs or libraries) or LentiCas9-Blast (Addgene 52962), and 0.9 μg each of the three packaging plasmids pMDLg/pRRE (Addgene 12251), pRSV-Rev (Addgene 12253), and pMD2.G (Addgene 12259). The medium was exchanged for fresh lentivirus packaging medium 6 h after the transfection. Supernatant containing viral particles was harvested at 24 h and 48 h, pooled, and passed through a 0.45 μm filter to remove cells. Further concentration of viral particles was not required. Virus was stored at -80 °C. One vial was subsequently thawed for estimating the virus titer.
Lentivirus titration for single gRNAs
HEK293T cells were seeded onto 24-well plates at 50,000 cells per well in 500 μl of culture medium (DMEM (Gibco cat. no. 10569010), 10% FCS, no antibiotics) and grown overnight to reach 30% to 50% confluence. The next day, medium was exchanged for 450 μl per well of fresh culture medium (prepared as described above) containing 8 μg/ml polybrene (Sigma Aldrich cat. no. H9268-5G), which was also used to dilute the viral stock. Lentivirus preparations were thawed from storage at -80 °C and titrated in a 1:5 dilution series ranging over ten wells (1:5 to 1:9,765,625). Each dilution was tested in duplicate by adding 50 μl per well to the 24-well plate. At least two wells per plate served as untransduced controls. 24 h after the transduction, the medium was exchanged for 500 μl per well of complete culture medium (DMEM (Gibco cat. no. 10569010), 10% FCS, penicillin/streptomycin). Starting at 48 h post-transduction, the medium was exchanged for selection medium every 2-3 d. Depending on the construct, the selection medium for HEK293T cells contained either 22.5 μg/ml of blasticidin (Invivogen cat. no. ant-bl-5) or 2.25 μg/ml of puromycin (Fisher Scientific cat. no. A1113803). As soon as all cells in the untransduced controls had died, medium was removed and the plate was washed once with 1x PBS. Resistant colonies were stained for 15 min in a solution of 1% (w/v) crystal violet (Sigma Aldrich cat. no. 61135-25G) in a 10% solution of ethanol in water. After washing the plate several more times with 1x PBS, colonies were counted, and the virus titer (transducing units per ml) was calculated as (no. of resistant colonies) x dilution factor) x (1,000 μl/50 μl).
Lentiviral transduction with gRNA libraries or single gRNAs
For suspension cell lines (Jurkat), cells were seeded in six-well plates at 5 million cells per well in 2 ml of complete culture medium (RPMI (Gibco cat. no. 21875-034) with penicillin/streptomycin and 10% FCS (Sigma)) containing 8 μg/ml polybrene (Sigma Aldrich cat. no. H9268-5G) and blasticidin (Invivogen cat. no. ant-bl-5) to maintain selection for LentiCas9-Blast. Wells 1 to 5 were transduced with 250 μl or 50 μl of the virus stock; or 50 μl of a 1:3, 1:6, or 1:12 dilution. Well 6 served as the untransduced control. Immediately after addition of the virus, cells were centrifuged at 37 °C and 1,200 RCF for 45 min, followed by overnight incubation at 37 °C, 5% CO2. After 24 h, cells were pelleted in 15 ml tubes, taken up in 30 ml of fresh complete culture medium containing blasticidin and transferred to T75 culture flasks. At 48 h post-transduction, selection with puromycin (Fisher Scientific cat. no. A1113803) was started. A flask with about 30% surviving cells was chosen and grown for 10 d to allow for efficient genome editing while renewing the selective medium (containing blasticidin and puromycin) every 2-3 d. Adherent cells (HEK293T) were plated at 5 million cells per 15 cm dish in complete culture medium (DMEM (Gibco cat. no. 41965-039), 10% FCS, penicillin/streptomycin), preparing six plates per library. After 24 h, the medium was replaced by complete culture medium containing 8 μg/ml polybrene, and different amounts of virus were added to the plates: 250 or 50 μl of the virus stock; or 50 μl of a 1:3, 1:6, or 1:12 dilution. The sixth plate remained untransduced. After 24 h, the virus-containing medium was replaced by fresh complete culture medium. Puromycin selection was started at 48 h post-transduction, and 2 d later a plate with about 30% surviving cells was selected and grown under blasticidin and puromycin selection for 10 d.
Preparation of lentivirus for the arrayed validation screen
We cloned 48 individual CROPseq-Guide-Puro plasmids, targeting 20 genes with two gRNAs each and including eight nontargeting controls. All plasmids were verified by Sanger sequencing with the primer 5’-TTGGGCACTGACAATTCCGT-3’. HEK293T cells were seeded onto a 96-well plate at 50,000 cells per well in 200 μl lentivirus packaging medium (Opti-MEM I (Gibco cat. no. 51985-034), 5% FCS (Sigma), 200 μM sodium pyruvate (Gibco cat. no. 11360-070), no antibiotics), and incubated overnight at 37 °C and 5% CO2. Virus production was initiated by transfection with Lipofectamine 3000 reagents (ThermoFisher cat. no. L3000015): transfection mix A (per reaction: 25 μl Opti-MEM I, 0.6 μl P3000 Enhancer Reagent, 90 ng each of the three packaging plasmids pMDLg/pRRE (Addgene 12251), pRSV-Rev (Addgene 12253), and pMD2.G (Addgene 12259)) was distributed in a 96-well plate, and 170 ng of the respective CROPseq-Guide-Puro transfer plasmid were added. Lipid complexes were formed by addition of transfection mix B (per reaction: 25 μl Opti-MEM I, 0.7 μl Lipofectamine 3000 Reagent) and incubating for 20 min at room temperature. 100 µl of medium was removed from the HEK293T cells, then 50 µl of the lipid complexes was added. After 6 h, the medium was exchanged for fresh lentivirus production medium. Viral supernatant was collected at 24 h and 48 h, combined in a sterile deepwell plate (Corning cat. no. 07-200-700), and filtered through a sterile 0.45 µm filter plate (Merck cat. no. MSHVS4510) to remove cells.
Production of single gene knockout cell lines for the arrayed validation screen
Jurkat cells stably expressing Cas9 (after transduction with LentiCas9-Blast, Addgene 52962) were seeded onto a 96-well plate at 250,000 cells per well in 100 µl of complete culture medium (RPMI (Gibco cat. no. 21875-034), 10% FCS (Sigma), penicillin-streptomycin) containing 25 µg/ml blasticidin (Invivogen cat. no. ant-bl-5) to maintain the selection pressure for Cas9 and 8 µg/ml polybrene (Sigma Aldrich cat. no. H9268-5G) to facilitate the interaction of viral particles with the cells. To transduce the cells, between 2.5 and 10 µl of the prepared lentiviral particles were added with a multichannel pipette, and the plate was centrifuged at 37 °C and 1,200 RCF for 45 min, followed by overnight incubation at 37 °C and 5% CO2. After 24 h, cells were pelleted by centrifugation at 300 RCF for 5 min, and the medium was exchanged to select for transduced cells expressing the gRNA construct (RPMI, 10% FCS, penicillin/streptomycin, 25 µg/ml blasticidin, 2 µg/ml puromycin (Fisher Scientific cat. no. A1113803)). 48 h post-transduction, the virus concentration yielding about 30% transduced cells was chosen for further expansion. Cells were subjected to continued selection over the course of 10 d, moving to 24-well and eventually to six-well plates.
T7 endonuclease assay
For easy-to-resolve cleavage products, PCR primers were designed such that the DNMT3B and MBD1 gRNA targets are located off center of the amplicons (sequences and annealing temperatures are provided in Supplementary Table 1). PCR reactions were set up in a reaction volume of 50 μl: 25 μl of Q5 Hot Start High-Fidelity 2x Master Mix (NEB cat. no. M0494L), 2.5 μl 10 μM FWD primer, 2.5 μl 10 μM REV primer, 100 ng of genomic DNA, and water, and incubated as follows: 98 °C for 30 s, 35x (98 °C for 5 s, Ta for 10 s, 72 °C for 20 s), 72 °C for 2 min and storage at 10 °C. PCR products were purified by a 2.0x AMPure XP bead clean-up (Beckman Coulter cat. no. A63880) and measured in a Qubit HS assay (Invitrogen cat. no. Q32854). 200 ng of each PCR product were taken up in 19 μl of 1x NEB buffer 2 and subjected to denaturation and reannealing: 95 °C for 5 min, 95 to 85 °C at -2 °C/s, 85 to 25 °C at -0.1 °C/s, hold at 4 °C. Mismatched DNA duplexes were then digested by addition of 1 μl T7 endonuclease I (NEB cat. no. M0302) followed by incubation at 37 °C for 15 min. The reaction was stopped with 1.5 μl of 0.25 M EDTA (Invitrogen cat. no. 15575020), and 1 μl was analyzed on an Agilent High Sensitivity DNA chip (Agilent cat. no. 5067-4626), yielding the molarity of digested and undigested fragments from which the editing efficiencies were calculated as: average(digested1, digested2)/sum(average(digested1, digested2), undigested).
Indel analysis by next-generation sequencing
Amplicons were designed to be smaller than 500 bp, with the gRNA target site at the center. PCR was performed as described for the T7 endonuclease assay. PCR products were purified by a 2.0x AMPure XP bead cleanup (Beckman Coulter cat. no. A63880) and used as input for Nextera XT (Illumina cat. no. 15032350) library preparations done according to the standard protocol. Libraries were sequenced on the Illumina MiSeq platform using a 150-cycle v3 flow cell with dual indexing. The machine was set to read lengths of 159 (read1) + 8 (i7) + 8 (i5) bases. To analyze the data, we first defined two 10 bp long ‘anchor’ sequences on both sides of the gRNA at a fixed distance of 30 bp. We then extracted reads spanning the gRNA target site from the BAM file via a grep operation for the pattern “<anchor_left>.*<anchor_right>” on the BAM file, using the -o option to return only the matching part of the sequence. For unedited fragments, this sequence is exactly 100 bp long: 10 bp anchor_left + 30 bp + 20 bp (gRNA) + 30 bp + 10 bp anchor_right. The size of insertions and deletions was calculated as the deviation from the unedited fragment length, summarized, and plotted.
Plasmid library complexity and gRNA dynamics
The gRNA expression cassette was amplified from either the plasmid library or the genomic DNA of cells at day 10 post-transduction. All functional sequences required for compatibility with Illumina sequencing were introduced in a single PCR reaction with specially designed primers (Supplementary Table 1). We included 8 bp long i7 barcodes and a stagger sequence to increase library complexity, resulting in a set of primer variants that can be freely combined. Purified PCR products were pooled and sequenced on an Illumina MiSeq machine, using a 150-cycle v3 flowcell with a read configuration of 159 (read1) + 8 (i7) + 8 (i5). Read1 covers the stagger sequence (variable length), the FWD primer binding site (23 bases), the gRNA target (20 bases), and the backbone (82 bases). gRNA read counts were normalized by the total number of sequenced reads. The number of cells assigned to a specific gRNA in CROP-seq (regardless of the number of genes per cell) was normalized to the total number of cells assigned to any gRNA. Fold change values were calculated between normalized cell counts from the CROP-seq screen and the normalized gRNA counts from the plasmid library.
Anti-CD3/CD28 stimulation of the T cell receptor pathway in Jurkat cells
Jurkat cells were serum starved for 3 h before stimulation. They were stimulated with 1 µg/ml anti-CD3 (eBioscience cat. no. 16-0037) and 1 µg/ml anti-CD28 antibody (eBioscience cat. no. 16-0289-81) for 4 h, while the unstimulated (naive) control was subjected to continued starvation. Samples collected for both conditions were subjected to CROP-seq.
Anti-CD3/CD28 stimulation of single gene knockout cells for the arrayed validation screen
Jurkat cells that were knockouts for single genes were grown in six-well plates and split into two equal parts of 900 µl in a sterile deepwell storage plate (Corning cat. no. 07-200-700). The cells were washed once with 800 µl of starvation medium (RPMI (Gibco cat. no. 21875-034), 0% FCS, penicillin/streptomycin) and then resuspended in 600 µl of starvation medium. After 3 h of serum starvation, one part was stimulated by addition of 1 µg/ml anti-CD3 (eBioscience cat. no. 16-0037) and 1 µg/ml anti-CD28 antibody (eBioscience cat. no. 16-0289-81) diluted in serum-free medium, while the other part was subjected to continued starvation. After another 4 h, we proceeded with extraction of genomic DNA and RNA (Qiagen cat. no. 80311) as well as flow cytometry.
Flow cytometry for the arrayed validation screen
Following treatment, anti-CD3/CD28 stimulated and unstimulated Jurkat T cells were transferred to 96-well V-bottom plates and washed twice with PBS containing 0.1% BSA and 5 mM EDTA (PBS + BSA + EDTA). Cells were then incubated with anti-CD16/CD32 (clone 93, Biolegend) to prevent nonspecific binding. Single-cell suspensions were stained with antibodies against CD25 (clone BC96), CD38 (clone HB-7), CD69 (clone FN50), CD82 (clone ASL-24), CD154 (clone 24-31), PD-1 (clone EH12.2H7), and Zombie NIR fixable viability dye (all from Biolegend) for 30 min at 4 °C. Cells were washed twice with PBS + BSA + EDTA and fixed in Biolegend fixation/permeabilization buffer for 1 h at room temperature followed by permeabilization using True-Nuclear Transcription Factor buffer (Biolegend) for 45 min in the presence of antibody against Ki-67 (clone Ki-67). Cells were washed twice with permeabilization buffer, acquired with a LSRFortessa (BD) cell analyzer, and data analysis was performed with FlowJo (Tree Star) software.
Drop-seq protocol for highly multiplexed single-cell RNA-seq in microfluidic droplets
Adherent cells were detached using Trypsin-EDTA (Gibco cat. no. 25300-054) following standard cell culture practices. Cells were collected by centrifugation at 300 RCF for 5 min, washed once with PBS-0.01% BSA (freshly prepared on the day of the run), and resuspended in 1 ml of PBS-0.01% BSA. Cells were filtered through a 40 µm cell strainer to obtain a single-cell suspension, and cell numbers were counted using a CASY device. Single cells were then coencapsulated with barcoded beads (ChemGenes cat. no. Macosko-2011-10) using an Aquapel-coated PDMS microfluidic device (FlowJEM) connected to syringe pumps (kdScientific) via polyethylene tubing with an inner diameter of 0.38 mm (Scicominc cat. no. BB31695-PE/2). Cells were supplied in PBS-0.01% BSA at a concentration of 220 cells/µl, while barcoded beads were resuspended in Drop-seq lysis buffer at a concentration of 150 beads/µl. The flow rates for cells and beads were set to 1.6 ml/h, while droplet generation oil (BioRad cat. no. 1864006) was provided at 8 ml/h. During the run, the barcoded bead solution was mixed by magnetic stirring with a mixing disk set to 1 jump/s. A typical run lasted between 35 and 40 min. In case of multiple runs per day, droplets were temporarily stored at 4 °C and processed together. Our most important modification to the Drop-seq protocol was an alternative way to break droplets, which recovered beads much more efficiently than in the original publication of Drop-seq7. After removing as much oil below the droplet layer as possible, we added 30 ml of 6x SSC buffer (Promega cat. no. V4261) and 1 ml of Perfluoroctanol (Sigma Aldrich cat. no. 370533-25G) and shook the tube forcefully six times to break the droplets. Based on their large diameter, beads were then collected by syringe filtering the solution through a 0.22 µm filter unit (Merck cat. no. SLGV033RS), washing 2x with 20 ml of 6x SSC buffer and eluting by turning the filter upside down and rinsing it with 3x 10 ml of 6x SSC buffer. Beads were collected by centrifugation at 1,250 RCF for 2 min, setting the brake speed to 50%. After washing a second time with 10 ml 6x SSC buffer, the pellet was taken up in 200 µl of 5x RT buffer and transferred to a 1.5 ml tube. Reverse transcription and exonuclease I treatment were performed as described in the original publication7, and the number of barcoded beads was estimated using a Fuchs-Rosenthal counting chamber (mixing the bead suspension with 6x DNA loading dye). Depending on the performance of the experiment, we prepared up to 24 PCR reactions per Drop-seq run, adding 4,400 beads (~220 cells) per PCR and enriching the cDNA for 4 + 10 cycles, using the already described reagents. We then prepared Drop-seq libraries using the Nextera XT kit (Illumina cat. no. 15032350), starting from 1 ng of cDNA pooled in equal amounts from all PCR reactions for a given run. We typically required an additional ten enrichment cycles using the Illumina Nextera XT i7 primers along with the Drop-seq New-P5 SMART-PCR hybrid oligonucleotide. The slightly increased cDNA input typically resulted in an average size distribution of about 575 bp. After quality control, libraries were sequenced with paired-end SBS chemistry on Illumina HiSeq 3000/4000 instruments. Drop-seq Custom Read1 Primer was spiked into the HP10 primer solution, located in column 11 of the cBot Reagent Plate at 1 µM final concentration. High sequence complexity needed for optimal base calling performance was achieved by adding 20-30% PhiX as spike in. Cluster generation and Read1 primer hybridization were completed using the Illumina cBot protocol “HiSeq_3000_4000_HD_Exclusion_Amp_v1.0.” Alternatively, libraries were sequenced on an Illumina NextSeq 500 instrument using the 75 cycle High Output v2 Kit (Illumina cat. no. FC-404-2005). We loaded 1.8 pM library and provided Drop-seq Custom Read1 Primer at 0.3 µM in position 7 of the reagent cartridge. On NextSeq machines, we sequenced without PhiX spike-in, using a read configuration of 20 bases (Read1), 8 bases (Index1) and 64 bases (Read2).
Preprocessing of single-cell sequencing data
Single-cell sequencing data were processed using the Drop-seq Tools v1.12 software7. Briefly, each transcriptome Read2 was tagged with the cell barcode (bases 1 to 12) and unique molecular identifier (UMI) barcode (bases 13 to 20) obtained from Read1, trimmed for sequencing adapters and poly-A sequences, and aligned using STAR v2.4.019 to a concatenation of the mouse and human genomes (for the species mixing experiment) or to the human reference genome assembly (Ensembl GRCh38 release), which was extended with artificial chromosomes that represent the CROPseq-Guide-Puro plasmid construct (250 bp common U6 promoter sequence, one gRNA sequence per artificial chromosome (20 bp), and the remaining 260 bp downstream plasmid backbone until the poly-A consensus sequence). Cell barcodes were corrected for possible bead synthesis errors, allowing the removal of up to four bases using the DetectBeadSynthesisErrors tool from the Drop-seq Tools v1.12 software. Reads aligning to exons were tagged with the respective gene name, and counts of UMI-deduplicated reads per gene within each cell were used to build a digital gene expression matrix comprising all cells with transcript counts for at least 500 genes. This matrix was converted to transcripts per million and log transformed for further analysis. We excluded weakly expressed genes and genes coding from ribosomal proteins from the analysis given that their widespread detection may be caused by solubilization of their mRNAs as noted previously7. For the assignment of gRNAs to cells, we quantified the overlap of UMI-deduplicated reads to the specific gRNA sequence within the CROPseq-Guide-Puro plasmid chromosomes and assigned the most abundant gRNA to the respective cell. For the analysis shown in Figure 1i, cells were classified as uniquely assigned if the dominant gRNA was three times more abundant than the sum of all other gRNAs; they were classified as containing multiple gRNAs where this difference was smaller than three; and they were classified as unassigned where no reads were overlapping with the gRNA sequence. For the Jurkat screen, we calculated the purity for each cell as the fraction of unique reads for the most abundant detected gRNA over the sum of all unique gRNA reads in the same cell, and we retained only cells with >0.9 purity for further analysis.
Preprocessing of bulk RNA sequencing data
Reads were trimmed with Trimmomatic20 and aligned to the GRCh38 assembly of the human genome using Bowtie121 with the following parameters: -q -p 6 -a -m 100 -minins 0 -maxins 5000 -fr -sam -chunkmbs 200. Duplicate reads were removed with Picard’s MarkDuplicates utility using standard parameters, followed by transcript quantification with BitSeq22 using the Markov chain Monte Carlo method and standard parameters. To obtain gene-level quantifications, we assigned to each gene the expression value of its most highly expressed transcript, calculated transcripts per million, and log transformed the expression matrix.
Transcriptome signature analysis
We first investigated the concordance between gRNAs across the whole transcriptome by measuring pairwise distances (L2-norm metric) between gRNAs. We used the median expression of single cells with the same gRNA assigned and assessed the significance of differences between the distributions of gRNAs targeting the same gene and gRNAs targeting different genes or between data types using the Mann-Whitney U test. Significant perturbations by a gRNA were assessed by grouping cell transcriptomes in the same manner (median) and comparing each gRNA with the respective stimulation condition, assessing significance using the Mann-Whitney U test. For gene-level quantifications, we combined the gRNA P values obtained from the previous step with Fisher’s method.
Sparse transcriptome signatures were used to position single cells sharing a putative perturbation (here, gene knockouts) within a multidimensional trajectory of cell states (here, measured activation of the TCR pathway in Jurkat cells). This method is reminiscent of a method for the assessment of cross-contamination between cell types in single cells23 or a recently described approach for monitoring cell fate conversion24. We identified genes associated with the TCR response (signature genes), selected cells assigned to a nontargeting control gRNA as representatives of the unperturbed stimulation (control cells), produced in silico mixtures of gene expression profiles of increasingly more stimulated cells based on the control cells from both conditions (mix profiles), and identified the mixed profile that best matched the expression profile of each single cell (signature position).
To determine a TCR-specific activation signature, we aggregated single cells by their assigned gRNA target genes and performed principal component analysis on all genes. This analysis detected a clear separation of genes by the TCR-activating anti-CD3/CD28 stimulation condition in principal component 1 (Supplementary Fig. 6c). Signature genes were defined as having an absolute loading higher than the 99th percentile for this principal component. Bioinformatic analysis of the signature gene function was performed with the gene set enrichment analysis tool Enrichr25, and the retrieved combined score (log[P value] * z-score) was displayed.
Based on the median expression level of cells assigned to nontargeting control gRNAs, we constructed for the signature genes a matrix Z of stepwise weighted linear mixtures of expression profiles between the unstimulated and stimulated conditions, which is given by:
where μa and μb are vectors with mean expression values of cells in the unstimulated and stimulated conditions, respectively; and i and j are indices of the signature genes (n) and the number of desired mixtures m. We selected m = 100; and to account for cell-to-cell variability as well as overshooting changes in the signature genes compared with the mean of the control cells, we generated an extended linear space of length m with boundaries -20 and 120. For m = 100, this is equivalent to μa and μb being placed at index j = 20 and j = 80 of the Z matrix, respectively.
We positioned each single cell in the matrix by retrieving the argmax of the Pearson correlation of each cell to each mixture in the matrix and visualized the relationship between the correlation argmax and the mean number of unique reads per cell, the minimum and (1 - maximum) correlation to ensure these variables were not confounding the signature positioning. Grouping cells by their gRNA assignment, we visualized the distribution of cell signature positions per group and calculated the mean group signature position for groups with more than ten cells. We then calculated the log fold deviation of each group of cells to the group of control cells within the respective stimulation condition to determine a deviation relative to genetically unperturbed cells.
The above procedure was repeated for each bulk RNA-seq sample on the same set of genes, but with theμa and μb of the signature matrix as the medians of the eight nontargeting control bulk cell lines. Signature values for gRNAs or genes obtained from the CROP-seq data were compared with those derived from bulk RNA-seq if available, and the Pearson and Spearman correlations were calculated. To assess the robustness of signature positions derived from the CROP-seq data, we subsampled 100 fractions of the data set and compared the Pearson correlation of the gRNA or gene aggregated signatures with those provided by the bulk RNA-seq as reference at each fraction for 100 iterations with random sampling.
Supplementary Material
Acknowledgements
We would like to thank J. Bigenzahn, A. Fauster, and M. Owusu (CeMM) for providing Cas9-expressing cell lines; M. Farlik for contributing to the Drop-seq setup; F. Müller and J. Menche for bioinformatic discussions; N. Winhofer for feedback on the illustrations; the Biomedical Sequencing Facility at CeMM for assistance with next generation sequencing, and all members of the Bock lab for their help and advice. C.S. is supported by a Feodor Lynen Fellowship of the Alexander von Humboldt Foundation. C.B. is supported by a New Frontiers Group award of the Austrian Academy of Sciences and by an ERC Starting Grant (European Union’s Horizon 2020 research and innovation programme, grant agreement no. 679146).
Footnotes
Author contributions
P.D., A.F.R., C.S., and C.B. conceptualized the project; P.D. designed and developed CROP-seq; P.D., C.S., P.T., and L.C.S. conducted CROP-seq experiments; D.A. optimized sequencing protocols; P.D., T.K., L.C.S., and A.K. performed the arrayed validation screen; A.F.R. and J.K. developed software; P.D., A.F.R., C.S., and T.K. analyzed data; P.D. and A.F.R. visualized data; P.D., A.F.R., C.S., and C.B. wrote the original draft; T.K., P.T., L.C.S., A.K., and D.A. reviewed the draft; and C.B. supervised the project.
Competing financial interests
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
Reagent and data availability
The CROPseq-Guide-Puro plasmid is available from Addgene (86708). Raw and processed CROP- seq data are provided via the NCBI GEO database (GSE92872). The source code of the computational analyses is included as Supplementary Software and maintained in an open Github repository (https://github.com/epigen/crop-seq).
Accession codes
Raw and processed CROP-seq data are available from NCBI GEO under accession number GSE92872.
References
- 1.Blomen VA, et al. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350:1092–1096. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- 2.Wang T, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marceau CD, et al. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature. 2016;535:159–163. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamb J. The Connectivity Map: a new tool for biomedical research. Nat Rev Cancer. 2007;7:54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- 6.Gapp BV, et al. Parallel reverse genetic screening in mutant human cells using transcriptomics. Mol Syst Biol. 2016;12:879. doi: 10.15252/msb.20166890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tiscornia G, Singer O, Verma IM. Design and Cloning of an shRNA into a Lentiviral Silencing Vector: Version A. CSH Protoc. 2008 doi: 10.1101/pdb.prot5010. [DOI] [PubMed] [Google Scholar]
- 10.de Kok S, et al. Rapid and reliable DNA assembly via ligase cycling reaction. ACS Synth Biol. 2014;3:97–106. doi: 10.1021/sb4001992. [DOI] [PubMed] [Google Scholar]
- 11.Guschin DY, et al. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 12.Brownlie RJ, Zamoyska R. T cell receptor signalling networks: branched, diversified and bounded. Nat Rev Immunol. 2013;13:257–269. doi: 10.1038/nri3403. [DOI] [PubMed] [Google Scholar]
- 13.Datlinger P, et al. Pooled CRISPR screening with single-cell transcriptome read-out. bioRxiv. 2016 doi: 10.1038/nmeth.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dixit A, et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell. 2016;167:1853–1866. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adamson B, et al. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell. 2016;167:1867–1882. doi: 10.1016/j.cell.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaitin DA, et al. Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell. 2016;167:1883–1896. doi: 10.1016/j.cell.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 17.Zheng GXY, et al. Massively parallel digital transcriptional profiling of single cells. bioRxiv. 2016 doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bock C, Farlik M, Sheffield NC. Multi-Omics of Single Cells: Strategies and Applications. Trends Biotechnol. 2016;34:605–608. doi: 10.1016/j.tibtech.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glaus P, Honkela A, Rattray M. Identifying differentially expressed transcripts from RNA-seq data with biological variation. Bioinformatics. 2012;28:1721–1728. doi: 10.1093/bioinformatics/bts260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, et al. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep. 2016;17:178–187. doi: 10.15252/embr.201540946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Treutlein B, et al. Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature. 2016;534:391–395. doi: 10.1038/nature18323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.